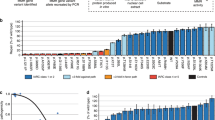
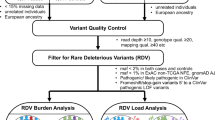
Abstract
Patients with the heritable cancer disease, Lynch syndrome, carry germline variants in the MLH1, MSH2, MSH6 and PMS2 genes, encoding the central components of the DNA mismatch repair system. Loss-of-function variants disrupt the DNA mismatch repair system and give rise to a detrimental increase in the cellular mutational burden and cancer development. The treatment prospects for Lynch syndrome rely heavily on early diagnosis; however, accurate diagnosis is inextricably linked to correct clinical interpretation of individual variants. Protein variant classification traditionally relies on cumulative information from occurrence in patients, as well as experimental testing of the individual variants. The complexity of variant classification is due to (1) that variants of unknown significance are rare in the population and phenotypic information on the specific variants is missing, and (2) that individual variant testing is challenging, costly and slow. Here, we summarise recent developments in high-throughput technologies and computational prediction tools for the assessment of variants of unknown significance in Lynch syndrome. These approaches may vastly increase the number of interpretable variants and could also provide important mechanistic insights into the disease. These insights may in turn pave the road towards developing personalised treatment approaches for Lynch syndrome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Pearlman R, Frankel WL, Swanson B, Zhao W, Yilmaz A, Miller K, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol. 2017;3:464–71.
Yurgelun MB, Kulke MH, Fuchs CS, Allen BA, Uno H, Hornick JL, et al. Cancer susceptibility gene mutations in individuals with colorectal cancer. J Clin Oncol. 2017;35:1086–95.
Dominguez-Valentin M, Sampson JR, Seppälä TT, ten Broeke SW, Plazzer J-P, Nakken S, et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the Prospective Lynch Syndrome Database. Genet Med. 2020;22:15–25.
Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med. 2005;352:1851–60.
Møller P, Seppälä T, Bernstein I, Holinski-Feder E, Sala P, Evans DG, et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut. 2017;66:464–72.
Boland PM, Yurgelun MB, Boland CR. Recent progress in lynch syndrome and other familial colorectal cancer syndromes. CA Cancer J Clin. 2018;68:217–31.
Seppälä TT, Latchford A, Negoi I, Sampaio Soares A, Jimenez-Rodriguez R, Sánchez-Guillén L, et al. European guidelines from the EHTG and ESCP for Lynch syndrome: an updated third edition of the Mallorca guidelines based on gene and gender. Br J Surg. 2021;108:484–98.
Crosbie EJ, Ryan NAJ, Arends MJ, Bosse T, Burn J, Cornes JM, et al. The Manchester International Consensus Group recommendations for the management of gynecological cancers in Lynch syndrome. Genet Med J Am Coll Med Genet. 2019;21:2390–400.
Grindedal EM, Renkonen-Sinisalo L, Vasen H, Evans G, Sala P, Blanco I, et al. Survival in women with MMR mutations and ovarian cancer: a multicentre study in Lynch syndrome kindreds. J Med Genet. 2010;47:99–102.
Schmeler KM, Lu KH. Gynecologic cancers associated with Lynch syndrome/HNPCC. Clin Transl Oncol. 2008;10:313–7.
Therkildsen C, Jensen LH, Rasmussen M, Bernstein I. An update on immune checkpoint therapy for the treatment of Lynch syndrome. Clin Exp Gastroenterol. 2021;14:181–97.
StCharles JA, Liberti SE, Williams JS, Lujan SA, Kunkel TA. Quantifying the contributions of base selectivity, proofreading and mismatch repair to nuclear DNA replication in Saccharomyces cerevisiae. DNA Repair. 2015;31:41–51.
Huang Y, Li G-M. DNA mismatch repair preferentially safeguards actively transcribed genes. DNA Repair. 2018;71:82–6.
Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol 2010;20:281–93.
Hitchins MP. Inheritance of epigenetic aberrations (constitutional epimutations) in cancer susceptibility. Adv Genet. 2010;70:201–43.
Hitchins MP, Ward RL. Constitutional (germline) MLH1 epimutation as an aetiological mechanism for hereditary non-polyposis colorectal cancer. J Med Genet. 2009;46:793–802.
Ward RL, Dobbins T, Lindor NM, Rapkins RW, Hitchins MP. Identification of constitutional MLH1 epimutations and promoter variants in colorectal cancer patients from the Colon Cancer Family Registry. Genet Med. 2013;15:25–35.
Ligtenberg MJL, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet. 2009;41:112–7.
Duraturo F, Liccardo R, Cavallo A, De Rosa M, Grosso M, Izzo P. Association of low-risk MSH3 and MSH2 variant alleles with Lynch syndrome: probability of synergistic effects. Int J Cancer. 2011;129:1643–50.
Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–46.
Dufner P, Marra G, Räschle M, Jiricny J. Mismatch recognition and DNA-dependent stimulation of the ATPase activity of hMutSα is abolished by a single mutation in the hMSH6 subunit *. J Biol Chem. 2000;275:36550–5.
Gupta S, Gellert M, Yang W. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat Struct Mol Biol. 2012;19:72–8.
Reyes GX, Schmidt TT, Kolodner RD, Hombauer H. New insights into the mechanism of DNA mismatch repair. Chromosoma. 2015;124:443–62.
Groothuizen FS, Winkler I, Cristóvão M, Fish A, Winterwerp HH, Reumer A, et al. MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. eLife. 2015;4:e06744.
Bowen N, Smith CE, Srivatsan A, Willcox S, Griffith JD, Kolodner RD. Reconstitution of long and short patch mismatch repair reactions using Saccharomyces cerevisiae proteins. Proc Natl Acad Sci USA. 2013;110:18472–7.
Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013;5:a012633.
Groothuizen FS, Sixma TK. The conserved molecular machinery in DNA mismatch repair enzyme structures. DNA Repair. 2016;38:14–23.
Gueneau E, Dherin C, Legrand P, Tellier-Lebegue C, Gilquin B, Bonnesoeur P, et al. Structure of the MutLα C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nat Struct Mol Biol 2013;20:461–8.
Goellner EM, Putnama CD, Kolodnera RD. Exonuclease 1-dependent and independent mismatch repair. DNA Repair. 2015;32:24–32.
Cannavo E, Sanchez A, Anand R, Ranjha L, Hugener J, Adam C, et al. Regulation of the MLH1–MLH3 endonuclease in meiosis. Nature. 2020;586:618–22.
Kadyrova LY, Gujar V, Burdett V, Modrich PL, Kadyrov FA. Human MutLγ, the MLH1–MLH3 heterodimer, is an endonuclease that promotes DNA expansion. Proc Natl Acad Sci USA. 2020;117:3535–42.
Campbell CS, Hombauer H, Srivatsan A, Bowen N, Gries K, Desai A, et al. Mlh2 is an accessory factor for DNA mismatch repair in Saccharomyces cerevisiae. PLOS Genet. 2014;10:e1004327.
Gupta D, Heinen CD. The mismatch repair-dependent DNA damage response: mechanisms and implications. DNA Repair. 2019;78:60–9.
Lynch HT, Snyder CL, Shaw TG, Heinen CD, Hitchins MP. Milestones of Lynch syndrome: 1895–2015. Nat Rev Cancer. 2015;15:181–94.
De’ Angelis GL, Bottarelli L, Azzoni C, De’ Angelis N, Leandro G, Di Mario F, et al. Microsatellite instability in colorectal cancer. Acta Bio-Med Atenei Parm. 2018;89:97–101.
Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–80.
Yamamoto H, Imai K. Microsatellite instability: an update. Arch Toxicol. 2015;89:899–921.
Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–5.
Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–6.
Umar A, Boland CR, Terdiman JP, Syngal S, Chapelle A, de la, Rüschoff J, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. JNCI J Natl Cancer Inst. 2004;96:261–8.
Kastrinos F, Allen JI, Stockwell DH, Stoffel EM, Cook EF, Mutinga ML, et al. Development and validation of a colon cancer risk assessment tool for patients undergoing colonoscopy. Am J Gastroenterol. 2009;104:1508–18.
Giardiello FM, Allen JI, Axilbund JE, Boland RC, Burke CA, Burt RW, et al. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-Society Task Force on colorectal cancer. J Am Coll Gastroenterol ACG. 2014;109:1159–79.
Win AK, Jenkins MA, Dowty JG, Antoniou AC, Lee A, Giles GG, et al. Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2017;26:404–12.
Moreira L, Balaguer F, Lindor N, de la Chapelle A, Hampel H, Aaltonen LA, et al. Identification of Lynch syndrome among patients with colorectal cancer. J Am Med Assoc. 2012;308:1555–65.
Peltomäki P. Update on Lynch syndrome genomics. Fam Cancer. 2016;15:385–93.
Dominguez-Valentin M, Plazzer J-P, Sampson JR, Engel C, Aretz S, Jenkins MA, et al. No difference in penetrance between truncating and missense/aberrant splicing pathogenic variants in MLH1 and MSH2: a prospective Lynch Syndrome Database Study. J Clin Med. 2021;10:2856.
Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA 1971;68:820–3.
Hemminki A, Peltomäki P, Mecklin J-P, Järvinen H, Salovaara R, Nyström-Lahti M, et al. Loss of the wild type MLH1 gene is a feature of hereditary nonpolyposis colorectal cancer. Nat Genet. 1994;8:405–10.
Zhang M, Xiang S, Joo H-Y, Wang L, Williams KA, Liu W, et al. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSα. Mol Cell. 2014;55:31–46.
Abildgaard AB, Stein A, Nielsen SV, Schultz-Knudsen K, Papaleo E, Shrikhande A, et al. Computational and cellular studies reveal structural destabilization and degradation of MLH1 variants in Lynch syndrome. eLife. 2019;8:e49138.
Hinrichsen I, Weßbecher IM, Huhn M, Passmann S, Zeuzem S, Plotz G, et al. Phosphorylation-dependent signaling controls degradation of DNA mismatch repair protein PMS2. Mol Carcinog. 2017;56:2663–8.
Mohd AB, Palama B, Nelson SE, Tomer G, Nguyen M, Huo X, et al. Truncation of the C-terminus of human MLH1 blocks intracellular stabilization of PMS2 and disrupts DNA mismatch repair. DNA Repair. 2006;5:347–61.
Rosty C, Clendenning M, Walsh MD, Eriksen SV, Southey MC, Winship IM, et al. Germline mutations in PMS2 and MLH1 in individuals with solitary loss of PMS2 expression in colorectal carcinomas from the Colon Cancer Family Registry Cohort. BMJ Open. 2016;6:e010293.
ten Broeke SW, van der Klift HM, Tops CMJ, Aretz S, Bernstein I, Buchanan DD, et al. Cancer risks for PMS2-associated Lynch syndrome. J Clin Oncol. 2018;36:2961–8.
Wang C, Wang Y, Hughes KS, Parmigiani G, Braun D. Penetrance of colorectal cancer among mismatch repair gene mutation carriers: a meta-analysis. JNCI Cancer Spectr. 2020;4:pkaa027.
Thompson BA, Spurdle AB, Plazzer J-P, Greenblatt MS, Akagi K, Al-Mulla F, et al. Application of a five-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants lodged on the InSiGHT locus-specific database. Nat Genet. 2014;46:107–15.
Williams PD, Pollock DD, Goldstein RA. Functionality and the evolution of marginal stability in proteins: inferences from lattice simulations. Evol Bioinforma Online. 2007;2:91–101.
Abildgaard AB, Gersing SK, Larsen-Ledet S, Nielsen SV, Stein A, Lindorff-Larsen K, et al. Co-chaperones in targeting and delivery of misfolded proteins to the 26S proteasome. Biomolecules. 2020;10:1141.
Clausen L, Abildgaard AB, Gersing SK, Stein A, Lindorff-Larsen K, Hartmann-Petersen R. Chapter two - protein stability and degradation in health and disease. In: Advances in protein chemistry and structural biology. Donev R, editor. Vol. 114. London, United Kingdom: Academic Press; 2019. p. 61–83.
Kohler V, Andréasson C. Hsp70-mediated quality control: should I stay or should I go? Biol Chem. 2020;401:1233–48.
Reinle K, Mogk A, Bukau B. The diverse functions of small heat shock proteins in the proteostasis network. J Mol Biol. 2022;434:167157.
Rosenzweig R, Nillegoda NB, Mayer MP, Bukau B. The Hsp70 chaperone network. Nat Rev Mol Cell Biol. 2019;20:665–80.
Hernandez-Pigeon H, Laurent G, Humbert O, Salles B, Lautier D. Degadration of mismatch repair hMutSα heterodimer by the ubiquitin-proteasome pathway. FEBS Lett. 2004;562:40–4.
Wu Q, Huang Y, Gu L, Chang Z, Li G-M. OTUB1 stabilizes mismatch repair protein MSH2 by blocking ubiquitination. J Biol Chem. 2021;296:100466.
Zhang M, Hu C, Tong D, Xiang S, Williams K, Bai W, et al. Ubiquitin-specific peptidase 10 (USP10) deubiquitinates and stabilizes MutS homolog 2 (MSH2) to regulate cellular sensitivity to DNA damage*. J Biol Chem. 2016;291:10783–91.
Jia X, Burugula BB, Chen V, Lemons RM, Jayakody S, Maksutova M, et al. Massively parallel functional testing of MSH2 missense variants conferring Lynch syndrome risk. Am J Hum Genet. 2021;108:163–75.
Nielsen SV, Hartmann-Petersen R, Stein A, Lindorff-Larsen K. Multiplexed assays reveal effects of missense variants in MSH2 and cancer predisposition. PLOS Genet. 2021;17:e1009496.
Nielsen SV, Stein A, Dinitzen AB, Papaleo E, Tatham MH, Poulsen EG, et al. Predicting the impact of Lynch syndrome-causing missense mutations from structural calculations. PLoS Genet. 2017;13:e1006739.
Ollodart AR, Yeh C-LC, Miller AW, Shirts BH, Gordon AS, Dunham MJ. Multiplexing mutation rate assessment: determining pathogenicity of Msh2 variants in Saccharomyces cerevisiae. Genetics. 2021;218:iyab058.
Arlow T, Scott K, Wagenseller A, Gammie A. Proteasome inhibition rescues clinically significant unstable variants of the mismatch repair protein Msh2. Proc Natl Acad Sci USA. 2013;110:246–51.
Kampmeyer C, Nielsen SV, Clausen L, Stein A, Gerdes A-M, Lindorff-Larsen K, et al. Blocking protein quality control to counter hereditary cancers. Genes Chromosomes Cancer. 2017;56:823–31.
Stein A, Fowler DM, Hartmann-Petersen R, Lindorff-Larsen K. Biophysical and mechanistic models for disease-causing protein variants. Trends Biochem Sci. 2019;44:575–88.
Livesey BJ, Marsh JA. Using deep mutational scanning to benchmark variant effect predictors and identify disease mutations. Mol Syst Biol. 2020;16:e9380.
Starita LM, Ahituv N, Dunham MJ, Kitzman JO, Roth FP, Seelig G, et al. Variant interpretation: functional assays to the rescue. Am J Hum Genet. 2017;101:315–25.
Ponti G, Castellsagué E, Ruini C, Percesepe A, Tomasi A. Mismatch repair genes founder mutations and cancer susceptibility in Lynch syndrome: mismatch repair genes founder mutations in Lynch syndrome. Clin Genet. 2015;87:507–16.
Foulkes WD, Thiffault I, Gruber SB, Horwitz M, Hamel N, Lee C, et al. The founder mutation MSH2*1906G->C is an important cause of hereditary nonpolyposis colorectal cancer in the Ashkenazi Jewish population. Am J Hum Genet. 2002;71:1395–412.
Brnich SE, Rivera‐Muñoz EA, Berg JS. Quantifying the potential of functional evidence to reclassify variants of uncertain significance in the categorical and Bayesian interpretation frameworks. Hum Mutat. 2018;39:1531–41.
Houlleberghs H, Goverde A, Lusseveld J, Dekker M, Bruno MJ, Menko FH, et al. Suspected Lynch syndrome associated MSH6 variants: a functional assay to determine their pathogenicity. PLOS Genet. 2017;13:e1006765.
Houlleberghs H, Dekker M, Lantermans H, Kleinendorst R, Dubbink HJ, Hofstra RMW, et al. Oligonucleotide-directed mutagenesis screen to identify pathogenic Lynch syndrome-associated MSH2 DNA mismatch repair gene variants. Proc Natl Acad Sci USA. 2016;113:4128–33.
Alim I, Loke J, Yam S, Templeton AS, Newcomb P, Lindor NM, et al. Cancer risk C (CR-C), a functional genomics test is a sensitive and rapid test for germline mismatch repair deficiency. Genet. Med. 2022;S1098360022007584. https://doi.org/10.1016/j.gim.2022.05.003.
Cagiada M, Johansson KE, Valanciute A, Nielsen SV, Hartmann-Petersen R, Yang JJ, et al. Understanding the origins of loss of protein function by analyzing the effects of thousands of variants on activity and abundance. Mol Biol Evol. 2021;38:3235–46.
Arora S, Huwe PJ, Sikder R, Shah M, Browne AJ, Lesh R, et al. Functional analysis of rare variants in mismatch repair proteins augments results from computation-based predictive methods. Cancer Biol Ther. 2017;18:519–33.
Niroula A, Vihinen M. Classification of amino acid substitutions in mismatch repair proteins using PON-MMR2. Hum Mutat. 2015;36:1128–34.
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7:Unit7.20.
Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–7.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81.
Hopf TA, Ingraham JB, Poelwijk FJ, Schärfe CPI, Springer M, Sander C, et al. Mutation effects predicted from sequence co-variation. Nat Biotechnol. 2017;35:128–35.
Laine E, Karami Y, Carbone A. GEMME: a simple and fast global epistatic model predicting mutational effects. Mol Biol Evol. 2019;36:2604–19.
Riesselman AJ, Ingraham JB, Marks DS. Deep generative models of genetic variation capture the effects of mutations. Nat Methods. 2018;15:816–22.
Frazer J, Notin P, Dias M, Gomez A, Min JK, Brock K, et al. Disease variant prediction with deep generative models of evolutionary data. Nature. 2021;599:91–5.
Park H, DiMaio F, Baker D. CASP11 refinement experiments with ROSETTA. Proteins Struct Funct Bioinforma. 2016;84:314–22.
Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–87.
Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–8.
Raevaara TE, Korhonen MK, Lohi H, Hampel H, Lynch E, Lönnqvist KE, et al. Functional significance and clinical phenotype of nontruncating mismatch repair variants of MLH1. Gastroenterology. 2005;129:537–49.
Takahashi M, Shimodaira H, Andreutti-Zaugg C, Iggo R, Kolodner RD, Ishioka C. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 2007;67:4595–604.
De Baets G, Van Durme J, Reumers J, Maurer-Stroh S, Vanhee P, Dopazo J, et al. SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 2012;40:D935–9.
Gerasimavicius L, Liu X, Marsh JA. Identification of pathogenic missense mutations using protein stability predictors. Sci Rep. 2020;10:15387.
Pey AL, Stricher F, Serrano L, Martinez A. Predicted effects of missense mutations on native-state stability account for phenotypic outcome in phenylketonuria, a paradigm of misfolding diseases. Am J Hum Genet. 2007;81:1006–24.
Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat Methods. 2014;11:801–7.
Raraigh KS, Han ST, Davis E, Evans TA, Pellicore MJ, McCague AF, et al. Functional assays are essential for interpretation of missense variants associated with variable expressivity. Am J Hum Genet. 2018;102:1062–77.
Sun S, Yang F, Tan G, Costanzo M, Oughtred R, Hirschman J, et al. An extended set of yeast-based functional assays accurately identifies human disease mutations. Genome Res. 2016;26:670–80.
Jepsen MM, Fowler DM, Hartmann-Petersen R, Stein A, Lindorff-Larsen K. Chapter 5 - Classifying disease-associated variants using measures of protein activity and stability. In: Protein homeostasis diseases. Pey AL, editor. London, United Kingdom: Academic Press; 2020, p. 91–107.
Matreyek KA, Starita LM, Stephany JJ, Martin B, Chiasson MA, Gray VE, et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat Genet. 2018;50:874–82.
Brnich SE, On behalf of the Clinical Genome Resource Sequence Variant Interpretation Working Group, Abou Tayoun AN, Couch FJ, Cutting GR, Greenblatt MS, et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2020;12:3.
Ernst C, Hahnen E, Engel C, Nothnagel M, Weber J, Schmutzler RK, et al. Performance of in silico prediction tools for the classification of rare BRCA1/2 missense variants in clinical diagnostics. BMC Med Genomics. 2018;11:35.
Miosge LA, Field MA, Sontani Y, Cho V, Johnson S, Palkova A, et al. Comparison of predicted and actual consequences of missense mutations. Proc Natl Acad Sci USA. 2015;112:E5189–98.
Grimm DG, Azencott C-A, Aicheler F, Gieraths U, MacArthur DG, Samocha KE, et al. The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum Mutat. 2015;36:513–23.
Wu Y, Li R, Sun S, Weile J, Roth FP. Improved pathogenicity prediction for rare human missense variants. Am J Hum Genet. 2021. https://doi.org/10.1016/j.ajhg.2021.08.012.
Høie MH, Cagiada M, Beck Frederiksen AH, Stein A, Lindorff-Larsen K. Predicting and interpreting large-scale mutagenesis data using analyses of protein stability and conservation. Cell Rep. 2022;38:110207.
Thompson S, Zhang Y, Ingle C, Reynolds KA, Kortemme T. Altered expression of a quality control protease in E. coli reshapes the in vivo mutational landscape of a model enzyme. eLife. 2020;9:e53476.
Brusa I, Sondo E, Falchi F, Pedemonte N, Roberti M, Cavalli A. Proteostasis regulators in cystic fibrosis: current development and future perspectives. J Med Chem. 2022. https://doi.org/10.1021/acs.jmedchem.1c01897.
Hutt DM, Herman D, Rodrigues APC, Noel S, Pilewski JM, Matteson J, et al. Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nat Chem Biol. 2010;6:25–33.
Okiyoneda T, Veit G, Dekkers JF, Bagdany M, Soya N, Xu H, et al. Mechanism-based corrector combination restores ΔF508-CFTR folding and function. Nat Chem Biol. 2013;9:444–54.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Fayer S, Horton C, Dines JN, Rubin AF, Richardson ME, McGoldrick K, et al. Closing the gap: systematic integration of multiplexed functional data resolves variants of uncertain significance in BRCA1, TP53, and PTEN. Am J Hum Genet. 2021;S0002-9297(21)00411–0. https://doi.org/10.1016/j.ajhg.2021.11.001.
Kuang D, Truty R, Weile J, Johnson B, Nykamp K, Araya C, et al. Prioritizing genes for systematic variant effect mapping. Bioinformatics. 2020;36:5448–55.
Manolio TA, Fowler DM, Starita LM, Haendel MA, MacArthur DG, Biesecker LG, et al. Bedside back to bench: building bridges between basic and clinical genomic research. Cell. 2017;169:6–12.
Funding
Our research in this area is funded by the Novo Nordisk Foundation Challenge programme PRISM (NNFOC180033950; to AS, KLL and RHP), the Lundbeck Foundation (R272-2017-4528 to AS), and the Danish Council for Independent Research, Natural Sciences (7014-00039B to RHP).
Author information
Authors and Affiliations
Contributions
ABA and SVN prepared the figures. ABA, SVN, IB, AS, KLL and RHP wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abildgaard, A.B., Nielsen, S.V., Bernstein, I. et al. Lynch syndrome, molecular mechanisms and variant classification. Br J Cancer 128, 726–734 (2023). https://doi.org/10.1038/s41416-022-02059-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-022-02059-z
This article is cited by
-
Comparison of standard mismatch repair deficiency and microsatellite instability tests in a large cancer series
Journal of Translational Medicine (2024)
-
Splice modulators target PMS1 to reduce somatic expansion of the Huntington’s disease-associated CAG repeat
Nature Communications (2024)