Abstract
Background
Head and neck squamous cell carcinoma (HNSCC) is a significant problem and is frequently resistant to current treatments. STAT1 is important in anti-tumour immune responses against HNSCC. However, the role of STAT1 expression by tumour cells and its regulation during HNSCC is unclear.
Methods
We determined the effects of STAT1 inhibition on tumour development and immunity in CAL27 and UMSCC22A HNSCC cell lines in vitro and in a HNSCC carcinogen-induced model in vivo.
Results
STAT1 siRNA knockdown in human HNSCC cells impaired their proliferation and expression of the immunosuppressive marker PD-L1. Stat1-deficient mice displayed increased oral lesion incidence and multiplicity during tumour carcinogenesis in vivo. Immunosuppressive markers PD-1 in CD8+ T cells and PD-L1 in monocytic MDSCs and macrophages were reduced in oral tumours and draining lymph nodes of tumour-bearing Stat1-deficient mice. However, STAT1 was required for anti-tumour functions of T cells during HNSCC in vivo. Finally, we identified TRIM24 to be a negative regulator of STAT1 that plays a similar tumorigenic function to STAT1 in vitro and thus may be a potential target when treating HNSCC.
Conclusion
Our findings indicate that STAT1 activity plays an important role in tumorigenicity and immunosuppression during HNSCC development.
Similar content being viewed by others
Background
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common form of cancer, affecting nearly one million people per year [1, 2]. With the annual burden of HNSCC expected to increase, there exists a dire need to identify and elucidate the context-dependent function of molecular targets that can be therapeutically exploited in cancer cells and associated cells of the HNSCC tumour microenvironment [3]. Signal transducer and activator of transcription 1 (STAT1) mediates the signalling of type I, II and III interferons (IFNs) and growth factors like epidermal growth factor and is involved in several biological processes, including apoptosis [4], inflammation [5] and cell cycle modulation [6, 7]. Not surprisingly, due to the wide range of biological activity mediated by STAT1, its activity is heavily altered in many HNSCC cases [8]. Interestingly, STAT1 downregulation has been implicated in oral carcinogenesis and immune escape, while STAT1 phosphorylation has been associated with increased survival [9,10,11].
Recent studies conducted by our group identified the critical anti-tumour role of STAT1 in the host immune response to HNSCC [12]. We used an orthotopic model of metastatic HNSCC cells injected into Stat1-deficient (Stat1−/−) or Stat1-sufficient (Stat1+/+) mice to show that mice deficient in Stat1 are highly susceptible to tumour growth and metastasis. This was associated with an exhausted T cell phenotype with increased programmed cell death protein 1 (PD-1) expression, and increased accumulation of immunosuppressive myeloid-derived suppressor cell (MDSC) populations in the tumours of Stat1-deficient mice. These results indicate that host derived STAT1 is critical for anti-tumour immune responses against HNSCC. However, questions remain regarding the role of STAT1 expression by oral epithelial and cancer cells during stages of HNSCC tumour development.
Although STAT1 has long been considered to be a tumour suppressor by promoting HNSCC cell death [10], recent reports show that STAT1 activation also mediates programmed cell death ligand 1 (PD-L1) expression in HNSCC cells thereby promoting tumour immunosuppression [13, 14]. STAT1 activation has also been shown to mediate resistance to cisplatin therapy [15] and radioresistance in HNSCC cells [16]. A limitation to these studies that have investigated the impact of STAT1 activation in HNSCC cells are that the methods mostly used are in vitro cell culture models, xenograft models into nude mice or correlative studies from HNSCC clinical samples. These approaches generally do not address (i) the influence of STAT1 on the tumour microenvironment, (ii) the interaction of STAT1 expressing HNSCC cells with immune cells, stromal cells, and fibroblasts and (iii) the impact of STAT1 activation by oral epithelial cells on HNSCC tumour initiation and promotion. This represents a major gap in our knowledge of STAT1 activation by HNSCC cells in the tumour microenvironment, which is essential to advance our mechanistic understanding of the role of STAT1 in HNSCC, and design therapies that exploit the STAT1 pathway in HNSCC treatment.
Identifying cell-specific regulators of STAT1 activation in HNSCC cells can facilitate the development of novel approaches to HNSCC treatment. In this study, we identify a novel potential negative regulator of STAT1 in HNSCC cells—tripartite motif (Trim) 24 protein, also known as transcriptional intermediary factor 1. TRIM24 is a multifunctional protein that binds and regulates nuclear receptors, such as retinoic acid and oestrogen receptors, and associates with chromatin- and heterochromatin-associated factors that result in inactivation of Stat1 transcription [17]. Thus, TRIM24 presents itself as a potential target for modulating STAT1 expression in the tumour microenvironment. Although TRIM24 has been shown to promote colorectal, ovarian and other cancers, [18,19,20,21], the effect of the TRIM24–STAT1 axis on HNSCC development still remains unclear.
To enhance our understanding of STAT1 activation and regulation in HNSCC initiation, growth and anti-tumour immune responses, we determined the effects of STAT1 small interfering RNA (siRNA) knockdown on the human papillomavirus (HPV)-negative oral cancer cell lines CAL27 and UM-SCC22A. We also used the 4-nitroquinoline-1-oxide (4NQO) carcinogen-induced HNSCC model in STAT1-competent and STAT1-deficient mice to determine the effect of STAT1 on tumour development and immune responses to HNSCC in vivo [22]. This 4NQO mouse model recapitulates the mutational profile and immune landscape of non-HPV-associated human HNSCC and mirrors its development from early dysplastic to invasive stages [23]. Specifically, high incidence mutations observed in human HNSCC, such as in Trp53, Notch1, Fat1, Lama3 and Syne2, were demonstrated to be associated with 4NQO-induced mouse tumours, while immune infiltration to tumours was maintained [23]. Indeed, STAT1 and IFN-γ activity are upregulated during PD-1 blockade in mice administered 4NQO [24]. Because of the similar mutational landscape of 4NQO-induced mouse oral carcinogenesis to human HNSCC, combined with the use of immunocompetent hosts, this model further allows for study of the immunologic determinants of HNSCC development [25]. Finally, we investigate the impact of TRIM24 siRNA knockdown on STAT1 expression and subsequent cell proliferation, apoptosis and expression of immunosuppressive biomarkers in CAL27 and UM-SCC22A HNSCC cells. The findings of our study shed light on the role of STAT1 expression and regulation by HNSCC cells on tumour development and the immune response during HNSCC.
Materials and methods
Mice
Stat1+/+ and Stat1−/− male and female BALB/c mice, 6–8 weeks old (N = 20, 10 males, 10 females per group) were used for experimental HNSCC studies. This sample size was selected to achieve 80% power to detect a 2.5-fold difference between groups (corresponding to an effect size of 2). Animals were kept according to regulations maintained by the University Laboratory Animal Resources. Experiments were approved by the Institutional Animal Care and Use Committee (Protocol #2018A00000054) and Institutional Biosafety Committee of The Ohio State University.
Cell lines
Authenticated human HNSCC cell lines CAL27 (ATCC Cat# CRL-2095, RRID:CVCL_1107) and UM-SCC22A (Millipore Cat# SCC076, RRID:CVCL_7731) were used for these studies.
siRNA knockdown
A reverse transfection mRNA knockdown cocktail containing STAT1 siRNA (Cell Signaling; Danvers, MA), TRIM24 siRNA (Life Technologies; Carlsbad, CA) or scrambled siRNA (Thermo Fisher; Waltham, MA) and 1 μL of Lipofectamine RNAiMAX in 100 μL OptiMEM serum-free media (Life Technologies; Carlsbad, CA) was prepared and plated at 100 μL per well. Following 15 min of room temperature incubation, cells were plated at 2.5 × 105 cells/well in 500 μL antibiotic-free complete media in each well. Cells were allowed to grow for 48 or 72 h before downstream analyses.
Western blot
In all, 30 μg protein was loaded into a 10% acrylamide SDS-PAGE gel (National Diagnostics; Atlanta, GA). Standard Western Blotting techniques were used as described previously [26]. Antibodies used included STAT1(14994T), phospho STAT1 (9167S), TRIM24 (79030S), MCM3 (4012S), cleaved CASP8 (8592T), GAPDH (2118S) (Cell Signaling), BCL2 (IMG-80093, IMGENEX) and KI67 (M7240, Agilent). Blots were incubated with goat anti-rabbit HRP-linked (31460, Thermo Fisher Scientific, Rockford, IL) or anti-mouse IgG HRP-linked (7076 S, Cell Signaling Technology) secondary antibodies and treated with Pierce ECL Western Blotting Substrate (Thermo Fisher; Waltham, MA).
Enzyme-linked immunosorbent assay (ELISA)
IFN-γ ELISA was performed as described previously [27], using capture antibody (Biolegend, 505702, San Diego, CA) and detection antibody (505804).
Carcinogen-induced HNSCC model
Stat1+/+ and Stat1−/− (n = 20; 6–8 weeks old) BALB/c mice were administered the oral carcinogen 4NQO (Sigma Aldrich; St. Louis, MO) at a concentration 100 μg/mL in their drinking water for 16 weeks, followed by drinking water for 8 weeks [28, 29]. Control mice of each genotype were administered drinking water for the entire duration of the experiment. At 24 weeks following initial 4NQO treatment, mice were sacrificed. Tongue lesions and tumours were counted macroscopically and microscopically.
Immunohistochemistry
Formalin-fixed-paraffin-embedded tissue sections were cut at a thickness of 5 µm and rehydrated. Heat-induced epitope retrieval was performed in sodium citrate antigen retrieval buffer. Tissues were stained with H&E or, for immunohistochemistry, Granzyme B (Abcam, ab4059; Cambridge, MA), Ki-67 (Abcam, Ab15580), PD1 (Cell Signaling 9664S), cleaved caspase 3 (Cell Signaling, 84651S), PD-L1 (Invitrogen, 14-5982-82; Waltham, MA), CD4 (Invitrogen, 14-0042-82), CD8 (Invitrogen, 14-0808-82) and CXCL9 (BioXCell, BE0309). Following the addition of primary antibody, the tissues were incubated with biotinylated goat anti-rabbit (Vector Laboratories, BA-1000; Burlingame, CA), biotinylated goat anti-hamster (Invitrogen, 31750), biotinylated goat anti-rat (Vector Laboratories, BA-9401) IgG (H + L) secondary antibodies or, for immunofluorescent staining, Alexa Fluor 488-conjugated goat anti-rabbit (Invitrogen, A11034) and Alexa Fluor 555 goat anti-rat (Invitrogen, A21434) with a DAPI counterstain. Regions of positive staining were quantified using ImageJ [26].
Flow cytometry
Cells were stained with extracellular antibodies prior to fixation in 4% neutral-buffered formalin and permeabilisation. Fluorochrome-conjugated antibodies against CD4, CD8, CD11b, Ly6C, Ly6G, PD-1 and PD-L1 were used for extracellular staining. Intracellular staining was performed with antibodies against Gzmb, IFN-γ, pSTAT1 Y701 (Biolegend) and TRIM24 (AbClonal; Woburn MA). Mouse IgG1κ and Rat IgG1κ were used as isotype controls for Gzmb and IFN-γ, respectively. TRIM24 primary antibodies were further incubated with goat anti-rabbit FITC-conjugated secondary antibodies (BD Biosciences; San Jose, CA) prior to acquisition. Other samples were treated with propidium iodide (PI) and FITC-conjugated Annexin V for 15 min for apoptosis analysis. Samples were analysed using a FACS Celesta flow cytometer (BD Biosciences; San Jose, CA). Analysis was performed using the FlowJo software (FlowJo; Ashland, OR).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
RNA from cell lines was isolated using acid/chloroform/guanidinium thiocyanate/phenol extraction. Tongue samples were homogenised using a Bead Mill (VWR; Radnor, PA) before RNA extraction with DirectZol RNA miniprep (Zymo Research; Irvine, CA). cDNA was prepared from extracted RNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). RT-qPCR was performed on a CFX384 Real-Time PCR system (BioRad; Hercules, CA) using SYBR Green Master Mix (Thermo Fisher; Waltham, MA). Sequences of primers for Bactin, Bcl2, Casp8, Cxcl9, Gapdh, Gzmb, Ifng, Ki67, Mcm3, Pd1 and Pdl1 were generated using PrimerBank (https://pga.mgh.harvard.edu/primerbank/).
Statistics
Analysis was performed in a blinded fashion. Statistical analyses were performed using GraphPad Prism v9.0 (GraphPad Software; San Diego, CA). Analysis of variance or Student’s t test (two-sided) was used to determine statistically significant differences between groups.
Results
STAT1 promotes expression of PD-L1 and proliferative biomarkers in HNSCC cells
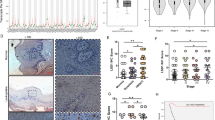
To explore the role of STAT1 in HNSCC, we performed siRNA mediated knockdown of Stat1 expression in the human HNSCC cells. We used the CAL27 and UM-SCC22A cell lines because they are HPV negative HNSCC cells that express PD-L1 and STAT1. We first confirmed downregulation of Stat1 mRNA transcripts and total STAT1 protein in both cell lines using RT-qPCR and western blot (Fig. 1a, b). Further, in CAL27 cells, STAT1 siRNA knockdown decreased STAT1 phosphorylation compared to scrambled RNA treated cells. We did not observe any differences in STAT1 phosphorylation between STAT1 siRNA and scrambled RNA treated UM-SCC22A cells (Fig. 1a, b). Next, we determined the effects of STAT1 knockdown on immune suppression, apoptosis and biomarkers of cell proliferation. We first examined the effects of Stat1 silencing on HNSCC cell PD-L1 expression by flow cytometry. We observed that PD-L1 surface expression was significantly downregulated following Stat1 knockdown in both cell lines, indicating an essential role for STAT1 in promoting PD-L1 surface expression on HNSCC cells (Fig. 1c, e). Notably, despite the low constitutive expression of PD-L1 by UM-SCC22A cells, STAT1 knockdown eliminated the expression of this immunosuppressive receptor on this HNSCC cell line (Fig. 1e). Our data demonstrate that STAT1 promotes PD-L1 expression on HPV negative HNSCC cells in vitro. We further characterised the relationship between STAT1 and PD-L1 expression using the publicly available cancer genome atlas (TCGA) database of HNSCC patients. We observed a striking positive correlation between STAT1 and PDL1 in tumours of HNSCC patients (Fig. 1g), further supporting our hypothesis of PD-L1 regulation by STAT1 in HNSCC.
a, b Gene expression of Stat1 determined by RT-qPCR, STAT1 and GAPDH protein levels, determined by western blot, and phospho STAT1 (pSTAT1) protein levels after IFNγ stimulation determined by flow cytometry, in Stat1 siRNA and scrambled RNA treated a CAL27 and b UM-SCC22A cells. STAT1 and GAPDH protein levels were detected after 72 h of siRNA treatment, and western blot band intensity of STAT1:GAPDH were calculated in scramble and Stat1 siRNA-treated cells. Numbers in histogram plots represent percentage of phospho STAT1 positive cells. Controls are unstimulated cells. c, e PD-L1 expression by c CAL27 and e UM-SCC22A cells after 72 h of siRNA treatment, determined by flow cytometry. d, f Flow gating and frequencies of AnnexinV+PI− populations among d CAL27 and f UM-SCC22A cells after 72 h of siRNA treatment. h, i Gene expression of Ki67 and Mcm3, determined by RT-qPCR in h CAL27 and i UM-SCC22A cells after 48 or 72 h siRNA treatment. Data are presented as mean +/− SEM. *p value <0.05; **p value <0.01; ***p value <0.001, ****p value <0.0001 between groups.
To examine the effects of STAT1 on apoptosis, we utilised Annexin-V/PI staining on cell populations 48 and 72 h following STAT1 siRNA knockdown. We observed a significantly higher percentage of early apoptotic Annexin-V+I− cells in STAT1-deficient compared to STAT1-sufficient CAL27 and UM-SCC22A cell lines (Fig. 1d, f). Finally, we examined the effects of STAT1 on biomarkers of cell proliferation (Ki67 and Mcm3) by RT-qPCR [30]. We observed decreased expression of both markers following STAT1 knockdown in CAL27 (Fig. 1h). In UM-SCC22A, we observed a similar downregulation of these proliferation markers at 48 h following STAT1 inhibition (Fig. 1i). Taken together, these data indicate that STAT1 expression by HNSCC cells promotes PDL1 expression, increases proliferative potential, and inhibits apoptosis, thereby enhancing the tumorigenicity of HNSCC cells.
Global STAT1 deficiency in a 4NQO model of HNSCC results in increased incidence of carcinogenesis
Previous work from our group demonstrated that in STAT1-sufficient murine HNSCC cells orthotopically injected into Stat1+/+ and Stat1−/− mice, tumour growth is significantly enhanced when Stat1 is deleted in host tissue [12]. Therefore, to determine the impact of tumour-derived STAT1 on HNSCC tumour development in vivo, we used the oral carcinogen 4NQO to induce oral carcinogenesis in both Stat1+/+ and Stat1−/− mice (Fig. 2a). We chose this model because it recapitulates the initiation, promotion and progression stages of carcinogenesis in clinical cases of non-HPV associated HNSCC. First, we observed increased mortality in carcinogen induced Stat1−/− compared to Stat1+/+ mice (Fig. 2b). At terminal sacrifice, we also noted higher incidence and multiplicity of both lesions and tumours in Stat1−/− mice (Fig. 2c–e). Our findings were confirmed by H&E staining of the carcinogen-induced tongue tissue (Fig. 2f). To determine whether global STAT1 deficiency affects proliferation and apoptosis of tumour cells in vivo, we measured Ki-67 and caspase 3 expression in tumours by RT-qPCR and immunohistochemistry. We also measured Aurka and Bcl2 gene expression levels. Overall, proliferative and apoptotic markers were not affected by STAT1 deficiency in vivo (Fig. 2g–i). Taken together, our results indicate that despite the pro-tumorigenic properties of STAT1 in HNSCC tumour cells, global STAT1 deficiency results in higher tumour burdens in the carcinogen-induced model of HNSCC carcinogenesis. Although these results mimic our observations in the orthotopic model, the difference in tumour development between Stat1+/+ and Stat1−/− mice in the 4NQO model was not as high as in the orthotopic model. These results demonstrate that, although STAT1 expression by immune cells is essential for an anti-tumour response, STAT1 expression by tumour cells might influence tumour development to a lesser extent than in immune cells, but in a manner that suggests a tumour promoting role in vivo.
a 4NQO model of carcinogen induced oral carcinogenesis showing time period of 4NQO administration and terminal sacrifice. b Kaplan–Meier curve depicting survival of 4NQO-treated Stat1+/+ and Stat1−/− mice through the duration of the experiment. c Counts of lesions and tumours on the tongues of Stat1+/+ and Stat1−/− 4NQO-treated mice determined at terminal sacrifice. d Representative images of mouse tongues containing tumours from 4NQO-treated Stat1+/+ and Stat1−/− at terminal sacrifice. e Tabular illustration of the incidence and multiplicity of lesions and tumours in experimental mice. f Representative H&E-stained images of dysplastic lesions in tongues from Stat1+/+ and Stat1−/− mice. g, h Representative immunohistochemistry images of oral lesions in tongues of Stat1+/+ and Stat1−/− mice stained with g KI-67 and h cleaved caspase 3. Bar graphs depict regions of positive staining from 10 representative fields each for n = 6 or 8 mice per group. i Fold induction of Ki67, Aurka, Casp3 and Bcl2 transcripts in the tongues of control and carcinogen-induced Stat1+/+ and Stat1−/− mice determined by RT-qPCR (n = 10 cancer-induced mice per group). Data are presented as mean +/− SEM. *p value <0.05; ***p value <0.005 between groups.
STAT1 promotes PD-1/PD-L1 expression in the tumour microenvironment during HNSCC
Given the observed role for STAT1 in PD-L1 expression by HNSCC cells in vitro, we next explored the impact of STAT1 on PD-1 and PD-L1 expression in the tumour microenvironment during HNSCC carcinogenesis in vivo. We observed significantly reduced gene expression and cell protein expression of Pd1 at the tongue tissues of carcinogen induced Stat1−/− mice, indicating decreased PD-1 immune checkpoint activity (Fig. 3a). We had previously observed that PD-1 was strongly induced in T-lymphocytes of tumour-bearing Stat1−/− mice orthotopically injected with Stat1 competent aggressive HNSCC [12]. Therefore, we examined PD-1 expression in T lymphocytes of carcinogen-induced Stat1+/+ and Stat1−/− mice. In contrast with Stat1−/− orthotopic tumour-bearing mice, PD-1 expression was reduced in CD8+ T cells in the tumours, lymph nodes and spleens of carcinogen-induced Stat1−/− mice compared to Stat1+/+ mice (Fig. 3b, d). PD-1 expression in CD4+T cells was reduced in spleens, but not tumours and draining lymph nodes of carcinogen-induced Stat1−/− mice compared to Stat1+/+ mice (Fig. 3c, e). Combined with previous studies, these data suggest that during HNSCC, induction of PD-1 expression on T cells is partly dependent on STAT1 expression by tumour cells.
a Gene and protein expression of Pd1 in the tongues of experimental mice determined by RT-qPCR and immunofluorescence microscopy. b, c Representative immunofluorescence staining of b PD1 (green), CD8 (red) and c PD1 (green), CD4 (red) in tongue tumours of carcinogen-induced Stat1+/+ and Stat1−/− mice. Bar graphs depict average numbers of PD1+CD8+ and PD1+CD4+ cells obtained from 10 representative fields each for n = 6 or 8 mice per group d, e PD-1 surface expression by d CD8+ cells and e CD4+ cells from the lymph nodes and spleens of experimental mice. Graph shows average percentage of PD1+CD8+ and PD1+CD4+ cells (n = 15 or 17 cancer induced mice per group). f Gene and protein expression of Pdl1 in the tongues of experimental mice determined by RT-qPCR and immunofluorescence microscopy. g Representative immunofluorescence staining of PDL1 (green) and CD11b (red) in tongue tumours of carcinogen-induced Stat1+/+ and Stat1−/− mice. Bar graph depicts average numbers of PDL1+CD11b+ cells obtained from 10 representative fields each for n = 6 or 8 mice per group h, i Comparative histograms of PD-L1 surface expression by h M-MDSCs and j M12 macrophages in carcinogen-induced Stat1+/+ and Stat1−/− mice. Graphs show mean fluorescence intensities (MFI) of PD-L1 surface expression by h M-MDSCs, i G-MDSCs, j M1 macrophages and k M2 macrophages in lymph nodes, spleens and bone marrow of experimental mice (n = 15 or 17 cancer-induced mice per group). Data are presented as mean +/− SEM. *p value <0.05; **p value <0.01; ***p value <0.001 between groups.
Pdl1 gene expression was similarly reduced in tongue tissues of carcinogen induced Stat1−/− mice (Fig. 3f). Because PD-L1 expression is induced in myeloid cells during HNSCC, we analysed the role of STAT1 in PD-L1 expression by CD11b+ myeloid populations including Ly6Cint Ly6G+ cells, Ly6Chi Ly6G− cells, classical M1 macrophages (Ly6C−F4/80+CD206−), and alternatively activated M2 macrophages (Ly6C−F4/80+CD206+) from the lymph nodes, spleens, and bone marrow of carcinogen induced Stat1−/− and Stat1+/+ mice. PD-L1 expression was reduced among CD11b+ cells in the tumours of Stat1−/− mice (Fig. 3g). Ly6Chi Ly6G− cells, M1, and M2 macrophages from the draining lymph nodes of Stat1−/− mice demonstrated lower PD-L1 expression compared to Stat1+/+ mice (Fig. 3h–k). In the spleen, we observed a decrease in PD-L1 among M1 macrophages in Stat1−/− mice, although we did not observe any differences among other immunosuppressive myeloid populations (Fig. 3j). Examination of myeloid cells in the bone marrow showed diminished PD-L1 expression by Ly6Cint Ly6G+ and Ly6Chi Ly6G− cells (Fig. 3h, i). Taken together, our findings suggest that tumour-derived STAT1 is required for PD-1/PD-L1 immune checkpoint expression and immune suppression during experimental HNSCC in vivo.
STAT1 is required for anti-tumour function by T-lymphocytes during HNSCC carcinogenesis
Despite the increased PD-1/PD-L1 expression observed in carcinogen induced Stat1 competent mice, tumour development was still lower than in Stat1−/− mice. Therefore, we next examined potential STAT1 mediated anti-tumour mechanisms. Specifically, we measured IFN-γ, GranzymeB, and CXCL9 [31, 32], markers known to be regulated by STAT1 and are associated with T cell anti-tumour activity, effector responses and chemotaxis of anti-tumour effector T cells. At the primary tumour site, we found that Gzmb, Ifng, and Cxcl9 transcripts were significantly reduced in carcinogen-induced Stat1−/− compared to Stat1+/+ mice (Fig. 4a). ELISA analysis of tongue lysates and immunohistochemical staining of tongue lesion sites further corroborated the decreased IFN-γ, GranzymeB and Cxcl9 protein expression in Stat1−/− mice (Fig. 4b–d). Given the decreased anti-tumour activity in the STAT1-deficient tumour microenvironment, we analysed T-cell populations of the oral draining lymph nodes and spleens of experimental mice. At the lymph nodes, Gzmb expression by CD4+ and CD8+ lymphocytes were decreased in Stat1−/− mice (Fig. 4e–g). IFN-γ production was decreased in splenic CD8+ T cells and NK cells of the lymph nodes and spleen of carcinogen-induced Stat1−/− mice compared to carcinogen-induced Stat1+/+ mice (Fig. 4h–j and Supplementary Fig. 1). These results demonstrate the importance of STAT1 in maintaining an effective anti-tumour immune response and immunosurveillance during HNSCC, and explain potential mechanisms underlying the increased tumour burden observed in Stat1−/− mice compared to their Stat1+/+ counterparts.
a Fold induction of Ifng, Gzmb and Cxcl9 transcripts in the tongues of experimental mice determined by RT-qPCR. b IFNγ protein expression in tongue lysates of carcinogen-induced Stat1+/+ and Stat1−/− mice as determined by ELISA. c, d Representative immunohistochemistry images of oral tumours in tongues of Stat1+/+ and Stat1−/− mice stained with c Gzmb and d Cxcl9. Bar graphs depict regions of positive staining from 10 representative fields each for n = 6 or 8 mice per group. e–j Representative dot plots of e Gzmb and h IFNγ in CD4+ T cells in draining lymph nodes of carcinogen-induced Stat1+/+ and Stat1−/− mice as determined by intracellular flow cytometry. Graphs depict average percentage of f Gzmb+ CD4+, (G) Gzmb+ CD4+, i IFN-γ+CD4+, and j IFN-γ+CD8+ T cells in draining lymph nodes and spleens of cancer induced and control Stat1+/+ and Stat1−/− mice (n = 15 or 17 cancer-induced mice per group). Data are presented as mean +/− SEM. *p value <0.05; **p value <0.01; ***p value <0.001 between groups; ****p value <0.0001.
TRIM24 is selectively expressed in HNSCC cells and negatively regulates STAT1
We next investigated potential STAT1 modulators that could selectively impact STAT1 signalling in HNSCC cells. TRIM24 has previously been shown to be over expressed in advanced HNSCC and is associated with HNSCC metastasis [19]. Pathway analysis of RNA sequencing data from tumours from Stat1+/+ and Stat1−/− mice orthotopically injected with LY2 HNSCC cells also identified TRIM24 as the top predicted negative transcriptional regulator of STAT1 during HNSCC in our data set (z score –5.804, p value 1.32 × 10−16) (Fig. 5a, b). Mutational analysis of these orthotopically injected tumours in Stat1+/+ and Stat1−/− mice from sequencing data are shown in Supplementary Fig. 2. Analysis of 519 HNSCC patients from the cancer genome atlas (TCGA) did not show a real inverse correlation between TRIM24 and STAT1 expression (r = −0.1764, p = 7.05 × 10−5) (Fig. 5c). However, in STAT1-knockdown CAL27 HNSCC cells, gene and protein expression of TRIM24 is slightly, but significantly increased (Fig. 5d, e). Interestingly, in our orthotopic HNSCC model, we found that TRIM24 expression is high in HNSCC cells but not in tumour-infiltrating T lymphocytes (Fig. 5f, g), suggesting the potential of targeting TRIM24 to modulate STAT1 signalling specifically in HNSCC cells but not T cells. Therefore, we investigated the effects of TRIM24 in human HNSCC cells using TRIM24 specific siRNA knockdown. We first verified knockdown of TRIM24 gene and protein expression by RT-qPCR and Western blot (Fig. 5h, i). Next, we determined the effect of TRIM24 on STAT1 expression, and we observed a marked increase in STAT1 transcripts (Fig. 5h), total STAT1 and phosphorylated STAT1 protein (Fig. 5i and Supplementary Fig. 3). These data strongly implicate TRIM24 as a negative regulator of STAT1 production and activity in HNSCC cells. These results were further corroborated in UMSCC22A cells, although p-STAT1 protein expression was unchanged in TRIM24 knockdown cells (Fig. 5j, k).
a Volcano plot showing top 100 upstream regulators based on Ingenuity pathway analysis of RNA sequencing data from oral tumour bearing Stat1+/+ and Stat1−/− mice (n = 4 per group). b Predicted mechanistic network analysis showing transcriptional targets of TRIM24 based on Ingenuity pathway analysis of RNA sequencing data from oral tumour-bearing Stat1+/+ and Stat1−/− mice. Genes in red are upregulated and blue are downregulated in the RNA sequencing data set. c Correlation of TRIM24 expression levels with STAT1 expression in tumours from HNSCC patients samples acquired from The Cancer Genome Atlas. Significance determined by Pearson correlation coefficient. d Gene expression of Trim24 determined by RT-qPCR in CAL27 cells treated with scramble or Stat1 siRNA for 72 h. e TRIM24 protein expression by CAL27 cells treated with scramble or Stat1 siRNA for 72 h, determined by flow cytometry. f Gene expression of TRIM24 in tumours of HNSCC tumour-bearing mice orthotopically injected with the LY2 cell line. g TRIM24 protein expression, determined by flow cytometry, in HNSCC cells of mice orthotopically injected with the LY2 cell line. h, j Gene expression of Trim24 and Stat1 determined by RT-qPCR in (H) CAL27 and (J) UMSCC22A cells treated with scramble or Trim24 siRNA for 48 or 72 h. i, k Protein expression of TRIM24, STAT1, pSTAT1 and GAPDH, determined by western blot, in i CAL27 and k UMSCC22A cells accompanied by the protein band intensities relative to GAPDH in scramble and Trim24 siRNA-treated cells after 48 and 72 h (n = 3 samples per group). Data are presented as mean +/− SEM. #p value <0.1, *p value <0.05; **p value <0.01; ***p value <0.001 between groups.
Effects of TRIM24 on HNSCC cells are mediated by STAT1-dependent and independent mechanisms
Given the co-inhibitory effects of TRIM24 and STAT1 expression in HNSCC cells, (Fig. 6a, b), we further explored the interaction between TRIM24 and STAT1 on HNSCC cell PD-L1 expression, apoptosis and cell proliferation. Interestingly, despite the increase in STAT1 induced by TRIM24 knockdown, we observed that TRIM24 silencing inhibited the tumour promoting effects and enhanced the tumour inhibitory effects of STAT1 activation. Specifically, while we showed STAT1 to be important for PD-L1 expression, we surprisingly saw that TRIM24 knockdown resulted in an even more significant decrease in PD-L1 surface expression in CAL27 cells compared to STAT1 siRNA treated cells (Fig. 6c). Similarly, TRIM24 knockdown significantly decreased the expression of the proliferation markers KI67 and MCM3, which we had showed to be decreased by STAT1 inhibition (Fig. 6d, e and Supplementary Fig. 3). This was also true of the increased expression of the apoptotic markers CASP8 and BCL2 in TRIM24 knockdown CAL27 and UMSCC22A cells (Fig. 6d, e and Supplementary Fig. 3). These results indicate that TRIM24, while a negative regulator of STAT1 in HNSCC cells, inhibits the tumour-promoting effects and enhances the tumour-inhibitory effects of STAT1 activation.
a, b Gene expression of a Stat1 and b Trim24 determined by RT-qPCR in CAL27 cells at 72 h post-siRNA treatment of cells treated with scramble, Stat1, Trim24 and Stat1/Trim24 siRNA. c PD-L1 protein expression by siRNA-treated CAL27 cells, determined by flow cytometry. d Gene and e protein expression of Ki67, Mcm3, Bcl2, and Casp8, determined by RT-qPCR and Western blot in siRNA-treated CAL27 cells at 48 or 72 h. f Graphical model demonstrating the influence of STAT1 and TRIM24 on gene/protein expression and cell function in myeloid cells, T cells and HNSCC cells. Statistical significance (p < 0.05) in comparison to a +Stat1+Trim24, b −Stat1+Trim24 and c +Stat1−Trim24 is noted above each group (n = 5 biological replicates per group for RT-PCR and 3 biological replicates per group for Western blot). Data are presented as mean +/− SEM.
Next, we performed double knockdown experiments of STAT1 and TRIM24 to determine mechanisms of TRIM24 effects on HNSCC cells and the involvement of STAT1 activity. We determined the expression of PDL1 on TRIM24 and STAT1 single or double knockdown CAL27 cells. We observed that knockdown of STAT1 abrogated the effects of TRIM24 on PDL1 expression in these cells. On the other hand, knockdown of TRIM24 did not significantly affect PDL1 expression on STAT1 knock down cells (Fig. 6c). This suggests that the effects of TRIM24 on PDL1 expression in CAL27 HNSCC cells are STAT1 dependent. We also examined the impact of TRIM24 and STAT1 double knockdown HNSCC cells on markers of apoptosis and cellular proliferation. We observed similar or enhanced reduction of the proliferation markers Ki67 and MCM3 in double knockdown compared to single knock down CAL27 and UMSCC cells (Fig. 6d, e and Supplementary Fig. 3). Taken together, our results demonstrate a unique interaction between TRIM24 and STAT1 in HNSCC cells. Based on these findings, we propose a model demonstrating the divergent roles of STAT1 signalling in cancer cells and immune cells of the tumour microenvironment (Fig. 6f). We also demonstrate the interactions of TRIM24 as an upstream regulator of STAT1, inhibiting STAT1 expression and phosphorylation in HNSCC cells. Conversely, STAT1 inhibits TRIM24 expression, although to a lesser extent (Fig. 6f). Further we demonstrate that the effects of TRIM24 on immunosuppression, apoptosis and proliferation of HNSCC cell are mediated by STAT1-dependent and independent mechanisms.
Discussion
This study highlights new mechanistic insights into the role of STAT1 expression by HNSCC cells and immune cells, as well as the function of a novel STAT1 negative regulator TRIM24, in HNSCC cells. Using RNAi silencing technology, we found that STAT1 plays an essential role in PD-L1 expression by HNSCC cells. We further found, using the 4NQO murine model of HNSCC, that STAT1 signalling is essential for PD-1/PD-L1 expression in the tumour microenvironment and by immune cells, including the accumulation of immunosuppressive populations during head and neck carcinogenesis. We further demonstrate an important role for STAT1 in anti-tumour immunity by T cells, which supports our previous reports [12]. Lastly, we show a negative co-regulatory function between STAT1 and TRIM24, which together induce PD-L1 expression in HNSCC cells (Fig. 6f).
The results of our study indicate a pro-tumour role for tumour-derived STAT1 expression in HNSCC based on its ability to enhance PD-L1-mediated immunosuppression and proliferation in both UM-SCC22A and CAL27 HNSCC cell lines. The role of STAT1 in regulating apoptosis, however, appears to be context-dependent. A downregulatory effect of STAT1 on the anti-apoptotic protein BCL2 has been previously established, although not in HNSCC cells [33]. We show that following STAT1 silencing in CAL27 cells, BCL2 expression is highly upregulated, showing that STAT1 maintains its downregulatory effect on BCL2 among the HNSCC cells tested. On the other hand, STAT1 activity appears to induce caspase-8 expression [34]. However, the non-canonical elevation of Casp8 expression we observed in CAL27 cells following STAT1 inhibition warrants further investigation, as this may be a driver behind tumour survival in cases of HNSCC with high expression of STAT1.
The role of tumour-derived STAT1 in promoting of PD-L1 expression, however, is more evident. Previously, our group observed that STAT1-competent HNSCC induced greater expression of both PD-1 and PD-L1 in immune cells of Stat1−/− mice compared to Stat1+/+ in both in situ and metastatic orthotopic models of HNSCC [12]. Our current results show that global STAT1 deficiency in a carcinogen-induced model of HNSCC leads to decreased PD-1/PD-L1 expression in the tumour microenvironment and by immune cells. In light of the finding that PD-1/PD-L1 expression STAT1-deficient immune cells were elevated in the context of STAT1-competent tumours but not STAT1-deficient tumours, it appears that STAT1 expression by tumour cells plays a direct role in inducing immune checkpoint signalling between immune cells. Moreover, host immune cells do not require STAT1 to express high levels of PD-1 or PD-L1. Taken together, these findings indicate that PD-1/PD-L1 activity in immune cells is dependent on STAT1 expression by tumour cells during HNSCC but may be independent of STAT1 expression by immune cells.
It was surprising to observe a reduced accumulation of immunosuppressive myeloid populations in carcinogen induced Stat1-deficient mice compared to Stat1-sufficient mice. This contrasts with our previous findings where these populations significantly accumulate in Stat1-deficient mice bearing Stat1 competent aggressive HSNCC tumours [12]. While it has been shown that STAT1 promotes M2 macrophage activity and development [35, 36], we previously observed that these populations also accumulate in Stat1-deficient mice. Our observation that these populations are reduced in the bone marrow of carcinogen induced Stat1−/− mice implies that tumour-derived STAT1 plays a major role. Notably, we observed a significant decrease in Ly6Chi Ly6G− cells, but not Ly6Cint Ly6G+ cells, in the lymph nodes and spleens of carcinogen induced Stat1-deficient mice, which is another population we previously observed to accumulate in Stat1-deficient mice bearing Stat1-competent aggressive HNSCC. Together, these results highlight the important role of tumour-derived STAT1 in promoting the accumulation of immunosuppressive myeloid populations, especially when these host derived immune cells are Stat1 deficient. Because M-MDSCs are known to facilitate the epithelial-to-mesenchymal transition that precedes metastasis [37], tumour-derived STAT1 may be a predictor of immune suppression and metastasis in HNSCC. Alternatively, MDSC populations have been shown to expand in response to chronic inflammation [38, 39], and their deficiency may be due to the failure of Stat1−/− mice to initiate a robust anti-tumour response during the early stages of head and neck carcinogenesis [40].
Despite the reduction in the immunosuppressive PD-1/PD-L1 axis and MDSC populations observed in carcinogen induced Stat1-deficient mice, they still displayed significantly higher mortality and burden of dysplasia and neoplasia compared to Stat1 competent mice. This finding may be explained by our findings of decreased expression of Gzmb and IFN-γ by lymphocytes in Stat1−/− mice, which are known to facilitate the anti-tumour immune response to HNSCC and inhibit tumour proliferation [41,42,43]. Initially, this observation may seem to contradict the heightened PD-1/PD-L1 signalling which is known to inhibit lymphocyte anti-tumour functions [14]. However, the global reduction of Gzmb in Stat1−/− mice can be explained by the role STAT1 plays in its production in CD8 T cells [44]. Alternatively, decreased PD-1 expression by T-cells may be resultant of decreased activation rather than a direct result of STAT1 deficiency. Further, absent IFN-γ signal transduction mediated by STAT1 leads to failure to prepare immune cells for sufficient anti-tumour immune activity, as evidenced by decreased IFN-γ expression in the tumour microenvironment. However, we also observed a corresponding decrease in PD-L1 in the tumour microenvironment, which corresponds to the role of IFN-γ and STAT1 signalling in the production of PD-L1. Thus, while immune cell STAT1 activity is strongly associated with HNSCC survival and can be considerably variable among patients [11], our data supports the hypothesis that STAT1 activation by HNSCC cells could simultaneously potentiate immunosuppressive and tumorigenic effects during HNSCC. These data taken together indicate that while impaired STAT1 signalling within lymphoid populations is likely to produce a deleterious effect in HNSCC, selective targeting of STAT1 signalling in HNSCC cells could potentially enhance anti-tumour immune responses and inhibit tumour cell growth.
TRIM24 is a transcriptional intermediary factor that we have found to negatively correlate with STAT1 expression. Thus TRIM24 presents itself as an interesting target for modulating STAT1 expression and downstream effects in HNSCC. Previously, TRIM24 has been shown to bind to retinoic acid responsive elements of the STAT1 promoter, thereby repressing STAT1 transcription [17, 19]. However, there has been a long-standing debate on whether TRIM24 functions as either a tumour suppressor or oncogene, and the answer appears to be context-dependent [18, 45]. Notably, TRIM24 expression has been shown correlate with both locally advanced and metastatic HNSCC, though the mechanisms underlying this effect remain unclear [19, 46]. Although we did not observe a strong correlation between STAT1 and TRIM24 based on TCGA data, our results demonstrate that TRIM24 negatively regulates STAT1, and we present novel evidence that STAT1 also negatively regulates TRIM24 in HNSCC cells.
Despite its inhibitory effect on STAT1 expression, we also found TRIM24 to have a significant impact on the immunosuppressive ability of HNSCC cells, indicating a complex regulatory interaction beyond simple co-inhibitory effects. For example, while STAT1 is involved in PD-L1 expression, TRIM24 knockdown in HNSCC cells significantly inhibited PD-L1 expression to an even greater extent than observed in STAT1 knockdown HNSCC cells. This finding was particularly surprising given the observed STAT1 mediated promotion of PD-L1 expression and the downregulatory effect of TRIM24 on STAT1 protein expression and phosphorylation. These results appear to highlight the complex crosstalk between TRIM24 and STAT1 to induce PD-L1 expression in HNSCC. This is further demonstrated by our observation that STAT1 depletion in TRIM24 knockdown cells partially restored PD-L1 expression levels, indicating that TRIM24 effects of PD-L1 expression on HNSCC cells are at least partly mediated by STAT1. Potential transcriptional, translational and post-translational mechanisms underlying the complex interplay between TRIM24 and STAT1 in mediating immunosuppression in HNSCC cells will require further investigation.
Given that TRIM24 was selectively expressed in tumour cells of the HNSCC microenvironment potentially provides an opportunity for selectively targeting STAT1 and mitigating its tumour promoting effects in HNSCC in vivo. This is particularly significant since STAT1 mediates tumour promoting and inhibitory roles in HNSCC cells despite playing a significant anti-tumour function in immune cells. Further, the tumour inhibitory effects mediated by TRIM24 inhibition were significantly greater than STAT1 inhibition. There is therefore a strong rationale to exploring TRIM24 as a target in HNSCC treatment, which can potentially leverage the STAT1 pathway to improve HNSCC tumour outcomes. It is possible that TRIM24 inhibition could directly or indirectly inhibit the tumorigenicity of HNSCC tumour cells. Additionally, while TRIM24 expression is low in tumour-infiltrating lymphocytes, its downregulation could, in turn, upregulate STAT1 anti-tumour activity. A small molecule inhibitor of TRIM24 has been reported with high potency and favourable pharmacokinetic properties—a therapeutic role in HNSCC could be further explored in future studies [47].
In conclusion, our findings indicate that tumour-derived STAT1 regulates HNSCC tumorigenicity and PD-L1 immune checkpoint signalling, while T-cell-derived STAT1 is necessary for immune activation and anti-tumour immune responses. We further demonstrate that TRIM24 potentiates the tumour promoting function of STAT1 in HNSCC cells.
Data availability
All raw data supporting the findings of this study are available upon request. Sequencing data are available via the Gene Expression Omnibus (GSE198790).
References
Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6:92.
Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–53.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Stephanou A, Latchman DS. STAT-1: a novel regulator of apoptosis. Int J Exp Pathol. 2003;84:239–44.
Kaplan MH. STAT signaling in inflammation. JAKSTAT. 2013;2:e24198.
Gao J, Wang F, Liu Y, Cai M, Xu H, Jiang J, et al. Revealing the cellular localization of STAT1 during the cell cycle by super-resolution imaging. Sci Rep. 2015;5:9045.
Ramana CV, Chatterjee-Kishore M, Nguyen H, Stark GR. Complex roles of Stat1 in regulating gene expression. Oncogene. 2000;19:2619–27.
Ni H, Sun H, Zheng M, Bian T, Liu J, Li X, et al. Mining database for the expression and clinical significance of STAT family in head and neck squamous cell carcinomas. Transl Oncol. 2021;14:100976.
Leibowitz MS, Andrade Filho PA, Ferrone S, Ferris RL. Deficiency of activated STAT1 in head and neck cancer cells mediates TAP1-dependent escape from cytotoxic T lymphocytes. Cancer Immunol Immunother. 2011;60:525–35.
Xi S, Dyer KF, Kimak M, Zhang Q, Gooding WE, Chaillet JR, et al. Decreased STAT1 expression by promoter methylation in squamous cell carcinogenesis. J Natl Cancer Inst. 2006;98:181–9.
Pappa E, Nikitakis N, Vlachodimitropoulos D, Avgoustidis D, Oktseloglou V, Papadogeorgakis N. Phosphorylated signal transducer and activator of transcription-1 immunohistochemical expression is associated with improved survival in patients with oral squamous cell carcinoma. J Oral Maxillofac Surg. 2014;72:211–21.
Ryan N, Anderson K, Volpedo G, Hamza O, Varikuti S, Satoskar AR, et al. STAT1 inhibits T-cell exhaustion and myeloid derived suppressor cell accumulation to promote antitumor immune responses in head and neck squamous cell carcinoma. Int J Cancer. 2020;146:1717–29.
Chen S, Crabill GA, Pritchard TS, McMiller TL, Wei P, Pardoll DM, et al. Mechanisms regulating PD-L1 expression on tumor and immune cells. J Immunother Cancer. 2019;7:305.
Ma H, Yang W, Zhang L, Liu S, Zhao M, Zhou G, et al. Interferon-alpha promotes immunosuppression through IFNAR1/STAT1 signalling in head and neck squamous cell carcinoma. Br J Cancer. 2019;120:317–30.
Chang WM, Chang YC, Yang YC, Lin SK, Chang PM, Hsiao M. AKR1C1 controls cisplatin-resistance in head and neck squamous cell carcinoma through cross-talk with the STAT1/3 signaling pathway. J Exp Clin Cancer Res. 2019;38:245.
Bharadwaj U, Eckols TK, Xu X, Kasembeli MM, Chen Y, Adachi M, et al. Small-molecule inhibition of STAT3 in radioresistant head and neck squamous cell carcinoma. Oncotarget. 2016;7:26307–30.
Tisserand J, Khetchoumian K, Thibault C, Dembélé D, Chambon P, Losson R. Tripartite motif 24 (Trim24/Tif1α) tumor suppressor protein is a novel negative regulator of interferon (IFN)/signal transducers and activators of transcription (STAT) signaling pathway acting through retinoic acid receptor α (Rarα) inhibition. J Biol Chem. 2011;286:33369–79.
Groner AC, Cato L, de Tribolet-Hardy J, Bernasocchi T, Janouskova H, Melchers D, et al. TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer Cell. 2016;29:846–58.
Cui Z, Cao W, Li J, Song X, Mao L, Chen W. TRIM24 overexpression is common in locally advanced head and neck squamous cell carcinoma and correlates with aggressive malignant phenotypes. PLoS ONE. 2013;8:e63887.
Xie W, Zhang Y, Wang B, Hu Y, Zhan B, Wei F, et al. Tripartite motif containing 24 regulates cell proliferation in colorectal cancer through YAP signaling. Cancer Med. 2020;9:6367–76.
Zhou HE, Pan SS, Han H. TRIM24 aggravates the progression of ovarian cancer through negatively regulating FOXM1 level. Eur Rev Med Pharmacol Sci. 2019;23:10647–56.
Knobloch TJ, Ryan NM, Bruschweiler-Li L, Wang C, Bernier MC, Somogyi A, et al. Metabolic regulation of glycolysis and AMP activated protein kinase pathways during black raspberry-mediated oral cancer chemoprevention. Metabolites. 2019;9:140.
Sequeira I, Rashid M, Tomas IM, Williams MJ, Graham TA, Adams DJ, et al. Genomic landscape and clonal architecture of mouse oral squamous cell carcinomas dictate tumour ecology. Nat Commun. 2020;11:5671.
Wang J, Xie T, Wang B, William WN Jr, Heymach JV, El-Naggar AK, et al. PD-1 blockade prevents the development and progression of carcinogen-induced oral premalignant lesions. Cancer Prev Res. 2017;10:684–93.
Li Q, Dong H, Yang G, Song Y, Mou Y, Ni Y. Mouse tumor-bearing models as preclinical study platforms for oral squamous cell carcinoma. Front Oncol. 2020;10:212.
Nedungadi D, Ryan N, Anderson K, Lamenza FF, Jordanides PP, Swingler MJ, et al. Modulation of the oral glucocorticoid system during black raspberry mediated oral cancer chemoprevention. Carcinogenesis. 2022;43:28–39.
Anderson K, Ryan N, Siddiqui A, Pero T, Volpedo G, Cooperstone JL, et al. Black raspberries and protocatechuic acid mitigate DNFB-induced contact hypersensitivity by down-regulating dendritic cell activation and inhibiting mediators of effector responses. Nutrients. 2020;12:1701.
Oghumu S, Casto BC, Ahn-Jarvis J, Weghorst LC, Maloney J, Geuy P, et al. inhibition of pro-inflammatory and anti-apoptotic biomarkers during experimental oral cancer chemoprevention by dietary black raspberries. Front Immunol. 2017;8:1325.
Oghumu S, Knobloch TJ, Terrazas C, Varikuti S, Ahn-Jarvis J, Bollinger CE, et al. Deletion of macrophage migration inhibitory factor inhibits murine oral carcinogenesis: potential role for chronic pro-inflammatory immune mediators. Int J Cancer. 2016;139:1379–90.
Valverde LF, de Freitas RD, Pereira TA, de Resende MF, Agra IMG, Dos Santos JN, et al. MCM3: a novel proliferation marker in oral squamous cell carcinoma. Appl Immunohistochem Mol Morphol. 2018;26:120–5.
Chang H, Lee YG, Ko YH, Cho JH, Choi JK, Park KU, et al. Prognostic value of CD200R1 mRNA expression in head and neck squamous cell carcinoma. Cancers. 2020;12:1777.
Chen J, Meng X, Zhou Q, Feng J, Zheng W, Wang Z, et al. Effect of CXCR5-positive cell infiltration on the immune contexture and patient prognosis in head and neck squamous cell carcinoma. Onco Targets Ther. 2020;13:5869–77.
Stephanou A, Brar BK, Knight RA, Latchman DS. Opposing actions of STAT-1 and STAT-3 on the Bcl-2 and Bcl-x promoters. Cell Death Differ. 2000;7:329–30.
Fulda S, Debatin KM. IFNgamma sensitizes for apoptosis by upregulating caspase-8 expression through the Stat1 pathway. Oncogene. 2002;21:2295–308.
Kemmner S, Bachmann Q, Steiger S, Lorenz G, Honarpisheh M, Foresto-Neto O, et al. STAT1 regulates macrophage number and phenotype and prevents renal fibrosis after ischemia-reperfusion injury. Am J Physiol Renal Physiol. 2019;316:F277–F91.
Kusmartsev S, Gabrilovich DI. STAT1 signaling regulates tumor-associated macrophage-mediated T cell deletion. J Immunol. 2005;174:4880–91.
Ouzounova M, Lee E, Piranlioglu R, El Andaloussi A, Kolhe R, Demirci MF, et al. Monocytic and granulocytic myeloid derived suppressor cells differentially regulate spatiotemporal tumour plasticity during metastatic cascade. Nat Commun. 2017;8:14979.
Anderson K, Ryan N, Volpedo G, Varikuti S, Satoskar AR, Oghumu S. Immune suppression mediated by STAT4 deficiency promotes lymphatic metastasis in HNSCC. Front Immunol. 2019;10:3095.
Varikuti S, Oghumu S, Elbaz M, Volpedo G, Ahirwar DK, Alarcon PC, et al. STAT1 gene deficient mice develop accelerated breast cancer growth and metastasis which is reduced by IL-17 blockade. Oncoimmunology. 2017;6:e1361088.
Chen WC, Lai CH, Chuang HC, Lin PY, Chen MF. Inflammation-induced myeloid-derived suppressor cells associated with squamous cell carcinoma of the head and neck. Head Neck. 2017;39:347–55.
Bisheshar SK, De Ruiter EJ, Devriese LA, Willems SM. The prognostic role of NK cells and their ligands in squamous cell carcinoma of the head and neck: a systematic review and meta-analysis. Oncoimmunology. 2020;9:1747345.
El Jamal SM, Taylor EB, Abd Elmageed ZY, Alamodi AA, Selimovic D, Alkhateeb A, et al. Interferon gamma-induced apoptosis of head and neck squamous cell carcinoma is connected to indoleamine-2,3-dioxygenase via mitochondrial and ER stress-associated pathways. Cell Div. 2016;11:11.
Anderson K, Ryan N, Alkhimovitch A, Siddiqui A, Oghumu S. Inhibition of PI3K isoform p110γ increases both anti-tumor and immunosuppressive responses to aggressive murine head and neck squamous cell carcinoma with low immunogenicity. Cancers. 2021;13:953.
Fortin C, Huang X, Yang Y. Both NK cell-intrinsic and -extrinsic STAT1 signaling are required for NK cell response against vaccinia virus. J Immunol. 2013;191:363–8.
Herquel B, Ouararhni K, Khetchoumian K, Ignat M, Teletin M, Mark M, et al. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc Natl Acad Sci USA 2011;108:8212–7.
Chang YC, Chi LH, Chang WM, Su CY, Lin YF, Chen CL, et al. Glucose transporter 4 promotes head and neck squamous cell carcinoma metastasis through the TRIM24-DDX58 axis. J Hematol Oncol. 2017;10:11.
Palmer WS, Poncet-Montange G, Liu G, Petrocchi A, Reyna N, Subramanian G, et al. Structure-guided design of IACS-9571, a selective high-affinity dual TRIM24-BRPF1 bromodomain inhibitor. J Med Chem. 2016;59:1440–54.
Funding
This study was funded by National Institute of Dental and Craniofacial Research (NIDCR - R56DE030093), National Cancer Institute (NCI - K01CA207599), National Institute of Drug Abuse (NIDA - DP1DA054344), and American Cancer Society (ACS - RSG-19-079-01-TBG), awarded to SO. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or American Cancer Society.
Author information
Authors and Affiliations
Contributions
SO designed the study. KA, NR, DN, FL, MS, AS, PU and SO conducted experiments. KA, NR, FL, AS, MP and SO analysed data. KA, NR and SO wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All animal experiments were approved by the Institutional Animal Care and Use Committee (Protocol #2018A00000054) of The Ohio State University and performed according to the committee’s guidelines.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Anderson, K., Ryan, N., Nedungadi, D. et al. STAT1 is regulated by TRIM24 and promotes immunosuppression in head and neck squamous carcinoma cells, but enhances T cell antitumour immunity in the tumour microenvironment. Br J Cancer 127, 624–636 (2022). https://doi.org/10.1038/s41416-022-01853-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-022-01853-z