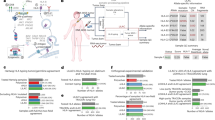
Abstract
Background
Intratumor heterogeneity (ITH) in microsatellite instability-high (MSI-H) colorectal cancer (CRC) has been poorly studied. We aimed to clarify how the ITH of MSI-H CRCs is generated in cancer evolution and how immune selective pressure affects ITH.
Methods
We reanalyzed public whole-exome sequencing data on 246 MSI-H CRCs. In addition, we performed a multi-region analysis from 6 MSI-H CRCs. To verify the process of subclonal immune escape accumulation, a novel computational model of cancer evolution under immune pressure was developed.
Results
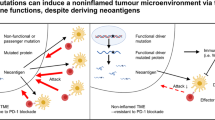
Our analysis presented the enrichment of functional genomic alterations in antigen-presentation machinery (APM). Associative analysis of neoantigens indicated the generation of immune escape mechanisms via HLA alterations. Multiregion analysis revealed the clonal acquisition of driver mutations and subclonal accumulation of APM defects in MSI-H CRCs. Examination of variant allele frequencies demonstrated that subclonal mutations tend to be subjected to selective sweep. Computational simulations of tumour progression with the interaction of immune cells successfully verified the subclonal accumulation of immune escape mutations and suggested the efficacy of early initiation of an immune checkpoint inhibitor (ICI) -based treatment.
Conclusions
Our results demonstrate the heterogeneous acquisition of immune escape mechanisms in MSI-H CRCs by Darwinian selection, providing novel insights into ICI-based treatment strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Raw sequencing data were deposited in the Japanese Genotype-phenotype Archive under the accession number JGAS000322.
References
Saito T, Niida A, Uchi R, Hirata H, Komatsu H, Sakimura S, et al. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nat Commun. 2018;9:2884.
Uchi R, Takahashi Y, Niida A, Shimamura T, Hirata H, Sugimachi K, et al. Integrated multiregional analysis proposing a new model of colorectal cancer evolution. PLoS Genet. 2016;12:e1005778.
Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–87.e2073.
Andre T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383:2207–18.
Kloor M, von Knebel Doeberitz M. The immune biology of microsatellite-unstable cancer. Trends Cancer. 2016;2:121–33.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumours with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20.
Friebel E, Kapolou K, Unger S, Nunez NG, Utz S, Rushing EJ, et al. Single-cell mapping of human brain cancer reveals tumour-specific instruction of tissue-invading leukocytes. Cell. 2020;181:1626–42.e1620.
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumours to PD-1 blockade. Science. 2017;357:409–13.
Grasso CS, Giannakis M, Wells DK, Hamada T, Mu XJ, Quist M, et al. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov. 2018;8:730–49.
Takatsuno Y, Mimori K, Yamamoto K, Sato T, Niida A, Inoue H, et al. The rs6983267 SNP is associated with MYC transcription efficiency, which promotes progression and worsens prognosis of colorectal cancer. Ann Surg Oncol. 2013;20:1395–402.
Shiraishi Y, Kataoka K, Chiba K, Okada A, Kogure Y, Tanaka H, et al. A comprehensive characterization of cis-acting splicing-associated variants in human cancer. Genome Res. 2018;28:1111–25.
Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Kurashige J, Hasegawa T, Niida A, Sugimachi K, Deng N, Mima K, et al. Integrated molecular profiling of human gastric cancer identifies ddr2 as a potential regulator of peritoneal dissemination. Sci Rep. 2016;6:22371.
Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878.
Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–61.
Iwasaki WM, Innan H. Simulation framework for generating intratumor heterogeneity patterns in a cancer cell population. PLoS One. 2017;12:e0184229.
Sato K, Kawazu M, Yamamoto Y, Ueno T, Kojima S, Nagae G, et al. Fusion kinases identified by genomic analyses of sporadic microsatellite instability-high colorectal cancers. Clin Cancer Res. 2019;25:378–89.
Jonchere V, Marisa L, Greene M, Virouleau A, Buhard O, Bertrand R, et al. Identification of positively and negatively selected driver gene mutations associated with colorectal cancer with microsatellite instability. Cell Mol Gastroenterol Hepatol. 2018;6:277–300.
Tikidzhieva A, Benner A, Michel S, Formentini A, Link KH, Dippold W, et al. Microsatellite instability and Beta2-Microglobulin mutations as prognostic markers in colon cancer: results of the FOGT-4 trial. Br J Cancer. 2012;106:1239–45.
de Miranda NF, van Dinther M, van den Akker BE, van Wezel T, ten Dijke P, Morreau H. Transforming growth factor β signaling in colorectal cancer cells with microsatellite instability despite biallelic mutations in TGFBR2. Gastroenterology. 2015;148:1427–37.e8.
Matsumoto A, Shimada Y, Nakano M, Oyanagi H, Tajima Y, Nakano M, et al. RNF43 mutation is associated with aggressive tumour biology along with BRAF V600E mutation in right-sided colorectal cancer. Oncol Rep. 2020;43:1853–62.
von Loga K, Woolston A, Punta M, Barber LJ, Griffiths B, Semiannikova M, et al. Extreme intratumor heterogeneity and driver evolution in mismatch repair deficient gastro-oesophageal cancer. Nat Commun. 2020;11:139.
Mitchell TJ, Turajlic S, Rowan A, Nicol D, Farmery JHR, O'Brien T, et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx Renal. Cell. 2018;173:611–23.e17.
Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–3.
Demeulemeester J, Dentro SC, Gerstung M, Van Loo P. Biallelic mutations in cancer genomes reveal local mutational determinants. Nat Genet. 2022;54:128–33.
Kohsaka S, Nagano M, Ueno T, Suehara Y, Hayashi T, Shimada N et al. A method of high-throughput functional evaluation of EGFR gene variants of unknown significance in cancer. Sci Transl Med. 2017;9;eaan6566.
Madsen RR, Knox RG, Pearce W, Lopez S, Mahler-Araujo B, McGranahan N, et al. Oncogenic PIK3CA promotes cellular stemness in an allele dose-dependent manner. Proc Natl Acad Sci USA. 2019;116:8380–9.
Saito Y, Koya J, Araki M, Kogure Y, Shingaki S, Tabata M, et al. Landscape and function of multiple mutations within individual oncogenes. Nature. 2020;582:95–99.
McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–71.e1211.
Aguiar VRC, Masotti C, Camargo AA, Meyer D. HLApers: HLA typing and quantification of expression with personalized index. Methods Mol Biol. 2020;2120:101–12.
Fayen J, Huang JH, Meyerson H, Zhang D, Getty R, Greenspan N, et al. Class I MHC alpha 3 domain can function as an independent structural unit to bind CD8 alpha. Mol Immunol. 1995;32:267–75.
Brusic V, Petrovsky N, Zhang G, Bajic VB. Prediction of promiscuous peptides that bind HLA class I molecules. Immunol Cell Biol. 2002;80:280–5.
Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–8.
Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A national cancer institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57.
Buhard O, Cattaneo F, Wong YF, Yim SF, Friedman E, Flejou JF, et al. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumours. J Clin Oncol. 2006;24:241–51.
Reiter JG, Makohon-Moore AP, Gerold JM, Bozic I, Chatterjee K, Iacobuzio-Donahue CA, et al. Reconstructing metastatic seeding patterns of human cancers. Nat Commun. 2017;8:14114.
Graham TA, Sottoriva A. Measuring cancer evolution from the genome. J Pathol. 2017;241:183–91.
Kawazu M, Ueno T, Saeki K, Sax N, Togashi Y, Kanaseki T, et al. HLA class I analysis provides insight into the genetic and epigenetic background of immune evasion in colorectal cancer with high microsatellite instability. Gastroenterology. 2022;162:799–812.
Montesion M, Murugesan K, Jin DX, Sharaf R, Sanchez N, Guria A, et al. Somatic HLA class I loss is a widespread mechanism of immune evasion which refines the use of tumour mutational burden as a biomarker of checkpoint inhibitor response. Cancer Discov. 2021;11:282–92.
Lakatos E, Williams MJ, Schenck RO, Cross WCH, Househam J, Zapata L, et al. Evolutionary dynamics of neoantigens in growing tumours. Nat Genet. 2020;52:1057–66.
Boudreau JE, Mulrooney TJ, Le Luduec JB, Barker E, Hsu KC. KIR3DL1 and HLA-B density and binding calibrate NK education and response to HIV. J Immunol. 2016;196:3398–410.
D'Amico S, D'Alicandro V, Compagnone M, Tempora P, Guida G, Romania P, et al. ERAP1 controls the interaction of the inhibitory receptor KIR3DL1 With HLA-B51:01 by affecting natural killer cell function. Front Immunol. 2021;12:778103.
Lipinski KA, Barber LJ, Davies MN, Ashenden M, Sottoriva A, Gerlinger M. Cancer evolution and the limits of predictability in precision cancer medicine. Trends Cancer. 2016;2:49–63.
Kakiuchi N, Yoshida K, Uchino M, Kihara T, Akaki K, Inoue Y, et al. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature. 2020;577:260–5.
Househam J, Heide T, Cresswell GD, Spiteri I, Kimberley C, Zapata L et al. Phenotypic plasticity and genetic control in colorectal cancer evolution. Nature. https://doi.org/10.1038/s41586-022-05311-x (2022).
Cercek A, Lumish M, Sinopoli J, Weiss J, Shia J, Lamendola-Essel M, et al. PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N Engl J Med. 2022;386:2363–76.
Gettinger S, Choi J, Hastings K, Truini A, Datar I, Sowell R, et al. Impaired HLA Class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov. 2017;7:1420–35.
D'Amico S, Tempora P, Melaiu O, Lucarini V, Cifaldi L, Locatelli F, et al. Targeting the antigen processing and presentation pathway to overcome resistance to immune checkpoint therapy. Front Immunol. 2022;13:948297.
Hazini A, Fisher K, Seymour L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J Immunother Cancer. 2021;9:e002899.
Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–29.
Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane JP, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun. 2017;8:1136.
Acknowledgements
We thank the members of the Institute for Department of Surgery of Kyushu University Beppu Hospital for sample collection and analysis. We also thank K. Oda, M. Kasagi, S. Sakuma, N. Mishima, T. Kawano, M. Oshiumi, and M. Utou for their technical assistance.
Funding
This project was supported by Science (JSPS) Grant-in-Aid for Science Research (15H05912, 19K09220, 22H04925:PAGS, 20H05039, 20K08930, 20K17556, 21K07179, 22K02903, 22K09006, 23K06765, and 23K08074), Priority Issue on Post-K computer (hp170227, hp160219), OITA Cancer Research Foundation (JP20cm0106475h0001), the Project for Cancer Research and Therapeutic Evolution (19 cm0106504h0004, 21 cm0106475h0002), AMED under Grant Number (23ck0106825h001, 23ck0106800h001, 22ama221501h0001, 21ck0106690s0201, 20ck0106547h0001, 20ck0106541h0001 and 22ama221XXXh0001), a research grant from the Takeda Science Foundation, and The Princess Takamatsu Cancer Research Fund. This study used the supercomputing resources provided by the Human Genome Center, Institute of Medical Science, University of Tokyo (http://sc.hgc.jp/shirokane.html).
Author information
Authors and Affiliations
Contributions
YK: Study design, sample collection, sample preparation, data analysis, and manuscript writing. AN, KSaeki, KT and HHaeno: Study design, project management, data analysis, and manuscript writing. SN: Study design, project management, sample collection, and sample preparation. TTobo: Histopathological diagnosis. SH, YO, HS, TH, and HN: Data analysis. AK, KSato, DS, HHirata, YH, TToshima, YY, TM, MO, SM, MU, MM, YD, and HE: Study design. TU, HM, and SI: Sample collection. YS: Sequence data assembly and study design. TS and KM: Project supervision, study design, and final approval of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study design was approved by the institutional review boards and ethics committees of hospitals where the study participants were admitted (Cancer Institute Hospital, Japanese Foundation for Cancer Research Institutional Review Board: Protocol Number 2011-1075, Kyushu University Institutional Review Board: Protocol Number 2020-74). The study was conducted in accordance with the principles of the Declaration of Helsinki. All patients provided informed consent.
Consent for publication
Consent was obtained from all participants to publish supporting data including Supplementary Table 1.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kobayashi, Y., Niida, A., Nagayama, S. et al. Subclonal accumulation of immune escape mechanisms in microsatellite instability-high colorectal cancers. Br J Cancer 129, 1105–1118 (2023). https://doi.org/10.1038/s41416-023-02395-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02395-8