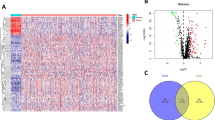
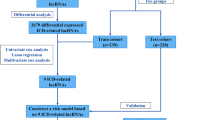
Abstract
Background
Long non-coding RNAs (lncRNAs)-related immune genes (lrRIGs) play a crucial role in the development and progression of lung adenocarcinoma (LUAD). However, reliable prognostic signatures based on lrRIGs have not yet been identified.
Methods
We screened lrRIGs associated with the prognosis of LUAD using The Cancer Genome Atlas (TCGA) database and then established a novel prognostic nine-gene signature composed of CD79A, INHA, SHC3, LIFR, TNFRSF11A, GPI, F2RL1, SEMA7A and WFDC2 through bioinformatic approaches. A risk score derived from this gene signature was used to divide LUAD patients into the low- and high-risk groups. The latter was confirmed to have markedly worse overall survival (O.S.). A nomogram was developed using the risk score and other independent prognostic elements, demonstrating excellent performance in predicting the O.S. rate of LUAD patients.
Results
We observed that the infiltration of diverse immune cell subtypes and response to immunotherapy and chemotherapy significantly differed between the low- and high-risk groups.
Conclusions
Overall, stratification based on this gene signature could be used to guide better therapeutic management and improve outcomes for LUAD patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Relevant research data have been presented in the text. All data will be provided upon request if necessary.
References
Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019;103:463–73. https://doi.org/10.1016/j.mcna.2018.12.006.
Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. https://doi.org/10.1158/1055-9965.Epi-15-0578.
Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–28. https://doi.org/10.1056/NEJMoa1501824.
Mazza F, Ferrari E, Maineri P, Dozin B, Ratto GB. Pleural lavage cytology predicts recurrence and survival, even in early non-small cell lung cancer. Surg Today. 2015;45:322–8. https://doi.org/10.1007/s00595-014-0915-3.
Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–71. https://doi.org/10.1038/nrc3611.
Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–84. https://doi.org/10.1101/gad.314617.118.
Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. https://doi.org/10.1056/NEJMoa1200690.
Lussier DM, O'Neill L, Nieves LM, McAfee MS, Holechek SA, Collins AW, et al. Enhanced T-cell immunity to osteosarcoma through antibody blockade of PD-1/PD-L1 interactions. J Immunother. 2015;38:96–106. https://doi.org/10.1097/cji.0000000000000065.
Gnjatic S, Bronte V, Brunet LR, Butler MO, Disis ML, Galon J, et al. Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer. 2017;5:44 https://doi.org/10.1186/s40425-017-0243-4.
Wang J, Ma X, Ma Z, Ma Y, Wang J, Cao B. Research progress of biomarkers for immune checkpoint inhibitors on digestive system cancers. Front Immunol. 2022;13:810539 https://doi.org/10.3389/fimmu.2022.810539.
Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118. https://doi.org/10.1038/s41580-020-00315-9.
Mathy NW, Chen XM. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J Biol Chem. 2017;292:12375–82. https://doi.org/10.1074/jbc.R116.760884.
Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18:962–72. https://doi.org/10.1038/ni.3771.
Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci USA. 2011;108:11381–6. https://doi.org/10.1073/pnas.1019711108.
Gao Y, Wang T, Li Y, Zhang Y, Yang R. Lnc-chop promotes immunosuppressive function of myeloid-derived suppressor cells in tumor and inflammatory environments. J Immunol. 2018;200:2603–14. https://doi.org/10.4049/jimmunol.1701721.
Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol. 2018;19:1112–25. https://doi.org/10.1038/s41590-018-0207-y.
Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q, et al. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 2019;18:115 https://doi.org/10.1186/s12943-019-1032-0.
Mohapatra S, Pioppini C, Ozpolat B, Calin GA. Non-coding RNAs regulation of macrophage polarization in cancer. Mol Cancer. 2021;20:24 https://doi.org/10.1186/s12943-021-01313-x.
Pi YN, Qi WC, Xia BR, Lou G, Jin WL. Long non-coding RNAs in the tumor immune microenvironment: biological properties and therapeutic potential. Front Immunol. 2021;12:697083 https://doi.org/10.3389/fimmu.2021.697083.
Li Y, Jiang T, Zhou W, Li J, Li X, Wang Q, et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat Commun. 2020;11:1000 https://doi.org/10.1038/s41467-020-14802-2.
Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014;15:R47 https://doi.org/10.1186/gb-2014-15-3-r47.
Zhang HJ, Chang WJ, Jia CY, Qiao L, Zhou J, Chen Q, et al. Destrin contributes to lung adenocarcinoma progression by activating Wnt/β-catenin signaling pathway. Mol Cancer Res. 2020;18:1789–802. https://doi.org/10.1158/1541-7786.Mcr-20-0187.
Sun S, Guo W, Wang Z, Wang X, Zhang G, Zhang H, et al. Development and validation of an immune-related prognostic signature in lung adenocarcinoma. Cancer Med. 2020;9:5960–75. https://doi.org/10.1002/cam4.3240.
Cao Y, Lu X, Li Y, Fu J, Li H, Li X, et al. Identification of a six-gene metabolic signature predicting overall survival for patients with lung adenocarcinoma. PeerJ. 2020;8:e10320 https://doi.org/10.7717/peerj.10320.
Zhang L, Zhang Z, Yu Z. Identification of a novel glycolysis-related gene signature for predicting metastasis and survival in patients with lung adenocarcinoma. J Transl Med. 2019;17:423 https://doi.org/10.1186/s12967-019-02173-2.
Li S, Xuan Y, Gao B, Sun X, Miao S, Lu T, et al. Identification of an eight-gene prognostic signature for lung adenocarcinoma. Cancer Manag Res. 2018;10:3383–92. https://doi.org/10.2147/cmar.S173941.
Ruf B, Heinrich B, Greten TF. Immunobiology and immunotherapy of HCC: spotlight on innate and innate-like immune cells. Cell Mol Immunol. 2021;18:112–27. https://doi.org/10.1038/s41423-020-00572-w.
Roh W, Chen PL, Reuben A, Spencer CN, Prieto PA, Miller JP, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9:eaah3560 https://doi.org/10.1126/scitranslmed.aah3560.
Kikuchi E, Yamazaki K, Torigoe T, Cho Y, Miyamoto M, Oizumi S, et al. HLA class I antigen expression is associated with a favorable prognosis in early stage non-small cell lung cancer. Cancer SCI. 2007;98:1424–30. https://doi.org/10.1111/j.1349-7006.2007.00558.
Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. https://doi.org/10.1126/science.aaa1348.
Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–1. https://doi.org/10.1056/NEJMc1713444.
Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. https://doi.org/10.1056/NEJMoa1406498.
McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–71. https://doi.org/10.1016/j.cell.2017.10.001.e11.
Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE. 2014;9:e107468 https://doi.org/10.1371/journal.pone.0107468.
Han J, Deng X, Sun R, Luo M, Liang M, Gu B, et al. GPI is a prognostic biomarker and correlates with immune infiltrates in lung adenocarcinoma. Front Oncol. 2021;11:752642 https://doi.org/10.3389/fonc.2021.752642.
Van Veen M, Matas-Rico E, van de Wetering K, Leyton-Puig D, Kedziora KM, De Lorenzi V, et al. Negative regulation of urokinase receptor activity by a GPI-specific phospholipase C in breast cancer cells. eLife 2017;6. https://doi.org/10.7554/eLife.23649.
Wu ST, Liu B, Ai ZZ, Hong ZC, You PT, Wu HZ, et al. Esculetin inhibits cancer cell glycolysis by binding tumor PGK2, GPD2, and GPI. Front Pharmacol. 2020;11:379 https://doi.org/10.3389/fphar.2020.00379.
Luo CT, Liao W, Dadi S, Toure A, Li MO. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature. 2016;529:532–6. https://doi.org/10.1038/nature16486.
Huang S, Houghton PJ. Targeting mTOR signaling for cancer therapy. Curr Opin Pharmacol. 2003;3:371–7. https://doi.org/10.1016/s1471-4892(03)00071-7.
Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. https://doi.org/10.1016/j.cell.2006.01.016.
Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. https://doi.org/10.1016/j.ccr.2007.05.008.
Shen S, Wang G, Zhang R, Zhao Y, Yu H, Wei Y, et al. Development and validation of an immune gene-set based prognostic signature in ovarian cancer. EBioMedicine. 2019;40:318–26. https://doi.org/10.1016/j.ebiom.2018.12.054.
Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157 https://doi.org/10.1186/gb-2007-8-8-r157.
Lin A, Hu Q, Li C, Xing Z, Ma G, Wang C, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P(3) to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol. 2017;19:238–51. https://doi.org/10.1038/ncb3473.
Sang LJ, Ju HQ, Liu GP, Tian T, Ma GL, Lu YX, et al. LncRNA CamK-A regulates Ca(2+)-signaling-mediated tumor microenvironment remodeling. Mol Cell. 2018;72:601 https://doi.org/10.1016/j.molcel.2018.10.024.
Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, Chu LW, et al. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis. 2011;32:1655–9. https://doi.org/10.1093/carcin/bgr187.
Li L, Jia F, Bai P, Liang Y, Sun R, Yuan F, et al. Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of gastric cancer. Tumour Biol. 2016;37:299–303. https://doi.org/10.1007/s13277-015-3750-2.
Li L, Wang Y, Song G, Zhang X, Gao S, Liu H. HOX cluster-embedded antisense long non-coding RNAs in lung cancer. Cancer Lett. 2019;450:14–21. https://doi.org/10.1016/j.canlet.2019.02.036.
Teng C, Huang G, Luo Y, Pan Y, Wang H, Liao X, et al. Differential long noncoding RNAs expression in cancer-associated fibroblasts of non-small-cell lung cancer. Pharmacogenomics. 2019;20:143–53. https://doi.org/10.2217/pgs-2018-0102.
Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol. 2019;20:835–51. https://doi.org/10.1038/s41590-019-0400-7.
Zhou WY, Zhang MM, Liu C, Kang Y, Wang JO, Yang XH. Long noncoding RNA LINC00473 drives the progression of pancreatic cancer via upregulating programmed death-ligand 1 by sponging microRNA-195-5p. J Cell Physiol. 2019;234:23176–89. https://doi.org/10.1002/jcp.28884.
Mason DY, Cordell JL, Brown MH, Borst J, Jones M, Pulford K, et al. CD79a: a novel marker for B-cell neoplasms in routinely processed tissue samples. Blood. 1995;86:1453–9.
Zomas AP, Matutes E, Morilla R, Owusu-Ankomah K, Seon BK, Catovsky D. Expression of the immunoglobulin-associated protein B29 in B cell disorders with the monoclonal antibody SN8 (CD79b). Leukemia. 1996;10:1966–70.
Viswanadhapalli S, Dileep KV, Zhang KYJ, Nair HB, Vadlamudi RK. Targeting LIF/LIFR signaling in cancer. Genes Dis. 2022;9:973–80. https://doi.org/10.1016/j.gendis.2021.04.003.
Pusapati RV, Daemen A, Wilson C, Sandoval W, Gao M, Haley B, et al. mTORC1-dependent metabolic reprogramming underlies escape from glycolysis addiction in cancer cells. Cancer Cell. 2016;29:548–62. https://doi.org/10.1016/j.ccell.2016.02.018.
Liotta LA, Mandler R, Murano G, Katz DA, Gordon RK, Chiang PK, et al. Tumor cell autocrine motility factor. Proc Natl Acad Sci USA. 1986;83:3302–6. https://doi.org/10.1073/pnas.83.10.3302.
Song Y, Wang L, Zhang L, Huang D. The involvement of semaphorin 7A in tumorigenic and immunoinflammatory regulation. J Cell Physiol. 2021;236:6235–48. https://doi.org/10.1002/jcp.30340.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. https://doi.org/10.1016/j.cell.2011.02.013.
Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–76. https://doi.org/10.1016/j.cell.2017.02.004.
Acknowledgements
We sincerely acknowledge the contributions from the TCGA, GEO, innateDB, GDSC and GSEA databases. This work was supported by the National Natural Science Foundation of China (Nos. 81372147, 81803575, 31902287), Henan University support grant CX3070A0780502, the Key Science and Technology Research and Development and Promotion Special Project of Henan Province (No. 232102311205), and the Key R&D and promotion projects of Kaifeng (No. 2203008).
Author information
Authors and Affiliations
Contributions
FL conceived and designed this article. ZY, JZ, TY, WT, and XZ participated in the experimental data collection; SJ provided technical assistance; FL drafted the manuscript; ZR revised the study draft. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The studies involving human participants were reviewed and approved by the Ethics Committee of Medical School of Henan University, China (HUSOM-2018-282). Informed consent was obtained from all subjects involved in the study.
Consent for publication
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41416_2023_2379_MOESM3_ESM.xlsx
Table S1. The clinicopathologic characteristics of 30 matched clinical LUAD samples employed for Western blot assay of protein expression of candidate genes.
41416_2023_2379_MOESM8_ESM.xlsx
Table S6. 27 lncRNAs-related immune genes were significantly associated with the OS of LUAD patients through univariate Cox regression analysis.
41416_2023_2379_MOESM9_ESM.xlsx
Table S7. Nine genes were screened to construct a significant prognostic signature by multivariate Cox regression analysis.
41416_2023_2379_MOESM10_ESM.xlsx
Table S8. Gene Set Enrichment Analysis (GSEA) was performed by comparing the high expression and low expression of the GPI gene based on the TCGA_LUAD, GSE31210, GSE68465 and GSE13213 databases.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, Z., Zhu, J., Yang, T. et al. Comprehensive analysis of the lncRNAs-related immune gene signatures and their correlation with immunotherapy in lung adenocarcinoma. Br J Cancer 129, 1397–1408 (2023). https://doi.org/10.1038/s41416-023-02379-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02379-8
This article is cited by
-
Immune check points in cancer treatment: current challenges and perspectives
British Journal of Cancer (2023)