Abstract
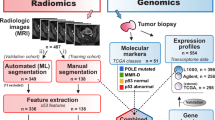
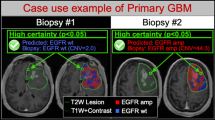
Radiogenomics, focusing on the relationship between genomics and imaging phenotypes, has been widely applied to address tumour heterogeneity and predict immune responsiveness and progression. It is an inevitable consequence of current trends in precision medicine, as radiogenomics costs less than traditional genetic sequencing and provides access to whole-tumour information rather than limited biopsy specimens. By providing voxel-by-voxel genetic information, radiogenomics can allow tailored therapy targeting a complete, heterogeneous tumour or set of tumours. In addition to quantifying lesion characteristics, radiogenomics can also be used to distinguish benign from malignant entities, as well as patient characteristics, to better stratify patients according to disease risk, thereby enabling more precise imaging and screening. Here, we have characterised the radiogenomic application in precision medicine using a multi-omic approach. we outline the main applications of radiogenomics in diagnosis, treatment planning and evaluations in the field of oncology with the aim of developing quantitative and personalised medicine. Finally, we discuss the challenges in the field of radiogenomics and the scope and clinical applicability of these methods.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Not applicable.
References
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–6.
van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e7.
Chaddad A, Daniel P, Sabri S, Desrosiers C, Abdulkarim B. Integration of radiomic and multi-omic analyses predicts survival of newly diagnosed IDH1 wild-type glioblastoma. Cancers. 2019;11:1148.
Ren Y, Zhang X, Rui W, Pang H, Qiu T, Wang J, et al. Noninvasive prediction of IDH1 mutation and ATRX expression loss in low-grade gliomas using multiparametric MR radiomic features. J Magn Reson Imaging. 2019;49:808–17.
Sakai Y, Yang C, Kihira S, Tsankova N, Khan F, Hormigo A, et al. MRI radiomic features to predict IDH1 mutation status in gliomas: a machine learning approach using gradient tree boosting. Int J Mol Sci. 2020;21:8004.
Bangalore Yogananda CG, Shah BR, Vejdani-Jahromi M, Nalawade SS, Murugesan GK, Yu FF, et al. A novel fully automated MRI-based deep-learning method for classification of IDH mutation status in brain gliomas. Neuro Oncol. 2020;22:402–11.
Choi YS, Bae S, Chang JH, Kang SG, Kim SH, Kim J, et al. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro Oncol. 2021;23:304–13.
Li Y, Liu X, Qian Z, Sun Z, Xu K, Wang K, et al. Genotype prediction of ATRX mutation in lower-grade gliomas using an MRI radiomics signature. Eur Radiol. 2018;28:2960–8.
Haubold J, Demircioglu A, Gratz M, Glas M, Wrede K, Sure U, et al. Non-invasive tumor decoding and phenotyping of cerebral gliomas utilizing multiparametric (18)F-FET PET-MRI and MR fingerprinting. Eur J Nucl Med Mol Imaging. 2020;47:1435–45.
Wu S, Zhang X, Rui W, Sheng Y, Yu Y, Zhang Y, et al. A nomogram strategy for identifying the subclassification of IDH mutation and ATRX expression loss in lower-grade gliomas. Eur Radiol. 2022;32:3187–98.
Wei J, Yang G, Hao X, Gu D, Tan Y, Wang X, et al. A multi-sequence and habitat-based MRI radiomics signature for preoperative prediction of MGMT promoter methylation in astrocytomas with prognostic implication. Eur Radiol. 2019;29:877–88.
Qian J, Herman MG, Brinkmann DH, Laack NN, Kemp BJ, Hunt CH, et al. Prediction of MGMT Status for Glioblastoma Patients Using Radiomics Feature Extraction From (18)F-DOPA-PET Imaging. Int J Radiat Oncol Biol Phys. 2020;108:1339–46.
Li ZC, Bai H, Sun Q, Li Q, Liu L, Zou Y, et al. Multiregional radiomics features from multiparametric MRI for prediction of MGMT methylation status in glioblastoma multiforme: a multicentre study. Eur Radiol. 2018;28:3640–50.
Kickingereder P, Neuberger U, Bonekamp D, Piechotta PL, Gotz M, Wick A, et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol. 2018;20:848–57.
Verduin M, Primakov S, Compter I, Woodruff HC, van Kuijk SMJ, Ramaekers BLT, et al. Prognostic and predictive value of integrated qualitative and quantitative magnetic resonance imaging analysis in glioblastoma. Cancers. 2021;13:722.
Mu W, Jiang L, Zhang J, Shi Y, Gray JE, Tunali I, et al. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat Commun. 2020;11:5228.
Chen BT, Jin T, Ye N, Mambetsariev I, Wang T, Wong CW, et al. Predicting survival duration with MRI radiomics of brain metastases from non-small cell lung cancer. Front Oncol. 2021;11:621088.
Song J, Wang L, Ng NN, Zhao M, Shi J, Wu N, et al. Development and validation of a machine learning model to explore tyrosine kinase inhibitor response in patients with stage IV EGFR variant-positive non-small cell lung cancer. JAMA Netw Open. 2020;3:e2030442.
Zhang T, Xu Z, Liu G, Jiang B, de Bock GH, Groen HJM, et al. Simultaneous identification of EGFR, KRAS, ERBB2, and TP53 mutations in patients with non-small cell lung cancer by machine learning-derived three-dimensional radiomics. Cancers. 2021;13:1814.
Wang J, Lv X, Huang W, Quan Z, Li G, Wu S, et al. Establishment and optimization of radiomics algorithms for prediction of KRAS gene mutation by integration of NSCLC gene mutation mutual exclusion information. Front Pharm. 2022;13:862581.
Agazzi GM, Ravanelli M, Roca E, Medicina D, Balzarini P, Pessina C, et al. CT texture analysis for prediction of EGFR mutational status and ALK rearrangement in patients with non-small cell lung cancer. Radio Med. 2021;126:786–94.
Song L, Zhu Z, Mao L, Li X, Han W, Du H, et al. Clinical, conventional CT and radiomic feature-based machine learning models for predicting ALK rearrangement status in lung adenocarcinoma patients. Front Oncol. 2020;10:369.
Chang C, Sun X, Wang G, Yu H, Zhao W, Ge Y, et al. A machine learning model based on PET/CT radiomics and clinical characteristics predicts ALK rearrangement status in lung adenocarcinoma. Front Oncol. 2021;11:603882.
Yang L, Dong D, Fang M, Zhu Y, Zang Y, Liu Z, et al. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur Radiol. 2018;28:2058–67.
Cui Y, Liu H, Ren J, Du X, Xin L, Li D, et al. Development and validation of a MRI-based radiomics signature for prediction of KRAS mutation in rectal cancer. Eur Radiol. 2020;30:1948–58.
Fathi Kazerooni A, Saxena S, Toorens E, Tu D, Bashyam V, Akbari H, et al. Clinical measures, radiomics, and genomics offer synergistic value in AI-based prediction of overall survival in patients with glioblastoma. Sci Rep. 2022;12:8784.
Shao L, Zuo X, Yang Y, Zhang Y, Yang N, Shen B, et al. The inherited variations of a p53-responsive enhancer in 13q12.12 confer lung cancer risk by attenuating TNFRSF19 expression. Genome Biol. 2019;20:103.
Crawford DC, Nickerson DA. Definition and clinical importance of haplotypes. Annu Rev Med. 2005;56:303–20.
Boettger LM, Salem RM, Handsaker RE, Peloso GM, Kathiresan S, Hirschhorn JN, et al. Recurring exon deletions in the HP (haptoglobin) gene contribute to lower blood cholesterol levels. Nat Genet. 2016;48:359–66.
Nativio R, Donahue G, Berson A, Lan Y, Amlie-Wolf A, Tuzer F, et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat Neurosci. 2018;21:497–505.
Kerns SL, Fachal L, Dorling L, Barnett GC, Baran A, Peterson DR, et al. Radiogenomics consortium genome-wide association study meta-analysis of late toxicity after prostate cancer radiotherapy. J Natl Cancer Inst. 2020;112:179–90.
Seibold P, Behrens S, Schmezer P, Helmbold I, Barnett G, Coles C, et al. XRCC1 polymorphism associated with late toxicity after radiation therapy in breast cancer patients. Int J Radiat Oncol Biol Phys. 2015;92:1084–92.
Tan WL, Jain A, Takano A, Newell EW, Iyer NG, Lim WT, et al. Novel therapeutic targets on the horizon for lung cancer. Lancet Oncol. 2016;17:e347–e62.
Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587–95.
Jiang M, Zhang X, Chen Y, Chen P, Guo X, Ma L, et al. A review of the correlation between epidermal growth factor receptor mutation status and (18)F-FDG metabolic activity in non-small cell lung cancer. Front Oncol. 2022;12:780186.
Lv Z, Fan J, Xu J, Wu F, Huang Q, Guo M, et al. Value of (18)F-FDG PET/CT for predicting EGFR mutations and positive ALK expression in patients with non-small cell lung cancer: a retrospective analysis of 849 Chinese patients. Eur J Nucl Med Mol Imaging. 2018;45:735–50.
Zhang J, Zhao X, Zhao Y, Zhang J, Zhang Z, Wang J, et al. Value of pre-therapy (18)F-FDG PET/CT radiomics in predicting EGFR mutation status in patients with non-small cell lung cancer. Eur J Nucl Med Mol Imaging. 2020;47:1137–46.
Zhang L, Chen B, Liu X, Song J, Fang M, Hu C, et al. Quantitative biomarkers for prediction of epidermal growth factor receptor mutation in non-small cell lung cancer. Transl Oncol. 2018;11:94–101.
Nair JKR, Saeed UA, McDougall CC, Sabri A, Kovacina B, Raidu BVS, et al. Radiogenomic models using machine learning techniques to predict EGFR mutations in non-small cell lung cancer. Can Assoc Radiol J. 2021;72:109–19.
Therkildsen C, Bergmann TK, Henrichsen-Schnack T, Ladelund S, Nilbert M. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: a systematic review and meta-analysis. Acta Oncol. 2014;53:852–64.
Feng Y, Bommer GT, Zhao J, Green M, Sands E, Zhai Y, et al. Mutant KRAS promotes hyperplasia and alters differentiation in the colon epithelium but does not expand the presumptive stem cell pool. Gastroenterology 2011;141:1003–13.
Nishihara R, Lochhead P, Kuchiba A, Jung S, Yamauchi M, Liao X, et al. Aspirin use and risk of colorectal cancer according to BRAF mutation status. J Am Med Assoc. 2013;309:2563–71.
Shin YR, Kim KA, Im S, Hwang SS, Kim K. Prediction of KRAS mutation in rectal cancer using MRI. Anticancer Res. 2016;36:4799–804.
Lubner MG, Stabo N, Lubner SJ, del Rio AM, Song C, Halberg RB, et al. CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging. 2015;40:2331–7.
Brooks SA, Brannon AR, Parker JS, Fisher JC, Sen O, Kattan MW, et al. ClearCode34: a prognostic risk predictor for localized clear cell renal cell carcinoma. Eur Urol. 2014;66:77–84.
Jamshidi N, Diehn M, Bredel M, Kuo MD. Illuminating radiogenomic characteristics of glioblastoma multiforme through integration of MR imaging, messenger RNA expression, and DNA copy number variation. Radiology. 2014;270:1–2.
Kwee SA, Tiirikainen M, Sato MM, Acoba JD, Wei R, Jia W, et al. Transcriptomics associates molecular features with (18)F-fluorocholine PET/CT imaging phenotype and its potential relationship to survival in hepatocellular carcinoma. Cancer Res. 2019;79:1696–704.
Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. AJR Am J Roentgenol. 2012;199:654–63.
Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–28.
Gao R, Ye M, Liu B, Wei M, Ma D, Dong K. m6A modification: a double-edged sword in tumor development. Front Oncol. 2021;11:679367.
Li W, Zhang X, Lu X, You L, Song Y, Luo Z, et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017;27:1243–57.
Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S, et al. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene. 2020;39:6906–19.
Cen D, Xu L, Zhang S, Chen Z, Huang Y, Li Z, et al. Renal cell carcinoma: predicting RUNX3 methylation level and its consequences on survival with CT features. Eur Radiol. 2019;29:5415–22.
Kanas VG, Zacharaki EI, Thomas GA, Zinn PO, Megalooikonomou V, Colen RR. Learning MRI-based classification models for MGMT methylation status prediction in glioblastoma. Comput Methods Prog Biomed. 2017;140:249–57.
Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–37.
Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017;16:41.
Kuo MD, Yamamoto S. Next generation radiologic-pathologic correlation in oncology: Rad-Path 2.0. AJR Am J Roentgenol. 2011;197:990–7.
Fan M, Xia P, Clarke R, Wang Y, Li L. Radiogenomic signatures reveal multiscale intratumour heterogeneity associated with biological functions and survival in breast cancer. Nat Commun. 2020;11:4861.
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–20.
Zhang B, Chang K, Ramkissoon S, Tanguturi S, Bi WL, Reardon DA, et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol. 2017;19:109–17.
Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, et al. Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res. 2018;24:1073–81.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Kyndi M, Sorensen FB, Knudsen H, Overgaard M, Nielsen HM, Overgaard J, et al. Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: the Danish Breast Cancer Cooperative Group. J Clin Oncol. 2008;26:1419–26.
Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–34.
Waks AG, Winer EP. Breast cancer treatment: a review. J Am Med Assoc. 2019;321:288–300.
Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI. Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology. 2014;273:365–72.
Leithner D, Bernard-Davila B, Martinez DF, Horvat JV, Jochelson MS, Marino MA, et al. Radiomic Signatures derived from diffusion-weighted imaging for the assessment of breast cancer receptor status and molecular subtypes. Mol Imaging Biol. 2020;22:453–61.
Xie T, Zhao Q, Fu C, Bai Q, Zhou X, Li L, et al. Differentiation of triple-negative breast cancer from other subtypes through whole-tumor histogram analysis on multiparametric MR imaging. Eur Radiol. 2019;29:2535–44.
Dilorenzo G, Telegrafo M, La Forgia D, Stabile Ianora AA, Moschetta M. Breast MRI background parenchymal enhancement as an imaging bridge to molecular cancer sub-type. Eur J Radiol. 2019;113:148–52.
Ramaswamy V, Remke M, Bouffet E, Bailey S, Clifford SC, Doz F, et al. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol. 2016;131:821–31.
Yan J, Liu L, Wang W, Zhao Y, Li KK, Li K, et al. Radiomic features from multi-parameter MRI combined with clinical parameters predict molecular subgroups in patients with medulloblastoma. Front Oncol. 2020;10:558162.
Mu W, Jiang L, Shi Y, Tunali I, Gray JE, Katsoulakis E, et al. Non-invasive measurement of PD-L1 status and prediction of immunotherapy response using deep learning of PET/CT images. J Immunother Cancer. 2021;9:e002118.
Sun R, Henry T, Laville A, Carre A, Hamaoui A, Bockel S, et al. Imaging approaches and radiomics: toward a new era of ultraprecision radioimmunotherapy? J Immunother Cancer. 2022;10:e004848.
Dall’Olio FG, Calabro D, Conci N, Argalia G, Marchese PV, Fabbri F, et al. Baseline total metabolic tumour volume on 2-deoxy-2-[18F]fluoro-d-glucose positron emission tomography-computed tomography as a promising biomarker in patients with advanced non-small cell lung cancer treated with first-line pembrolizumab. Eur J Cancer. 2021;150:99–107.
Kerns SL, Ostrer H, Rosenstein BS. Radiogenomics: using genetics to identify cancer patients at risk for development of adverse effects following radiotherapy. Cancer Discov. 2014;4:155–65.
Peng Z, Wang Y, Wang Y, Jiang S, Fan R, Zhang H, et al. Application of radiomics and machine learning in head and neck cancers. Int J Biol Sci. 2021;17:475–86.
Kim JH, Ko ES, Lim Y, Lee KS, Han BK, Ko EY, et al. Breast cancer heterogeneity: MR imaging texture analysis and survival outcomes. Radiology. 2017;282:665–75.
van Willigen WW, Gerritsen WR, Aarntzen E. 18F-FDG PET/CT of multiorgan sarcoid-like reaction during anti-PD-1 treatment for melanoma. Clin Nucl Med. 2019;44:905–6.
Chen R, Zhou X, Liu J, Huang G. Relationship between the expression of PD-1/PD-L1 and (18)F-FDG uptake in bladder cancer. Eur J Nucl Med Mol Imaging. 2019;46:848–54.
Xie Y, Liu C, Zhao Y, Gong C, Li Y, Hu S, et al. Heterogeneity derived from (18) F-FDG PET/CT predicts immunotherapy outcome for metastatic triple-negative breast cancer patients. Cancer Med. 2022;11:1948–55.
Mittendorf EA, Jeruss JS, Tucker SL, Kolli A, Newman LA, Gonzalez-Angulo AM, et al. Validation of a novel staging system for disease-specific survival in patients with breast cancer treated with neoadjuvant chemotherapy. J Clin Oncol. 2011;29:1956–62.
Nakashoji A, Matsui A, Nagayama A, Iwata Y, Sasahara M, Murata Y. Clinical predictors of pathological complete response to neoadjuvant chemotherapy in triple-negative breast cancer. Oncol Lett. 2017;14:4135–41.
Kickingereder P, Bonekamp D, Nowosielski M, Kratz A, Sill M, Burth S, et al. Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology 2016;281:907–18.
Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–91.
Kang J, Rancati T, Lee S, Oh JH, Kerns SL, Scott JG, et al. Machine learning and radiogenomics: lessons learned and future directions. Front Oncol. 2018;8:228.
Giraud P, Giraud P, Gasnier A, El Ayachy R, Kreps S, Foy JP, et al. Radiomics and machine learning for radiotherapy in head and neck cancers. Front Oncol. 2019;9:174.
Marcu LG, Forster JC, Bezak E. The potential role of radiomics and radiogenomics in patient stratification by tumor hypoxia status. J Am Coll Radiol. 2019;16:1329–37.
Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Inter Radiol. 2007;18:821–31.
Mazurowski MA. Radiogenomics: what it is and why it is important. J Am Coll Radiol. 2015;12:862–6.
Banerjee S, Wang DS, Kim HJ, Sirlin CB, Chan MG, Korn RL, et al. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62:792–800.
Buus R, Sestak I, Kronenwett R, Ferree S, Schnabel CA, Baehner FL, et al. Molecular drivers of oncotype DX, prosigna, endopredict, and the breast cancer index: a TransATAC study. J Clin Oncol. 2021;39:126–35.
Schaafsma E, Zhang B, Schaafsma M, Tong CY, Zhang L, Cheng C. Impact of oncotype DX testing on ER+ breast cancer treatment and survival in the first decade of use. Breast Cancer Res. 2021;23:74.
Nicolini A, Ferrari P, Duffy MJ. Prognostic and predictive biomarkers in breast cancer: past, present and future. Semin Cancer Biol. 2018;52:56–73.
Cheng R, Kong X, Wang X, Fang Y, Wang J. Oncotype DX breast recurrence score distribution and chemotherapy benefit among women of different age groups with HR-positive, HER2-negative, node-negative breast cancer in the SEER database. Front Oncol. 2020;10:1583.
Kondo M, Hoshi SL, Ishiguro H, Yoshibayashi H, Toi M. Economic evaluation of 21-gene reverse transcriptase-polymerase chain reaction assay in lymph-node-negative, estrogen-receptor-positive, early-stage breast cancer in Japan. Breast Cancer Res Treat. 2008;112:175–87.
Ashraf AB, Daye D, Gavenonis S, Mies C, Feldman M, Rosen M, et al. Identification of intrinsic imaging phenotypes for breast cancer tumors: preliminary associations with gene expression profiles. Radiology. 2014;272:374–84.
Ashraf AB, Gavenonis SC, Daye D, Mies C, Rosen MA, Kontos D. A multichannel Markov random field framework for tumor segmentation with an application to classification of gene expression-based breast cancer recurrence risk. IEEE Trans Med Imaging. 2013;32:637–48.
Woodard GA, Ray KM, Joe BN, Price ER. Qualitative radiogenomics: association between oncotype DX test recurrence score and BI-RADS mammographic and breast MR imaging features. Radiology. 2018;286:60–70.
Li H, Zhu Y, Burnside ES, Drukker K, Hoadley KA, Fan C, et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of mammaprint, oncotype DX, and PAM50 gene assays. Radiology. 2016;281:382–91.
Sutton EJ, Oh JH, Dashevsky BZ, Veeraraghavan H, Apte AP, Thakur SB, et al. Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. J Magn Reson Imaging. 2015;42:1398–406.
Dialani V, Gaur S, Mehta TS, Venkataraman S, Fein-Zachary V, Phillips J, et al. Prediction of low versus high recurrence scores in estrogen receptor-positive, lymph node-negative invasive breast cancer on the basis of radiologic-pathologic features: comparison with oncotype DX test recurrence scores. Radiology. 2016;280:370–8.
Ohnstad HO, Borgen E, Falk RS, Lien TG, Aaserud M, Sveli MAT, et al. Prognostic value of PAM50 and risk of recurrence score in patients with early-stage breast cancer with long-term follow-up. Breast Cancer Res. 2017;19:120.
Jamshidi N, Jonasch E, Zapala M, Korn RL, Aganovic L, Zhao H, et al. The radiogenomic risk score: construction of a prognostic quantitative, noninvasive image-based molecular assay for renal cell carcinoma. Radiology. 2015;277:114–23.
Jamshidi N, Jonasch E, Zapala M, Korn RL, Brooks JD, Ljungberg B, et al. The radiogenomic risk score stratifies outcomes in a renal cell cancer phase 2 clinical trial. Eur Radiol. 2016;26:2798–807.
Botta F, Raimondi S, Rinaldi L, Bellerba F, Corso F, Bagnardi V, et al. Association of a CT-based clinical and radiomics score of non-small cell lung cancer (NSCLC) with lymph node status and overall survival. Cancers. 2020;12:1432.
Fathi Kazerooni A, Bagley SJ, Akbari H, Saxena S, Bagheri S, Guo J, et al. Applications of radiomics and radiogenomics in high-grade gliomas in the era of precision medicine. Cancers. 2021;13:5921.
Jena B, Saxena S, Nayak GK, Balestrieri A, Gupta N, Khanna NN, et al. Brain tumor characterization using radiogenomics in artificial intelligence framework. Cancers. 2022;14:4052.
Saxena S, Jena B, Gupta N, Das S, Sarmah D, Bhattacharya P, et al. Role of artificial intelligence in radiogenomics for cancers in the era of precision medicine. Cancers. 2022;14:2860.
Berenguer R, Pastor-Juan MDR, Canales-Vazquez J, Castro-Garcia M, Villas MV, Mansilla Legorburo F, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288:407–15.
Pfaehler E, Zhovannik I, Wei L, Boellaard R, Dekker A, Monshouwer R, et al. A systematic review and quality of reporting checklist for repeatability and reproducibility of radiomic features. Phys Imaging Radiat Oncol. 2021;20:69–75.
Survarachakan S, Prasad PJR, Naseem R, Perez de Frutos J, Kumar RP, Lango T, et al. Deep learning for image-based liver analysis—a comprehensive review focusing on malignant lesions. Artif Intell Med. 2022;130:102331.
Kalendralis P, Traverso A, Shi Z, Zhovannik I, Monshouwer R, Starmans MPA, et al. Multicenter CT phantoms public dataset for radiomics reproducibility tests. Med Phys. 2019;46:1512–8.
Zwanenburg A, Vallieres M, Abdalah MA, Aerts H, Andrearczyk V, Apte A, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–38.
Zwanenburg AAM, Ashrafinia S. Results from the image biomarker standardisation initiative. Radiother Oncol. 2018;127:S543–S4.
Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–62.
Shi R, Chen W, Yang B, Qu J, Cheng Y, Zhu Z, et al. Prediction of KRAS, NRAS and BRAF status in colorectal cancer patients with liver metastasis using a deep artificial neural network based on radiomics and semantic features. Am J Cancer Res. 2020;10:4513–26.
Feng Z, Zhang L, Qi Z, Shen Q, Hu Z, Chen F. Identifying BAP1 mutations in clear-cell renal cell carcinoma by CT radiomics: preliminary findings. Front Oncol. 2020;10:279.
Li ZC, Zhai G, Zhang J, Wang Z, Liu G, Wu GY, et al. Differentiation of clear cell and non-clear cell renal cell carcinomas by all-relevant radiomics features from multiphase CT: a VHL mutation perspective. Eur Radiol. 2019;29:3996–4007.
Zeng H, Chen L, Wang M, Luo Y, Huang Y, Ma X. Integrative radiogenomics analysis for predicting molecular features and survival in clear cell renal cell carcinoma. Aging. 2021;13:9960–75.
Thakur SB, Durando M, Milans S, Cho GY, Gennaro L, Sutton EJ, et al. Apparent diffusion coefficient in estrogen receptor-positive and lymph node-negative invasive breast cancers at 3.0T DW-MRI: a potential predictor for an oncotype Dx test recurrence score. J Magn Reson Imaging. 2018;47:401–9.
Ha R, Chang P, Mutasa S, Karcich J, Goodman S, Blum E, et al. Convolutional neural network using a breast MRI tumor dataset can predict oncotype Dx recurrence score. J Magn Reson Imaging. 2019;49:518–24.
Li H, Wang J, Li Z, Dababneh M, Wang F, Zhao P, et al. Deep learning-based pathology image analysis enhances magee feature correlation with oncotype DX breast recurrence score. Front Med. 2022;9:886763.
Ha R, Mutasa S, Karcich J, Gupta N, Pascual Van Sant E, Nemer J, et al. Predicting breast cancer molecular subtype with MRI dataset utilizing convolutional neural network algorithm. J Digit Imaging. 2019;32:276–82.
Iima M, Kataoka M, Honda M, Ohashi A, Ohno Kishimoto A, Ota R, et al. The rate of apparent diffusion coefficient change with diffusion time on breast diffusion-weighted imaging depends on breast tumor types and molecular prognostic biomarker expression. Invest Radiol. 2021;56:501–8.
Tsai WC, Chang KM, Kao KJ. Dynamic contrast enhanced MRI and intravoxel incoherent motion to identify molecular subtypes of breast cancer with different vascular normalization gene expression. Korean J Radiol. 2021;22:1021–33.
Zhang Y, Chen JH, Lin Y, Chan S, Zhou J, Chow D, et al. Prediction of breast cancer molecular subtypes on DCE-MRI using convolutional neural network with transfer learning between two centers. Eur Radiol. 2021;31:2559–67.
Wang W, Cao K, Jin S, Zhu X, Ding J, Peng W. Differentiation of renal cell carcinoma subtypes through MRI-based radiomics analysis. Eur Radiol. 2020;30:5738–47.
Gao Y, Wang X, Wang S, Miao Y, Zhu C, Li C, et al. Differential diagnosis of type 1 and type 2 papillary renal cell carcinoma based on enhanced CT radiomics nomogram. Front Oncol. 2022;12:854979.
Dasgupta A, Gupta T, Pungavkar S, Shirsat N, Epari S, Chinnaswamy G, et al. Nomograms based on preoperative multiparametric magnetic resonance imaging for prediction of molecular subgrouping in medulloblastoma: results from a radiogenomics study of 111 patients. Neuro Oncol. 2019;21:115–24.
Buda M, Saha A, Mazurowski MA. Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm. Comput Biol Med. 2019;109:218–25.
Garcia Vicente AM, Perez-Beteta J, Amo-Salas M, Pena Pardo FJ, Villena Martin M, Sandoval Valencia H, et al. 18F-Fluorocholine PET/CT in the prediction of molecular subtypes and prognosis for gliomas. Clin Nucl Med. 2019;44:e548–e58.
Nam YK, Park JE, Park SY, Lee M, Kim M, Nam SJ, et al. Reproducible imaging-based prediction of molecular subtype and risk stratification of gliomas across different experience levels using a structured reporting system. Eur Radiol. 2021;31:7374–85.
Zhao K, Sun G, Wang Q, Xue Z, Liu G, Xia Y, et al. The diagnostic value of conventional MRI and CT features in the identification of the IDH1-mutant and 1p/19q co-deletion in WHO grade II gliomas. Acad Radiol. 2021;28:e189–98.
Li Y, Wei D, Liu X, Fan X, Wang K, Li S, et al. Molecular subtyping of diffuse gliomas using magnetic resonance imaging: comparison and correlation between radiomics and deep learning. Eur Radiol. 2022;32:747–58.
Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, et al. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70:1133–44.
He M, Zhang P, Ma X, He B, Fang C, Jia F. Radiomic feature-based predictive model for microvascular invasion in patients with hepatocellular carcinoma. Front Oncol. 2020;10:574228.
Liu P, Tan XZ, Zhang T, Gu QB, Mao XH, Li YC, et al. Prediction of microvascular invasion in solitary hepatocellular carcinoma </= 5 cm based on computed tomography radiomics. World J Gastroenterol. 2021;27:2015–24.
Funding
XWH is supported by the Major Science and Technology projects of Henan Province (Grant No. 221100310100).
Author information
Authors and Affiliations
Contributions
ZQL, XWH and YYZ provided direction and guidance throughout the preparation of this manuscript. TD, YYZ and ZQL wrote and edited the manuscript. YYZ and ZYZ reviewed and made significant revisions to the manuscript. SYW, YQR and HX collected and prepared the related papers. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Z., Duan, T., Zhang, Y. et al. Radiogenomics: a key component of precision cancer medicine. Br J Cancer 129, 741–753 (2023). https://doi.org/10.1038/s41416-023-02317-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02317-8
This article is cited by
-
MRI radiomics in head and neck cancer from reproducibility to combined approaches
Scientific Reports (2024)
-
Developing a Radiomics Atlas Dataset of normal Abdominal and Pelvic computed Tomography (RADAPT)
Journal of Imaging Informatics in Medicine (2024)