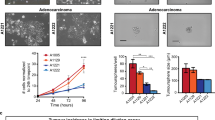
Abstract
Breast cancer (BCa) is a complex and heterogeneous disease, with different intrinsic molecular subtypes that have distinct clinical outcomes and responses to therapy. Although intrinsic subtyping provides guidance for treatment decisions, it is now widely recognised that, in some cases, the switch of the BCa intrinsic subtype (which embodies cellular plasticity), may be responsible for therapy failure and disease progression. Aberrant FGFR4 signalling has been implicated in various cancers, including BCa, where it had been shown to be associated with aggressive subtypes, such as HER2-enriched BCa, and poor prognosis. More importantly, FGFR4 is also emerging as a potential driver of BCa intrinsic subtype switching, and an essential promoter of brain metastases, particularly in the HER2-positive BCa. Although the available data are still limited, the findings may have far-reaching clinical implications. Here, we provide an updated summary of the existing both pre- and clinical studies of the role of FGFR4 in BCa, with a special focus on its contribution to subtype switching during metastatic spread and/or induced by therapy. We also discuss a potential clinical benefit of targeting FGFR4 in the development of new treatment strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci. 2001;98:10869.
Perou CM, Sørlie T, Eisen MB, Van De Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Hoon Tan P, Ellis I, Allison K, Brogi E, Fox SB, Lakhani S, et al. The 2019 WHO classification of tumours of the breast. Histopathology. 2020;77:181–5.
Lüönd F, Tiede S, Christofori G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br J Cancer. 2021;125:164–75.
Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J, et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5:2929–43.
Chen R, Goodison S, Sun Y. Molecular profiles of matched primary and metastatic tumor samples support a linear evolutionary model of breast cancer. Cancer Res. 2020;80:170–4.
Wang Y, Armstrong SA. Cancer: inappropriate expression of stem cell programs? Cell Stem Cell. 2008;2:297–9.
Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007.
Zardavas D, Irrthum A, Swanton C, Piccart M. Clinical management of breast cancer heterogeneity. Nat Rev Clin Oncol. 2015;12:381–94.
Brasó-Maristany F, Griguolo G, Pascual T, Paré L, Nuciforo P, Llombart-Cussac A, et al. Phenotypic changes of HER2-positive breast cancer during and after dual HER2 blockade. Nat Commun. 2020;11:385.
Falato C, Schettini F, Pascual T, Brasó-Maristany F, Prat A. Clinical implications of the intrinsic molecular subtypes in hormone receptor-positive and HER2-negative metastatic breast cancer. Cancer Treat Rev. 2023;112:102496.
Zhang R, Tu J, Liu S. Novel molecular regulators of breast cancer stem cell plasticity and heterogeneity. Semin Cancer Biol. 2022;82:11–25.
Burr R, Gilles C, Thompson EW, Maheswaran S. Epithelial-mesenchymal plasticity in circulating tumor cells, the precursors of metastasis. Adv Exp Med Biol. 2020;1220:11–34.
Mao X, Jin F. The exosome and breast cancer cell plasticity. Onco Targets Ther. 2019;12:9817–25.
Wang QA, Scherer PE. Remodeling of murine mammary adipose tissue during pregnancy, lactation, and involution. J Mammary Gland Biol Neoplasia. 2019;24:207–12.
Kong D, Hughes CJ, Ford HL. Cellular plasticity in breast cancer progression and therapy. Front Mol Biosci. 2020;7:72.
Turner KM, Yeo SK, Holm TM, Shaughnessy E, Guan JL. Heterogeneity within molecular subtypes of breast cancer. Am J Physiol Cell Physiol. 2021;321:C343–54.
Place AE, Jin Huh S, Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Res. 2011;13:227.
Drago JZ, Formisano L, Juric D, Niemierko A, Servetto A, Wander SA, et al. FGFR1 gene amplification mediates endocrine resistance but retains TORC sensitivity in metastatic hormone receptor positive (HR+) breast cancer. Clin Cancer Res. 2019;25:6443–51.
Formisano L, Lu Y, Servetto A, Hanker AB, Jansen VM, Bauer JA, et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat Commun. 2019;10:1–14.
Giltnane JM, Hutchinson KE, Stricker TP, Formisano L, Young CD, Estrada MV, et al. Genomic profiling of ER+ breast cancers after short-term estrogen suppression reveals alterations associated with endocrine resistance. Sci Transl Med. 2017;9:eaai7993.
Braun M, Piasecka D, Tomasik B, Mieczkowski K, Stawiski K, Zielinska A, et al. Hormonal receptor status determines prognostic significance of FGFR2 in invasive breast carcinoma. Cancers. 2020;12:2713.
Mieczkowski K, Kitowska K, Braun M, Galikowska-Bogut B, Gorska-Arcisz M, Piasecka D, et al. FGF7/FGFR2–JunB signalling counteracts the effect of progesterone in luminal breast cancer. Mol Oncol. 2022;16:2823–42.
Piasecka D, Kitowska K, Czaplinska D, Mieczkowski K, Mieszkowska M, Turczyk L, et al. Fibroblast growth factor signalling induces loss of progesterone receptor in breast cancer cells. Oncotarget. 2016;7:86011–25.
Turczyk L, Kitowska K, Mieszkowska M, Mieczkowski K, Czaplinska D, Piasecka D, et al. FGFR2-driven signaling counteracts tamoxifen effect on ERalpha-positive breast cancer cells. Neoplasia. 2017;19:791–804.
Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, et al. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9:R23.
Krishnamurti U, Silverman JF. HER2 in breast cancer: a review and update. Adv Anat Pathol. 2014;21:100–7.
Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–94.
Hanker AB, Garrett JT, Estrada MV, Moore PD, Ericsson PG, Koch JP, et al. HER2-overexpressing breast cancers amplify FGFR signaling upon acquisition of resistance to dual therapeutic blockade of HER2. Clin Cancer Res. 2017;23:4323–34.
Gaibar M, Novillo A, Romero-Lorca A, Malón D, Antón B, Moreno A, et al. FGFR1 Amplification and response to neoadjuvant Anti-HER2 treatment in early HER2-positive breast cancer. Pharmaceutics. 2022;14:242.
Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–66.
Levine KM, Ding K, Chen L, Oesterreich S. FGFR4: a promising therapeutic target for breast cancer and other solid tumors. Pharmacol Ther. 2020;214:107590.
Kostrzewa M, Müller U. Genomic structure and complete sequence of the human FGFR4 gene. Mamm Genome. 1998;9:131–5.
Holzmann K, Grunt T, Heinzle C, Sampl S, Steinhoff H, Reichmann N, et al. Alternative splicing of fibroblast growth factor receptor IgIII Loops in cancer. J Nucleic Acids. 2012;2012:950508.
Garcia-Recio S, Thennavan A, East MP, Parker JS, Cejalvo JM, Garay JP, et al. FGFR4 regulates tumor subtype differentiation in luminal breast cancer and metastatic disease. J Clin Investig. 2020;130:4871–87.
Liu R, Li J, Xie K, Zhang T, Lei Y, Chen Y, et al. FGFR4 promotes stroma-induced epithelial-to-mesenchymal transition in colorectal cancer. Cancer Res. 2013;73:5926–35.
Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Biol. 2013;14:166–80.
Urakawa I, Yamazaki Y, Shimada T, Iijima K, Hasegawa H, Okawa K, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–4.
Kurosu H, Choi M, Ogawa Y, Dickson AS, Goetz R, Eliseenkova AV, et al. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J Biol Chem. 2007;282:26687–95.
Li SA, Watanabe M, Yamada H, Nagai A, Kinuta M, Takei K. Immunohistochemical localization of Klotho protein in brain, kidney, and reproductive organs of mice. Cell Struct Funct. 2004;29:91–9.
Lin BC, Wang M, Blackmore C, Desnoyers LR. Liver-specific activities of FGF19 require Klotho beta. J Biol Chem. 2007;282:27277–84.
Chen Z, Jiang L, Liang L, Koral K, Zhang Q, Zhao L, et al. The role of fibroblast growth factor 19 in hepatocellular carcinoma. Am J Pathol. 2021;191:1180–92.
Buhmeida A, Dallol A, Merdad A, Al-Maghrabi J, Gari MA, Abu-Elmagd MM, et al. High fibroblast growth factor 19 (FGF19) expression predicts worse prognosis in invasive ductal carcinoma of breast. Tumour Biol. 2014;35:2817–24.
Dallol A, Buhmeida A, Merdad A, Al-Maghrabi J, Gari MA, Abu-Elmagd MM, et al. Frequent methylation of the KLOTHO gene and overexpression of the FGFR4 receptor in invasive ductal carcinoma of the breast. Tumour Biol. 2015;36:9677–83.
Ligumsky H, Rubinek T, Merenbakh-Lamin K, Yeheskel A, Sertchook R, Shahmoon S, et al. Tumor suppressor activity of Klotho in breast cancer is revealed by structure-function analysis. Mol Cancer Res. 2015;13:1398–407.
Rubinek T, Shulman M, Israeli S, Bose S, Avraham A, Zundelevich A, et al. Epigenetic silencing of the tumor suppressor klotho in human breast cancer. Breast Cancer Res Treat. 2012;133:649–57.
Wolf I, Levanon-Cohen S, Bose S, Ligumsky H, Sredni B, Kanety H, et al. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene. 2008;27:7094–105.
Weinstein M, Xu X, Ohyama K, Deng CX. FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development. 1998;125:3615–23.
Korhonen J, Partanen J, Alitalo K. Expression of FGFR-4 mRNA in developing mouse tissues. Int J Dev Biol. 1992;36:323–9.
Liu Y, Cao M, Cai Y, Li X, Zhao C, Cui R. Dissecting the role of the FGF19-FGFR4 signaling pathway in cancer development and progression. Front Cell Dev Biol. 2020;8:95.
Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419.
Levine KM, Priedigkeit N, Basudan A, Tasdemir N, Sikora MJ, Sokol ES, et al. FGFR4 overexpression and hotspot mutations in metastatic ER+ breast cancer are enriched in the lobular subtype. NPJ Breast Cancer. 2019;5:19.
Wei W, You Z, Sun S, Wang Y, Zhang X, Pang D, et al. Prognostic implications of fibroblast growth factor receptor 4 polymorphisms in primary breast cancer. Mol Carcinogen. 2018;57:988–96.
Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, et al. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 2002;62:840–7.
Xiong SW, Ma J, Feng F, Fu W, Shu SR, Ma T, et al. Functional FGFR4 Gly388Arg polymorphism contributes to cancer susceptibility: evidence from meta-analysis. Oncotarget. 2017;8:25300–9.
Xu W, Li Y, Wang X, Chen B, Wang Y, Liu S, et al. FGFR4 transmembrane domain polymorphism and cancer risk: a meta-analysis including 8555 subjects. Eur J Cancer. 2010;46:3332–8.
Agarwal D, Pineda S, Michailidou K, Herranz J, Pita G, Moreno LT, et al. FGF receptor genes and breast cancer susceptibility: results from the Breast Cancer Association Consortium. Br J Cancer. 2014;110:1088–100.
Jézéquel P, Campion L, Joalland MP, Millour M, Dravet F, Classe JM, et al. G388R mutation of the FGFR4 gene is not relevant to breast cancer prognosis. Br J Cancer. 2004;90:189–93.
Becker N, Nieters A, Chang-Claude J. The fibroblast growth factor receptor gene Arg388 allele is not associated with early lymph node metastasis of breast cancer. Cancer Epidemiol Biomark Prev. 2003;12:582–3.
Ulaganathan VK, Sperl B, Rapp UR, Ullrich A. Germline variant FGFR4 p.G388R exposes a membrane-proximal STAT3 binding site. Nature. 2015;528:570–4.
Seitzer N, Mayr T, Streit S, Ullrich A. A single nucleotide change in the mouse genome accelerates breast cancer progression. Cancer Res. 2010;70:802–12.
Gu W, Yang J, Wang Y, Xu J, Wang X, Du F, et al. Comprehensive identification of FGFR1-4 alterations in 5 557 Chinese patients with solid tumors by next-generation sequencing. Am J Cancer Res. 2021;11:3893–906.
Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res. 2016;22:259–67.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Frullanti E, Berking C, Harbeck N, Jézéquel P, Haugen A, Mawrin C, et al. Meta and pooled analyses of FGFR4 Gly388Arg polymorphism as a cancer prognostic factor. Eur J Cancer Prev. 2011;20:340–7.
Marmé F, Werft W, Benner A, Burwinkel B, Sinn P, Sohn C, et al. FGFR4 Arg388 genotype is associated with pathological complete response to neoadjuvant chemotherapy for primary breast cancer. Ann Oncol. 2010;21:1636–42.
Cejalvo JM, Martínez de Dueñas E, Galván P, García-Recio S, Burgués Gasión O, Paré L, et al. Intrinsic subtypes and gene expression profiles in primary and metastatic breast cancer. Cancer Res. 2017;77:2213–21.
Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7.
Nguyen B, Fong C, Luthra A, Smith SA, DiNatale RG, Nandakumar S, et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 2022;185:563–75.e11.
Freitag CE, Mei P, Wei L, Parwani AV, Li Z. Genetic alterations and their association with clinicopathologic characteristics in advanced breast carcinomas: focusing on clinically actionable genetic alterations. Hum Pathol. 2020;102:94–103.
Zou Y, Zheng S, Xie X, Ye F, Hu X, Tian Z, et al. N6-methyladenosine regulated FGFR4 attenuates ferroptotic cell death in recalcitrant HER2-positive breast cancer. Nat Commun. 2022;13:2672.
Varešlija D, Priedigkeit N, Fagan A, Purcell S, Cosgrove N, O’Halloran PJ, et al. Transcriptome Characterization of matched primary breast and brain metastatic tumors to detect novel actionable targets. J Natl Cancer Inst. 2019;111:388–98.
Aftimos P, Oliveira M, Irrthum A, Fumagalli D, Sotiriou C, Gal-Yam EN, et al. Genomic and transcriptomic analyses of breast cancer primaries and matched metastases in AURORA, the Breast International Group (BIG) molecular screening initiative. Cancer Discov. 2021;11:2796–811.
Jordan NV, Bardia A, Wittner BS, Benes C, Ligorio M, Zheng Y, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102–6.
Dai LJ, Ma D, Xu YZ, Li M, Li YW, Xiao Y, et al. Molecular features and clinical implications of the heterogeneity in Chinese patients with HER2-low breast cancer. Nat Commun. 2023;14:5112.
Priedigkeit N, Hartmaier RJ, Chen Y, Vareslija D, Basudan A, Watters RJ, et al. Intrinsic subtype switching and acquired ERBB2/HER2 amplifications and mutations in breast cancer brain metastases. JAMA Oncol. 2017;3:666–71.
Zhao X, Xu F, Dominguez NP, Xiong Y, Xiong Z, Peng H, et al. FGFR4 provides the conduit to facilitate FGF19 signaling in breast cancer progression. Mol Carcinog. 2018;57:1616–25.
Luo Y, Yang C, Ye M, Jin C, Abbruzzese JL, Lee MH, et al. Deficiency of metabolic regulator FGFR4 delays breast cancer progression through systemic and microenvironmental metabolic alterations. Cancer Metab. 2013;1:21.
Turunen SP, von Nandelstadh P, Öhman T, Gucciardo E, Seashore-Ludlow B, Martins B, et al. FGFR4 phosphorylates MST1 to confer breast cancer cells resistance to MST1/2-dependent apoptosis. Cell Death Differ. 2019;26:2577–93.
Tiong KH, Tan BS, Choo HL, Chung FF, Hii LW, Tan SH, et al. Fibroblast growth factor receptor 4 (FGFR4) and fibroblast growth factor 19 (FGF19) autocrine enhance breast cancer cells survival. Oncotarget. 2016;7:57633–50.
Roidl A, Berger HJ, Kumar S, Bange J, Knyazev P, Ullrich A. Resistance to chemotherapy is associated with fibroblast growth factor receptor 4 up-regulation. Clin Cancer Res. 2009;15:2058–66.
Xu M, Chen S, Yang W, Cheng X, Ye Y, Mao J, et al. FGFR4 links glucose metabolism and chemotherapy resistance in breast cancer. Cell Physiol Biochem. 2018;47:151–60.
Boise LH, González-García M, Postema CE, Ding L, Lindsten T, Turka LA, et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The cancer cell line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Tomasich E, Steindl A, Paiato C, Hatziioannou T, Kleinberger M, Berchtold L, et al. Frequent overexpression of HER3 in brain metastases from breast and lung cancer. Clin Cancer Res. 2023;29:3225–36.
Hong CS, Sun EG, Choi JN, Kim DH, Kim JH, Ryu KH, et al. Fibroblast growth factor receptor 4 increases epidermal growth factor receptor (EGFR) signaling by inducing amphiregulin expression and attenuates response to EGFR inhibitors in colon cancer. Cancer Sci. 2020;111:3268–78.
Chew NJ, Lim Kam Sian TCC, Nguyen EV, Shin S-Y, Yang J, Hui MN, et al. Evaluation of FGFR targeting in breast cancer through interrogation of patient-derived models. Breast Cancer Res. 2021;23:82.
Kim JH, Jeong SY, Jang HJ, Park ST, Kim HS. FGFR4 Gly388Arg polymorphism reveals a poor prognosis, especially in asian cancer patients: a meta-analysis. Front Oncol. 2021;11:762528.
Thussbas C, Nahrig J, Streit S, Bange J, Kriner M, Kates R, et al. FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J Clin Oncol. 2006;24:3747–55.
Meijer D, Sieuwerts AM, Look MP, van Agthoven T, Foekens JA, Dorssers LC. Fibroblast growth factor receptor 4 predicts failure on tamoxifen therapy in patients with recurrent breast cancer. Endocr Relat Cancer. 2008;15:101–11.
Joensuu H, Bono P, Kataja V, Alanko T, Kokko R, Asola R, et al. Fluorouracil, epirubicin, and cyclophosphamide with either docetaxel or vinorelbine, with or without trastuzumab, as adjuvant treatments of breast cancer: final results of the FinHer Trial. J Clin Oncol. 2009;27:5685–92.
Dieci MV, Prat A, Tagliafico E, Paré L, Ficarra G, Bisagni G, et al. Integrated evaluation of PAM50 subtypes and immune modulation of pCR in HER2-positive breast cancer patients treated with chemotherapy and HER2-targeted agents in the CherLOB trial. Ann Oncol. 2016;27:1867–73.
Chen X, Huang Y, Chen B, Liu H, Cai Y, Yang Y. Insight into the design of FGFR4 selective inhibitors in cancer therapy: prospects and challenges. Eur J Med Chem. 2024;263:115947.
Facchinetti F, Hollebecque A, Bahleda R, Loriot Y, Olaussen KA, Massard C, et al. Facts and new hopes on selective FGFR Inhibitors in Solid Tumors. Clin Cancer Res. 2020;26:764–74.
Sung-Young S, Nicole JC, Milad G, Anderly CC, Lan KN, Roger JD. Integrative modelling of signalling network dynamics identifies cell type-selective therapeutic strategies for FGFR4-driven Cancers. bioRxiv. 2021:2021.11.03.467180.
De Luca A, Esposito Abate R, Rachiglio AM, Maiello MR, Esposito C, Schettino C, et al. FGFR fusions in cancer: from diagnostic approaches to therapeutic intervention. Int J Mol Sci. 2020;21:6856
Krook MA, Reeser JW, Ernst G, Barker H, Wilberding M, Li G, et al. Fibroblast growth factor receptors in cancer: genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br J Cancer. 2021;124:880–92.
Tao Z, Cui Y, Xu X, Han T. FGFR redundancy limits the efficacy of FGFR4-selective inhibitors in hepatocellular carcinoma. Proc Natl Acad Sci USA. 2022;119:e2208844119.
Wei W, Cao S, Liu J, Wang Y, Song Q, A L, et al. Fibroblast growth factor receptor 4 as a prognostic indicator in triple-negative breast cancer. Transl Cancer. Res. 2020;9:6881–8.
Acknowledgements
Figure 1 was created with BioRender.com.
Funding
This research was funded by the Polish National Science Centre grant OPUS no. 2020/39/B/NZ4/02696 (to HMR), PRELUDIUM no. 2018/29/N/NZ4/02384, LIDER no. 0188/L-13/2022 (to MB), Sonata Bis no. UMO-2018/30/E/NZ3/00222 (to RS).
Author information
Authors and Affiliations
Contributions
MB and DP reviewed literature, drafted the manuscript and prepared the figures. MB, DP, RS and HMR conceived the study. RS and HMR supervised the study and revised the manuscript. All Authors read and approved the final version of manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Braun, M., Piasecka, D., Sadej, R. et al. FGFR4-driven plasticity in breast cancer progression and resistance to therapy. Br J Cancer (2024). https://doi.org/10.1038/s41416-024-02658-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41416-024-02658-y