Abstract
The evolutionarily conserved Hippo signaling pathway is a key regulator of stem cell self-renewal, differentiation, and organ size. While alterations in Hippo signaling are causally linked to uncontrolled cell growth and a broad range of malignancies, genetic mutations in the Hippo pathway are uncommon and it is unclear how the tumor suppressor function of the Hippo pathway is disrupted in human cancers. Here, we report a novel epigenetic mechanism of Hippo inactivation in the context of hepatocellular carcinoma (HCC). We identify a member of the microrchidia (MORC) protein family, MORC2, as an inhibitor of the Hippo pathway by controlling upstream Hippo regulators, neurofibromatosis 2 (NF2) and kidney and brain protein (KIBRA). Mechanistically, MORC2 forms a complex with DNA methyltransferase 3A (DNMT3A) at the promoters of NF2 and KIBRA, leading to their DNA hyper-methylation and transcriptional repression. As a result, NF2 and KIBRA are crucial targets of MORC2 to regulate confluence-induced activation of Hippo signaling and contact inhibition of cell growth under both physiological and pathological conditions. The MORC2-NF2/KIBRA axis is critical for maintaining self-renewal, sorafenib resistance, and oncogenicity of HCC cells in vitro and in nude mice. Furthermore, MORC2 expression is elevated in HCC tissues, associated with stem-like properties of cancer cells, and disease progression in patients. Collectively, MORC2 promotes cancer stemness and tumorigenesis by facilitating DNA methylation-dependent silencing of Hippo signaling and could be a potential molecular target for cancer therapeutics.
Similar content being viewed by others
Introduction
Molecular pathways that regulate stem cell homeostasis during organ growth are often hijacked to promote tumorigenesis. One such pathway dictating stem cell self-renewal, differentiation, and organ size is the evolutionarily conserved Hippo pathway [1,2,3]. The core components of the mammalian Hippo pathway are two protein kinase complexes: (i) the mammalian sterile 20-like 1/2 (MST1/2) and adapter Salvador homolog 1 (SAV1) complex, (ii) the large tumor suppressor homolog 1/2 (LATS1/2) and adapter MOB kinase activator 1A/B (MOB1A/B) [1,2,3]. When Hippo signaling is on, MST1/2 phosphorylates and activates LATS1/2, which in turn phosphorylates the Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ) protein, to promote their cytoplasmic retention and inactivation [4]. When Hippo signaling is off, YAP/TAZ enters the nucleus to function as co-activators for the TEAD family of transcription factors (TEAD1-4) [4]. The Hippo signaling pathway is controlled by a number of upstream regulators stimulated by extracellular cues, including cell polarity, cell confluence, mechanical stimulation, and growth factors [4]. Notably, Merlin (encoded by neurofibromatosis type II (NF2) gene), KIBRA, and Willin form a complex and recruit MST1/2 and LATS1/2 to the apical plasma membrane to dictate spatial activation of LATS1/2 by MST1/2 [5]. It is now appreciated that several peripheral components of Hippo signaling, including Merlin and KIBRA, are critical regulators of contact inhibition and tissue regeneration under physiological conditions [6,7,8].
Genetic evidence has established important roles for Hippo signaling in the control of stem cell maintenance and tumorigenesis in a variety of tissues, particularly the liver [9]. YAP overexpression in mice leads to hepatomegaly and hepatocellular carcinoma (HCC), which is reversible after restoring endogenous YAP levels [10,11,12]. Remarkably, acute YAP activation in vivo is sufficient to confer undifferentiated progenitor cell phenotypes in hepatocytes [13], while YAP inhibition induces hepatocyte differentiation and tumor regression in established HCC models [12]. Moreover, Hippo inactivation by depleting NF2, WW45, MST1/2, SAV1, or MOB1A/B in mice consistently induces liver cancer and a portion of tumor cells exhibit stem cell properties, suggesting that Hippo signaling and its effector YAP critically regulate cancer stem cell (CSC) self-renewal and expansion [9, 12, 14, 15]. Furthermore, the Hippo pathway is deregulated and overexpression/nuclear enrichment of YAP/TAZ is frequently observed in many types of cancers [1, 9, 16, 17]. However, genetic mutation rates of Hippo pathway genes and YAP amplification are relatively low in HCC [1, 9]. The underlying mechanism responsible for disruption of Hippo signaling in cancers remains to be fully elucidated.
Epigenetic alterations contribute to tumorigenesis primarily by interfering with oncogenic or tumor-suppressive signaling pathways [18]. In this regard, the microrchidia (MORC) protein family is emerging as an important epigenetic modifier and reader that facilitate DNA methylation [19,20,21], histone deacetylation [22], and chromatin remodeling [23, 24], and recognizes histone H3K4me3 [25]. The human MORC family is composed of five members, MORC1–4 and the divergent SMCHD1 (structural maintenance of chromosomes flexible hinge domain containing 1) [26]. MORC proteins are characterized by a GHKL (gyrase, Hsp90, histidine kinase, and MutL)-type ATPase domain and a CW-type zinc finger domain that cooperate to execute epigenetic regulatory functions [26]. Although previous studies have implicated MORC2 and MORC3 in tumor-promoting roles of liver cancer [27, 28], it remains unknown how MORC family members contribute to tumorigenesis.
Here, we reveal an epigenetic regulatory role for MORC2 in controlling Hippo signaling in HCC models. MORC2 interacts with the methyltransferase DNMT3A to facilitate DNA hyper-methylation of the NF2 and KIBRA promoters to drive tumorigenesis. More importantly, targeting GHKL-type ATPase activity of MORC2 suppresses uncontrolled growth, self-renewal, and tumorigenicity of HCC cells, representing a potential therapeutic approach to treat liver cancer.
Results
MORC2 is elevated in hepatocellular carcinoma and associated with disease progression
We observed that mRNA levels of MORC2 were increased in HCC tissues compared to adjacent non-tumor tissue (Fig. 1a and Supplementary Table 1). Upregulation of MORC2 transcript levels was validated in cancer cohorts from TCGA and Oncomine databases (Supplementary Figure 1a–b). Consistently, the abundance of MORC2 protein was elevated in HCC tissues (Fig. 1a and Supplementary Table 2), suggesting that MORC2 is specifically elevated in liver cancer.
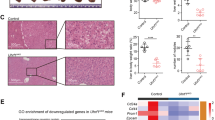
MORC2 inhibits the activity of Hippo signaling in HCC cells. a MORC2 is specifically elevated in liver cancer tissues. Analysis of MORC family member transcripts by qRT-PCR (upper panel) and MORC2 protein levels by western blot (lower panel) in HCC tumor and corresponding adjacent non-tumor tissues (N, non-tumor; T, tumor, n = 24 patients for qRT-PCR and n = 7 patients for western blot). b Immunohistochemistry (IHC) of MORC2 expression in liver cancer tissues. Left panel: representative IHC images showing low or high levels of anti-MORC2 staining (MORC2low or MORC2high). Right panel: quantitative analysis of anti-MORC2 staining revealing elevated expression of MORC2 in HCC samples. c Left panel: Heat maps illustrating differentially gene expression profiles of human immortalized hepatocyte L02 cells that were infected with control (Lv-ctrl) or MORC2-expressing (Lv-MORC2) lentivirus (n = 3 per biological repeats). Right panel: pathway analysis of downregulated genes in MORC2-expressing cells, as listed by enrichment score. d MORC2 suppresses Hippo signaling in HCC cells. Western blot analysis of Hippo core kinase activity in control or MORC2-expressing L02 and HepG2 cells, or various PLC cells (MORC2+/+ or MORC2−/− or MORC2−/− cells expressing ectopic MORC2). e Representative confocal microscopy images showing that MORC2 promotes YAP nuclear translocation in HCC cells. Quantitative analysis of YAP subcellular localization was provided in the lower panel. f MORC2 expression status correlates with YAP nuclear localization, as indicated by anti-MORC2 and anti-YAP IHC staining, in HCC tissues. Quantitative analysis of anti-MORC2 staining and YAP nuclear staining was provided in the lower panel (n = 100 patients). Data are presented as the mean ± SD from three separate experiments (a). Representative images are shown in a, b, d–f. *P < 0.05
We detected varying levels of MORC2 protein in the nuclei of liver cancer cells by IHC and found elevated expression of MORC2 in HCC samples (Fig. 1b and Supplementary Figure 1c, d). Tumor tissues with high levels of MORC2 (MORC2high) accounted for ~44% of HCC samples. MORC2high tumors were associated with increased tumor size, higher histological grades (III), and tumor cell invasion into lymph node and blood vessels (Supplementary Table 3), suggesting that upregulation of MORC2 was associated with HCC progression.
MORC2 inhibits Hippo tumor suppressor signaling during hepatocellular tumorigenesis
Given a role for MORC proteins in controlling epigenetic silencing, we analyzed the expression profile regulated by MORC2. Ectopic expression of MORC2 repressed the levels of 20,748 out of 45,033 transcripts (Fig. 1c, d). KEGG pathway analysis categorized these repressed genes into multiple developmental pathways, with the Hippo pathway the highest ranked (Fig. 1c).
We next examine whether MORC2 regulated Hippo signaling, and found that ectopic expression of MORC2 in both non-tumorigenic L02 cells and HCC HepG2 cells decreased levels of phosphorylated Thr1079-LATS1 and its substrate, pS127-YAP (Fig. 1d). Conversely, depleting MORC2 in PLC cells, an HCC cell line with high endogenous MORC2 expression, enhanced pT1079-LATS1 and pS127-YAP levels, while reintroducing MORC2 in MORC2−/− cells (Supplementary Figure 1c) suppressed Hippo signaling (Fig. 1d).
Phosphorylation of YAP by the Hippo pathway promotes its cytoplasmic sequestration and turns off YAP transcriptional activity. We found that deleting MORC2 promoted cytoplasmic sequestration of YAP to inhibit the activity of a YAP-TEAD responsive reporter, which was reversed following reconstitution of MORC2 in MORC2−/− cells (Fig. 1e and Supplementary Figure 1e).
To assess how MORC2 might affect Hippo signaling under physiological conditions, we used a spontaneous HCC model driven by hepatitis B virus X antigen (HBx) in C57BL/6 mice (HBx-transgenic, HBx-Tg). The abundance of MORC2 in livers of HBx-Tg mice was elevated in pre-malignant (50 weeks) and tumorous states (70 weeks), compared to non-transformed (10–30 weeks) tissues (Supplementary Figure 1f). Hippo signaling activity inversely correlated with MORC2 upregulation during HBx-driven liver tumorigenesis. Consistently, in human HCC samples, MORC2high tumors displayed enhanced YAP nuclear localization, while MORC2low tumors exhibited increased YAP cytoplasmic retention (Fig. 1f).
MORC2 promotes DNA hyper-methylation at NF2 and KIBRA loci
We found that NF2 and KIBRA, two upstream activators of Hippo signaling, but not other Hippo pathway components, were markedly suppressed upon MORC2 expression (Fig. 2a and Supplementary Figure 2a–b). Moreover, depleting MORC2 upregulated the mRNA levels of NF2 and KIBRA, while reintroducing MORC2 in MORC2-depleted cells restored their transcript levels (Fig. 2a).
MORC2 suppresses the transcription of NF2 and KIBRA by promoting DNA methylation. a MORC2 decreases expression of NF2 and KIBRA at transcriptional level. qRT-PCR analysis of NF2 and KIBRA mRNA levels in HepG2 cells infected with control or MORC2-expressing lentivirus (left panel) or indicated PLC cells (right panel). b MORC2-mediated suppression of NF2 and KIBRA expression is DNA methylation dependent. qRT-PCR analysis of NF2 and KIBRA mRNA levels in HepG2 cells infected with control or MORC2-expressing lentivirus, with or without 5-Aza treatment. c Endogenous MORC2 enhances DNA methylation of NF2 and KIBRA promoters. Bisulfite-sequencing PCR (BSP) analysis of the NF2 and KIBRA loci amplified from MORC2+/+ and MORC2−/− cells (MI, methylation index). Each row indicates one sequenced clone. Open circles indicate unmethylated CpG sites, and filled circles indicate methylated CpG sites. d MeDIP-qPCR analysis of DNA methylation status of the NF2 and KIBRA loci in HCC tissues and paired non-tumor livers (n = 50 patients). e Correlation analysis of DNA methylation status of NF2 or KIBRA locus with the levels of anti-MORC2 staining in HCC tissues (n = 50 patients). f Expression of NF2 and KIBRA is inversely associated MORC2 expression in HCC tissues. Upper panel: representative IHC images of anti-NF2 and anti-KIBRA staining in MORC2low or MORC2high liver cancers. Lower panel: quantification data (n = 100 patients). The specificity of MORC2 antibody was validated in Supplementary Figure 1d. Data are presented as the mean ± SD from triplicate experiments (a, b, d). Representative images are shown in f. *P < 0.05
Histone acetylation and methylation, as well as DNA methylation on promoter CpG islands, are well-established regulatory mechanisms for transcriptional activity [18]. We did not detect significant changes in the levels of histone H3 acetylation and histone lysine methylation at the NF2 and KIBRA loci (Supplementary Figure 2c–e). However, 5-azacytidine, a DNA methyltransferase (DNMT) inhibitor, reversed MORC2-mediated repression of NF2 and KIBRA expression (Fig. 2b). MORC2 overexpression significantly increased DNA methylation levels at both NF2 and KIBRA promoters (Supplementary Figure 2f). Conversely, depleting MORC2 resulted in hypo-methylation of the NF2 and KIBRA loci and an increase in mRNA levels, which was restored by reintroducing MORC2 (Supplementary Figure 2g). Bisulfite-sequencing analysis confirmed that depleting MORC2 reduced DNA methylation levels by more than 50% at both NF2 and KIBRA promoters (Fig. 2c). Finally, we observed that endogenous MORC2 bound to the NF2 and KIBRA promoters (Supplementary Figure 2h), suggesting that MORC2 is recruited to these promoters to facilitate DNA methylation and suppress their transcriptional activity.
We next examined whether MORC2-mediated DNA methylation and silencing of NF2 and KIBRA promoters is relevant to hepatocellular tumorigenesis. We found that DNA methylation of both promoters was elevated in HCC, compared to the adjacent normal liver tissue (Fig. 2d and Supplementary Table 4). Moreover, MORC2high tumors displayed elevated levels of DNA methylation, while MORC2low tumors exhibited DNA hypo-methylation, at NF2 and KIBRA promoters (Fig. 2e). Consequently, MORC2 expression levels inversely correlated with NF2 and KIBRA protein in tumor cells (Fig. 2f).
MORC2 binds the DNA methyltransferase DNMT3A at the NF2 and KIBRA loci
MORC proteins do not have DNA methyltransferase activity, suggesting that MORC2 might cooperate with a DNA methyltransferase/demethylase to promote DNA hyper-methylation. MORC2-mediated decrease in NF2 and KIBRA mRNA were mitigated by knockdown specifically of DNMT3A, but not DNMT1, DNMT3B, or TET1-3 (Fig. 3a and Supplementary Figure 3a–b).
MORC2 recruits DNMT3A to promote DNA methylation at the NF2 and KIBRA loci. a DNMT3A is required for MORC2 to suppress NF2 and KIBRA mRNA levels. HepG2 cells were infected with control or MORC2-expressing (Lv-MORC2) lentivirus, together with scramble siRNA or siRNAs targeting DNMT1 (si-DNMT1), DNMT3A (si-DNMT3A), or DNMT3b (si-DNMT3b) for 48 h. The relative levels of NF2 and KIBRA mRNA were determined by qRT-PCR (n = 3 per biological repeats). b Western blot analysis of anti-MORC2 and anti-DNMT3A immunoprecipitate demonstrates interaction between MORC2 and DNMT3A. c MORC2 is required for DNMT3A to interact with NF2 and KIBRA promoters. The occupancy of DNMT3A at the promoter regions of NF2 and KIBRA loci was determined by ChIP-qPCR. d–e MeDIP-qPCR analysis of DNA methylation status of the NF2 and KIBRA loci in control or MORC2-expressing HepG2 cells transfected scramble siRNA or si-DNMT3A (d), or in MORC2+/+ or MORC2−/− PLC cells infected with control or DNMT3A-expressing lentivirus (e). f DNMT3A is required for MORC2 to suppress expression of NF2 and KIBRA. Western blot analysis of NF2 and KIBRA in control or MORC2-expressing HepG2 cells, or cells that were further transfected with scramble siRNA or siRNAs targeting DNMT1, DNMT3A, and DNMT3B. Normal IgG was included as negative control for ChIP-qPCR and MeDIP-qPCR. β-actin was used as a loading control. Data are shown as the mean ± SD (a, c, d, e) of triplicates and representative images (b, f) are presented. *P < 0.05
MORC2 interacted specifically with DNMT3A in cells (Fig. 3b and Supplementary Figure 3c). And similar to MORC2, DNMT3A was bound to both NF2 and KIBRA promoters (Fig. 3c). Depletion of MORC2 impaired, while reintroducing MORC2 restored DNMT3A targeting to these promoters (Fig. 3c).
The increase in NF2 and KIBRA promoter methylation in MORC2-expressing cells were attenuated by depletion of DNMT3A (Fig. 3d and Supplementary Figure 3d). Conversely, DNMT3A-induced hyper-methylation of NF2 and KIBRA promoters were abolished in MORC2-depleted cells (Fig. 3e and Supplementary Figure 3e–f). Consequently, MORC2 repression of NF2 and KIBRA was diminished by depleting DNMT3A (Fig. 3f). These results suggest that MORC2 serves as a scaffold for DNMT3A to facilitate DNA hyper-methylation and transcriptional silencing of NF2 and KIBRA in HCC cells.
NF2 and KIBRA are essential for MORC2 inactivation of Hippo signaling
MORC2-mediated inhibition of pT1079-LATS1 and pS127-YAP, and the activity of a YAP-TEAD responsive reporter, was rescued by ectopic expression of either NF2 or KIBRA (Fig. 4a and Supplementary Figure 4a–b). Furthermore, depletion of either NF2 or KIBRA reduced pT1079-LATS1 and pS127-YAP levels, and enhanced nuclear retention and transcriptional activity of YAP in MORC2−/− cells (Fig. 4b–d). These results suggest that MORC2 inhibits expression of NF2 and KIBRA to suppress Hippo signaling, thus turning on the YAP transcriptional co-activator. Consistently, MORC2 expression status was inversely correlated with the levels of NF2 and KIBRA and Hippo signaling activity in a panel of hepatocytes (Fig. 4e).
MORC2 inhibits Hippo signaling activity through downregulating NF2 and KIBRA. a Control (Lv-ctrl) and MORC2-expressing (Lv-MORC2) HepG2 cells were infected with lentivirus encoding NF2 or KIBRA. Western blot was performed to examine the levels of Hippo signaling activity and YAP target gene CTGF. bMORC2+/+ and MORC2−/− PLC cells infected with lentivirus delivering scramble shRNA (sh-ctrl), NF2- or KIBRA-specific shRNA. Whole cell lysates of the stable cell lines were collected for western blot analysis. c MORC2 promotes YAP translocation from cytoplasm to nucleus through downregulating NF2 and KIBRA. Cells were treated as in b and immune-stained with YAP antibody (red) and DAPI (nuclei, blue), respectively. YAP subcellular localization from at least 100 randomly selected cells was quantified as shown in the right panel. C, cytoplasm; N, nucleus. d MORC2 promotes the activity of YAP transcriptional co-activation through reducing NF2 and KIBRA expression. Cells were treated as in b. The luciferase activities of 8XGTIIC-TEAD reporter in these cells were determined and presented as mean ± SD. e MORC2 abundance correlates with NF2 and KIBRA expression, Hippo signaling activity, and CTGF levels, as revealed by western blot analysis of cell lysates from L02 immortalized hepatocyte and a panel of HCC cell lines. β-actin was used as a loading control. Representative images from triplicate experiments are presented (a-ce). *P < 0.05
The MORC2-NF2/KIBRA axis regulates Hippo pathway activation induced by contact inhibition
Consistent with a role for the Hippo pathway in sensing cell confluence to control proliferation and growth [2, 3, 7], phosphorylation of LATS1 and YAP was increased in untransformed L02 hepatocytes at high density (Fig. 5a and Supplementary Figure 5a). Ectopic expression of MORC2 inhibited upregulation of pT1079-LATS1 and pS127-YAP levels upon cell confluence, coupled with continued proliferation (Fig. 5a and Supplementary Figure 5a). Furthermore, MORC2−/− PLC cells showed increased LATS1 activity at high cell density, whereas control cells did not elevate Hippo signaling upon reaching confluence (Fig. 5a). Moreover, introducing NF2 or KIBRA into MORC2-expressing L02 cells rendered them sensitive to confluence-induced activation of Hippo signaling, and inhibition of proliferation (Fig. 5b–c and Supplementary Figure 5b). Depleting either NF2 or KIBRA in MORC2−/− PLC cells reduced pT1079-LATS1 and pS127-YAP levels when cells reached confluence, allowing continued proliferation (Fig. 5b–c and Supplementary Figure 5c).
MORC2 modulates confluence-induced activation of Hippo signaling and growth arrest by suppressing NF2 and KIBRA expression. a MORC2 suppresses confluence-induced activation of Hippo signaling. Control (Lv-ctrl) and MORC2-expressing (Lv-MORC2) L02 cells, or MORC2+/+ and MORC2−/− PLC cells were seeded at different degree of confluence for 8 h. Cell lysates were collected for western blot analysis. b MORC2-expressing (Lv-MORC2) L02 cells were infected with control or NF2-encoding lentivirus. MORC2−/− PLC cells were infected with lentivirus delivering scramble shRNA or NF2-specific shRNA. The activity of Hippo signaling at low or high confluence was examined by western blot. c–e NF2 and KIBRA are critical downstream targets for MORC2 to override confluence-induced growth arrest. Control (Lv-ctrl) and MORC2-expressing (Lv-MORC2) cells were infected with empty vector or lentivirus encoding NF2 or KIBRA. Alternatively, MORC2+/+ and MORC2−/− cells were infected with lentivirus delivering scramble shRNA (sh-ctrl), NF2-, or KIBRA-specific shRNA. The morphologies of cells at low or high confluence were monitored by microscopy (c). The anchorage-dependent (d) and independent (e) growth capacities of these cells were examined and presented as mean ± SD. L02 cells and PLC cells were used for monitoring anchorage-dependent growth (c, d), while HepG2 and PLC cells were used for soft agar colony formation assay (e). Representative images from triplicate experiments are presented (a, b, c, e). *P < 0.05
We next set out to quantitatively measure contact inhibition and anchorage-independent proliferation capacity regulated by MORC2. Compared to naive cells, cells ectopically expressing MORC2 were refractory to contact inhibition, which was coupled with enhanced colony formation in soft agar (Fig. 5d–e). However, introducing NF2 or KIBRA into MORC2-expressing cells restored contact inhibition and reduced their anchorage-independent growth rate (Fig. 5d–e). Moreover, MORC2−/− PLC cells gained the ability to undergo contact inhibition and formed fewer colonies in soft agar. Whereas depleting either NF2 or KIBRA, but not TP53 or SMAD4, in MORC2−/− PLC cells, restored proliferation after confluence and anchorage-independent growth in soft agar (Fig. 5d–e and Supplementary Figure 5d–e). Thus, MORC2 restrains expression of NF2 and KIBRA to evade contact inhibition, thereby promoting cell growth and malignant transformation.
The MORC2-NF2/KIBRA axis is critical for maintaining stem-like properties and sorafenib resistance of hepatocellular carcinoma cells
The Hippo/YAP signaling pathway is a regulator of stem cell homeostasis [2, 3, 14, 29,30,31,32], suggesting that MORC2 could have a role in regulating cancer stemness. Ectopic expression of MORC2 induced expansion of HCC cells expressing Lgr5, a liver cancer stem cell marker [33], while introducing NF2 or KIBRA into MORC2-expressing cells reduced the pool of Lgr5+ cells (Fig. 6a and Supplementary Figure 6a). Moreover, depletion of MORC2 decreased Lgr5+ cells that were restored by depletion of NF2 or KIBRA (Fig. 6a and Supplementary Figure 6a). Furthermore, ectopically expressed MORC2 promoted growth of tumorspheres by HCC cells in stem cell medium, leading to elevated numbers of primary and secondary sphere-initiating cells, an indicator for self-renewing capacity in vitro, which was dependent on MORC2 suppression of NF2 and KIBRA (Fig. 6b–c and Supplementary Figure 6b). To demonstrate a role for endogenous MORC2 in sustaining cancer stemness, MORC2−/− PLC cells were deficient in sphere formation and contained significantly less primary and secondary sphere-initiating cells, which was partially rescued by depletion of NF2 and KIBRA (Fig. 6b-c and Supplementary Figure 6b).
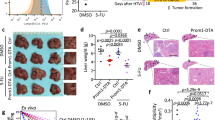
MORC2 promotes stemness, chemoresistance, and tumorigenicity of HCC cells by inhibiting NF2 and KIBRA. a–d Control (Lv-ctrl) and MORC2-expressing (Lv-MORC2) HepG2 cells were infected with empty vectors or lentivirus encoding NF2 or KIBRA. MORC2+/+ and MORC2−/− PLC cells were infected with lentivirus delivering scramble shRNA, or NF2-, or KIBRA- specific shRNA. The percentages of liver cancer stem cells (Lgr5+ cells) were examined by flow cytometry (a). Single cells were seeded in stem cell medium to grow as tumorspheres and representative images were shown (b). The single cells from primary and secondary tumorspheres were cultured in stem cell medium in a limiting dilution manner for 14 days to calculate the frequencies of sphere-initiating cells. Tumorspheres with a diameter larger than 50 μm were counted (c). Cells were treated with Sorafenib (4 μM for HepG2 cells and 8 μM for PLC cells) for 48 h and cultured in normal medium for 10 days to examine colony formation capacity (d). Colonies with a diameter greater than 75 μm were counted. Representative images were shown. e The growth kinetics of subcutaneous tumors generated by indicated PLC cells in nude mice (1 × 106 cells per injection, n = 5 mice). Xenograft tumors were dissected and tumor weight was presented as the mean ± SD. f The frequencies of cancer-initiating cells (CICs) of indicated PLC cells were analyzed by extreme limiting dilution assays (ELDAs) in NOD/SCID mice. Representative images are shown (b, e). Data are presented as the mean ± SD (a, d, e) or mean ± 95% confidence interval of each group from triple replicate (c). *P < 0.05
We found that MORC2 promoted cell survival and increased colony formation in the presence of sorafenib (Fig. 6d and Supplementary Figure 6c), which is consistent with enhanced cancer stemness contributing to drug resistance. The above-mentioned phenotypes in MORC2−/− cells could not be rescued by depleting TP53 or SMAD4 (Supplementary Figure 6d–f), further suggesting that MORC2 promotes stemness and drug resistance of HCC cells primarily through suppressing Hippo signaling pathway.
MORC2 promotes tumorigenicity and stemness of hepatocellular carcinoma cells through inhibiting NF2 or KIBRA in vivo
Subcutaneous xenograft tumors formed by MORC2−/− PLC cells displayed reduced growth and anti-Ki-67 staining, which was dependent on NF2 or KIBRA (Fig. 6e and Supplementary Figure 6g), suggesting that MORC2 promotes tumor growth primarily by suppressing NF2 and KIBRA expression.
To assess whether endogenous MORC2 is required for stemness of HCC cells in vivo, we carried out an in vivo limiting dilution xenotransplantation study [33,34,35]. To this end, depleting MORC2 led to a 8.7-fold decrease in the frequency of cancer-initiating cells (Fig. 6f). Moreover, knockdown of NF2, and to a lesser extent KIBRA, in MORC2−/− PLC cells efficiently restored the frequency of cancer-initiating cells (Fig. 6f). These results were consistent with the finding that expression of Nanog, a putative stem cell marker, was significantly reduced in the MORC2−/− tumors in a NF2- or KIBRA-dependent manner (Supplementary Figure 6g). Thus, the MORC2 is critical for maintaining stem-like properties and tumorigenicity of HCC cells in vivo.
YAP1 is required for the oncogenic functions of MORC2
Depleting YAP1, and TAZ to a lesser extent, in MORC2-expressing cells reduced the pool of Lgr5+ cells (Supplementary Figure 7a–b), leading to attenuated growth of tumorspheres and decreased numbers of sphere-initiating cells in HCC cells (Supplementary Figure 7c–d), indicating impaired self-renewing capacity. Consistently, loss of YAP1 in MORC2-expressing cells inhibited anchorage-independent cell growth and resistance to sorafenib (Supplementary Figure 7e–f). Therefore, MORC2 depends at least in part on YAP signaling activity to promote cancer stemness and sorafenib insensitivity.
The abundance of MORC2 correlates with stem-like properties of human hepatocellular carcinoma cells
We found MORC2 expression in HCC tissues correlated with expression levels of cancer stem cell markers (Fig. 7a–e). Lgr5, CD133, and Nanog expression was upregulated in MORC2high tumors, as compared to MORC2low HCC (Fig. 7b–d). Importantly, Sox9, a downstream target of the Hippo pathway, was also correlated with MORC2 expression (Fig. 7a, e), indicating that MORC2 may regulate stem-like properties of human HCC through the Hippo signaling pathway.
MORC2 overexpression is associated with stem cell traits and malignant progression of HCC. a Representative IHC staining images of MORC2, as well as cancer stem cell markers Lgr5, Nanog, CD133, and Sox9, in HCC and adjacent normal liver tissues (n = 100 patients). b–d Quantitative analysis of the levels of anti-Lgr5 (b), anti-CD133 (c), anti-Nanog (d), and anti-Sox9 (e) staining in sections of MORC2low or MORC2high liver cancer tissues. f The overall survival of patients with MORC2low or MORC2high HCC was analyzed by log-rank test (n = 200 patients). g A schematic of the underlying mechanisms by which MORC2, in cooperation with DNMT3A, facilitates DNA hyper-methylation at the NF2 and KIBRA loci to suppress Hippo signaling, thereby promoting stemness and tumorigenicity of HCC cells
We identified MORC2 expression as an independent prognostic indicator for shorter survival of HCC patients in multivariate analysis (Supplementary Table 5). Patients with MORC2high tumors had shorter overall survival (the median ± SD, 16.0 ± 2.1 months), compared to those with MORC2low tumors (27.0 ± 2.8 months, Fig. 7f). Together, upregulation of MORC2 in tumor cells is associated with disease progression, possibly due to enhanced cancer stemness (Fig. 7g).
The GHKL-ATPase activity is essential for oncogenic functions of MORC2
The GHKL-type ATPase activity of MORC family members is necessary for their biological functions [23, 25, 36]. We substituted an aspartate at residue 68 with an alanine (D68A), which disrupts ATPase activity of MORC2 (Supplementary Figure 8a) [23]. Consistent with the ATPase activity being necessary for MORC function [23, 25, 36], MORC2-D68A was deficient in association with DNMT3A (Supplementary Figure 8b), thus failing to recruit DNMT3A to NF2 and KIBRA promoters (Supplementary Figure 8c–d) and resulting in DNA promoter hypo-methylation (Fig. 8a and Supplementary Figure 8e) and elevated expression of NF2 and KIBRA (Fig. 8b and Supplementary Figure 8f). Consequently, MORC2-D68A-reconstituted cells had activated Hippo signaling to promote YAP nuclear exclusion (Fig. 8b) and suppress its transcriptional activity (Supplementary Figure 8g). Therefore, the GHKL-type ATPase activity is required for MORC2 to interact with DNMT3A and recruit it to the NF2 and KIBRA promoters to suppress Hippo signaling.
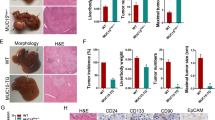
MORC2 promotes stemness and chemoresistance of HCC cells in an ATPase activity-dependent manner. a ATPase-deficient MORC2 fails to promote DNA methylation of the NF2 and KIBRA promoter. MeDIP-qPCR analysis of DNA methylation status of the NF2 and KIBRA loci in MORC2−/− PLC cells reconstituted with wild-type (WT) or ATPase-deficient MORC2 mutant (D68A). Normal IgG was included as negative control for MeDIP-qPCR. b MORC2-D68A does not inhibit the expression of NF2 and KIBRA to inactivate Hippo signaling, thus cannot promote YAP nucleus translocation. Hippo signaling activity and expression of CTGF were examined by western blot and YAP subcellular localization was visualized by confocal microscopy. β-actin was used as a loading control. c–d ATPase activity of MORC2 is essential for maintaining a pool of Lgr5+ CSCs, and the self-renewal of HCC cells in vitro. MORC2−/− PLC cells expressing MORC2-WT or MORC2-D68A were used. The percentages of Lgr5+ CSCs were examined by FACS (c). The frequencies of sphere-initiating cells were determined by an extreme limiting dilution assay of serially passaged tumorspheres in stem cell medium. Tumorspheres with a diameter greater than 50 μm were counted (d). e–f ATPase-deficient MORC2 fails to maintain self-renewal and tumorigenicity of HCC cells in vivo. Growth curves, representative images, and tumor weights of subcutaneous tumors formed by indicated PLC cells in nude mice (1 × 106 cells per injection, n = 5 mice) (e). The frequencies of cancer-initiating cells were determined by extreme limiting dilution assays (ELDAs) in NOD/SCID mice (n = 6 mice per group) (f). Representative images are shown (b, e). Data are presented as the mean ± SD (a, c, e) or mean ± 95% confidence interval (d, f) of each group from triple replicate. *P < 0.05
Phenotypically, compared to MORC2-WT-expressing cells, MORC2-D68A-expressing cells displayed reduced Lgr5+ cells and impaired self-renewing capacity, and were also more sensitive to sorafenib and less efficient in anchorage-independent growth and tumor growth (Fig. 8c–e and Supplementary Figure 8h–k). Moreover, MORC2-D68A tumors showed lower levels of Nanog staining and exhibited more than a 4.78-fold decrease in the frequency of cancer-initiating cells (Fig. 8f and Supplementary Figure 8l). Together, the GHKL-type ATPase activity of MORC2 is essential for the stemness and tumorigenicity of HCC cells.
Discussion
Here, we identify MORC2 as an upstream suppressor of Hippo signaling. In response to intrinsic and extrinsic signals, the Hippo pathway regulates the activity of YAP/TAZ to dictate their transcriptional co-activator activity [4]. We demonstrate that two components involved in Hippo signaling, NF2 and KIBRA, are directly repressed by MORC2 at the transcriptional level. As such, MORC2 is inversely correlated with the levels of Merlin and KIBRA and positively associated with YAP activation in HCC tissues. The Merlin/KIBRA/Willin protein complex has been shown to directly recruit MST1/2 and LATS1/2 to the plasma membrane, thereby dictating the phosphorylation and spatial activation of LATS1/2 by MST1/2 [5, 37, 38]. Our data define MORC2 as an upstream regulator of the Hippo pathway in HCC, and possibly in a broader spectrum of malignancies.
Our studies reveal a DNA methylation-dependent mechanism for Hippo inactivation. In human cancers, the abundance and nuclear localization of YAP/TAZ is frequently elevated, supporting an important role of Hippo signaling dysregulation in tumorigenesis. However, loss-of-function mutations in Hippo pathway components are uncommon in human cancers (except NF2 mutation in neurofibromatosis). For example, although loss of NF2 induces HCC in mice [9], the mutation rate of NF2, as well as KIBRA, is very low in human HCC (<5%) (cBioportal cancer genomics datasets) [39]. These results suggest that Hippo signaling may be inactivated by other regulatory mechanisms, such as epigenetic alterations. In this study, we show that DNA methylation at the promoters of NF2 and KIBRA are increased in HCC tissue, resulting in their decreased expression levels. Previous studies have reported hyper-methylation of the NF2 and KIBRA promoters in schwannoma and meningioma [40, 41] and chronic lymphocytic leukemia [42]. These results, together with previous reports, support the notion that multiple mechanisms, including DNA methylation, contribute to dysregulation of Hippo signaling in human cancers.
We find that MORC2 displays robust regulatory activity to promote DNA methylation status of NF2 and KIBRA loci. MORC2 binds to DNMT3A and recruits it to the NF2 and KIBRA promoters to promote DNA methylation. Previous studies have shown that during mouse germ cell development, MORC1 mutant cells exhibit locus- and stage-specific DNA methylation defects [20]. In Arabidopsis, AtMORC4 and AtMORC7 are both required for nuclear body formation and DNA methylation [19, 21]. Thus, MORC family members may regulate locus-specific DNA methylation, as well as other modes of epigenetic changes in a highly conserved manner [22,23,24,25].
Moreover, we found that MORC2 may promote YAP activation to drive CSC expansion and liver cancer initiation (Fig. 7g). These results are consistent with a recent report that YAP signaling is essential for expansion of Lgr5+ stem cells and tumorigenesis in the intestine [43]. Similarly, loss of NF2 and SAV1, or YAP overexpression, induces immature phenotypes on adult hepatocytes [9, 12, 14, 15]. Likewise, recent reports have shown additional roles of MORC family members in tumorigenesis. MORC2 regulates DNA damage response and survival of gastric cancer cells [22, 23, 44] and MORC3 expression affects p53 subnuclear localization and cellular senescence [23, 25]. Thus, MORC family members, especially MORC2, may play an essential role in the development of human malignancies.
Finally, chemical compounds that inhibit MORC2 GHKL-ATPase activity could be novel Hippo-reactivating agents to suppress oncogenicity and sorafenib resistance of HCC cells. Further studies are warranted to identify novel GHKL-ATPase inhibitors through high-throughout screening. Additionally, protein structures of HSP90 and GHKL-ATPase are highly conserved. Several HSP90 antagonists such as STA-9090 (Ganetespib) [45], AT-13387 (Onalespib) [46], or NVP-AUY922 (Luminespib) [47] are under clinical development for solid and hematopoietic tumors. It will be interesting if these HSP90 antagonists also inhibit GHKL-ATPase activity, thereby exerting anti-HCC activity. Taken together, MORC2 is an epigenetic modifier of Hippo signaling and MORC2 GHKL-ATPase inhibitors may be a promising approach for HCC therapy.
Material and methods
For details of Materials and Methods, please refer to the Supplementary information.
CRISPR-Cas9-mediated generation of MORC2 knockout cells
MORC2 knockout (MORC2−/−) cells were generated using CRISPR-Cas9 gene editing technology [48]. A CRISPR sgRNA (5′-ACACCTGAGTCTACTCAGAT-3′) targeting MORC2 were designed using the online tool (http://crispr.mit.edu). The DNA oligo was synthesized and ligated into pLentiCRISPR v2 (addgene plasmid # 52961) to generate the MORC2 knockout vector. Following transfection and puromycin selection, clonal wild type and MORC2-deleted cells were selected.
Limiting dilution assay in vitro and in vivo
For in vitro limiting dilution assay, decreasing cell numbers were seeded in stem cell medium in 96-well plates for 2 weeks. The numbers of wells with at least one tumorsphere (diameter >75 µm) were counted. For serial passage, tumorspheres were dissociated and single cells were re-seeded in a similar manner.
For in vivo limiting dilution assay, a defined dose of cells was injected subcutaneously into the flanks of NOD/SCID mice. For all experiments, six injection sites were tested. Three months later, the number of mice with xenografts in each group was counted. The frequency of sphere-initiating cells or cancer-initiating cells were calculated by extreme limiting dilution assays using ELDA website (http://bioinf.wehi.edu.au/software/elda/) [33,34,35].
Statistical analysis
Data are presented as mean ± SD, and analyzed with Student’s test or χ2-test by SPSS 18.0 software. Cox regression was performed to analyze univariate or multivariate variants that were associated with prognosis. Survival analysis was performed using the Kaplan–Meier method and estimated by log-rank test. Differences were considered as statistically significant (*) when P < 0.05, and not significant (n.s.) when P > 0.05.
References
Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–57.
Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–28.
Hong AW, Meng Z, Guan KL. The Hippo pathway in intestinal regeneration and disease. Nat Rev Gastroenterol Hepatol. 2016;13:324–37.
Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17.
Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–55.
Lallemand D, Curto M, Saotome I, Giovannini M, McClatchey AI. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 2003;17:1090–1100.
Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61.
Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci USA. 2011;108:11930–5.
Yimlamai D, Fowl BH, Camargo FD. Emerging evidence on the role of the Hippo/YAP pathway in liver physiology and cancer. J Hepatol. 2015;63:1491–501.
Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–60.
Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–33.
Fitamant J, Kottakis F, Benhamouche S, Tian HS, Chuvin N, Parachoniak CA, et al. YAP inhibition restores hepatocyte differentiation in advanced HCC, leading to tumor regression. Cell Rep. 2015;10:1692–707.
Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–38.
Benhamouche S, Curto M, Saotome I, Gladden AB, Liu CH, Giovannini M, et al. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718–30.
Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:8248–53.
Perra A, Kowalik MA, Ghiso E, Ledda-Columbano GM, Di Tommaso L, Angioni MM, et al. YAP activation is an early event and a potential therapeutic target in liver cancer development. J Hepatol. 2014;61:1088–96.
Sohn BH, Shim JJ, Kim SB, Jang KY, Kim SM, Kim JH, et al. Inactivation of Hippo pathway is significantly associated with poor prognosis in hepatocellular carcinoma. Clin Cancer Res. 2016;22:1256–64.
Feinberg AP, Koldobskiy MA, Gondor A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17:284–99.
Lorkovic ZJ, Naumann U, Matzke AJ, Matzke M. Involvement of a GHKL ATPase in RNA-directed DNA methylation in Arabidopsis thaliana. Curr Biol. 2012;22:933–8.
Pastor WA, Stroud H, Nee K, Liu W, Pezic D, Manakov S, et al. MORC1 represses transposable elements in the mouse male germline. Nat Commun. 2014;5:5795.
Harris CJ, Husmann D, Liu W, Kasmi FE, Wang H, Papikian A, et al. Arabidopsis AtMORC4 and AtMORC7 form nuclear bodies and repress a large number of protein-coding genes. PLoS Genet. 2016;12:e1005998.
Shao Y, Li Y, Zhang J, Liu D, Liu F, Zhao Y, et al. Involvement of histone deacetylation in MORC2-mediated down-regulation of carbonic anhydrase IX. Nucleic Acids Res. 2010;38:2813–24.
Li DQ, Nair SS, Ohshiro K, Kumar A, Nair VS, Pakala SB, et al. MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep. 2012;2:1657–69.
Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–51.
Li S, Yen L, Pastor WA, Johnston JB, Du J, Shew CJ, et al. Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin. Proc Natl Acad Sci USA. 2016;113:E5108–5116.
Li DQ, Nair SS, Kumar R. The MORC family: new epigenetic regulators of transcription and DNA damage response. Epigenetics. 2013;8:685–93.
Bard-Chapeau EA, Nguyen AT, Rust AG, Sayadi A, Lee P, Chua BQ, et al. Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat Genet. 2014;46:24–32.
Abbott KL, Nyre ET, Abrahante J, Ho YY, Isaksson Vogel R, Starr TK. The candidate cancer gene database: a database of cancer driver genes from forward genetic screens in mice. Nucleic Acids Res. 2015;43:D844–848.
Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–72.
Panciera T, Azzolin L, Fujimura A, Di Biagio D, Frasson C, Bresolin S, et al. Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell. 2016;19:725–37.
Qin H, Hejna M, Liu Y, Percharde M, Wossidlo M, Blouin L, et al. YAP induces human naive pluripotency. Cell Rep. 2016;14:2301–12.
Britschgi A, Duss S, Kim S, Couto JP, Brinkhaus H, Koren S, et al. The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERα. Nature. 2017;541:541–5.
Lei ZJ, Wang J, Xiao HL, Guo Y, Wang T, Li Q, et al. Lysine-specific demethylase 1 promotes the stemness and chemoresistance of Lgr5+ liver cancer initiating cells by suppressing negative regulators of b-catenin signaling. Oncogene. 2015;34:3188–98.
Shan J, Shen J, Liu L, Xia F, Xu C, Duan G, et al. Nanog regulates self-renewal of cancer stem cells through the insulin-like growth factor pathway in human hepatocellular carcinoma. Hepatology. 2012;56:1004–14.
Kreso A, van Galen P, Pedley NM, Lima-Fernandes E, Frelin C, Davis T, et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat Med. 2014;20:29–36.
Andrews FH, Tong Q, Sullivan KD, Cornett EM, Zhang Y, Ali M, et al. Multivalent chromatin engagement and inter-domain crosstalk regulate MORC3 ATPase. Cell Rep. 2016;16:3195–207.
Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–8.
Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–99.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Gonzalez-Gomez P, Bello MJ, Alonso ME, Lomas J, Arjona D, Campos JM, et al. CpG island methylation in sporadic and neurofibromatis type 2-associated schwannomas. Clin Cancer Res. 2003;9:5601–6.
Wang B, Cui Z, Zhong Z, Sun Y, Yang GY, Sun Q, et al. The role and regulatory mechanism of IL-1b on the methylation of the NF2 gene in benign meningiomas and leptomeninges. Mol Carcinog. 2016;55:2268–77.
Shinawi T, Hill V, Dagklis A, Baliakas P, Stamatopoulos K, Agathanggelou A, et al. KIBRA gene methylation is associated with unfavorable biological prognostic parameters in chronic lymphocytic leukemia. Epigenetics. 2012;7:211–5.
Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature. 2015;526:715–8.
Zhang Q, Song Y, Chen W, Wang X, Miao Z, Cao L, et al. By recruiting HDAC1, MORC2 suppresses p21 Waf1/Cip1 in gastric cancer. Oncotarget. 2015;6:16461–70. 16461-70
Goyal L, Wadlow RC, Blaszkowsky LS, Wolpin BM, Abrams TA, McCleary NJ, et al. A phase I and pharmacokinetic study of ganetespib (STA-9090) in advanced hepatocellular carcinoma. Invest New Drugs. 2015;33:128–37.
Shapiro GI, Kwak E, Dezube BJ, Yule M, Ayrton J, Lyons J, et al. First-in-human phase I dose escalation study of a second-generation non-ansamycin HSP90 inhibitor, AT13387, in patients with advanced solid tumors. Clin Cancer Res. 2015;21:87–97.
Sessa C, Shapiro GI, Bhalla KN, Britten C, Jacks KS, Mita M, et al. First-in-human phase I dose-escalation study of the HSP90 inhibitor AUY922 in patients with advanced solid tumors. Clin Cancer Res. 2013;19:3671–80.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Acknowledgements
We thank Ms. Qing-hua Ma for her assistance in FACS analysis and sorting (Institute of Pathology and Southwest Cancer Center, Southwest Hospital, Third Military Medical University). We also thank Prof. Jiang Zheng and Ms. Fang-fang Li for their technical assistance in immunofluorescence staining and confocal microscopy. This work was supported by the grants from the National Natural Science Foundation of China (NSFC Nos. 81472294 to BW and 81672463 to D-fC).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by R. De Maria
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wang, T., Qin, Zy., Wen, Lz. et al. Epigenetic restriction of Hippo signaling by MORC2 underlies stemness of hepatocellular carcinoma cells. Cell Death Differ 25, 2086–2100 (2018). https://doi.org/10.1038/s41418-018-0095-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0095-6
This article is cited by
-
Role of Hippo pathway dysregulation from gastrointestinal premalignant lesions to cancer
Journal of Translational Medicine (2024)
-
E3 ubiquitin ligases in cancer stem cells: key regulators of cancer hallmarks and novel therapeutic opportunities
Cellular Oncology (2023)
-
O-GlcNAcylation of MORC2 at threonine 556 by OGT couples TGF-β signaling to breast cancer progression
Cell Death & Differentiation (2022)
-
MORC2/β-catenin signaling axis promotes proliferation and migration of breast cancer cells
Medical Oncology (2022)
-
Predictive biomarkers and potential drug combinations of epi-drugs in cancer therapy
Clinical Epigenetics (2021)