Abstract
Background
SPOP, an E3 ubiquitin ligase adaptor, can act either as a tumour suppressor or a tumour promoter. In prostate cancer (PCa), it inhibits tumorigenesis by degrading several oncogenic substrates. SPOP is the most altered gene in PCa (~15%), which renders it ineffective, promoting cancer. The remaining PCa tumours, which retain WT-SPOP, still progress to castration-resistant (CRPC) stage, indicating that other critical mechanisms exist for downregulating SPOP. SPOP is reduced in ~94% of WT-SPOP-bearing prostate tumours; however, no molecular mechanism is known for its downregulation.
Methods
SPOP was identified as a direct target of LIMK2 using an innovative technique. The reciprocal relationship between SPOP and LIMK2 and its consequences on oncogenicity were analysed using a variety of biochemical assays. To probe this relationship in vivo, xenograft studies were conducted.
Results
LIMK2 degrades SPOP by direct phosphorylation at three sites. SPOP promotes LIMK2’s ubiquitylation, creating a feedback loop. SPOP’s degradation stabilises AR, ARv7 and c-Myc promoting oncogenicity. Phospho-resistant SPOP completely suppresses tumorigenesis in vivo, indicating that LIMK2-mediated SPOP degradation is a key event in PCa progression.
Conclusions
While genomically altered SPOP-bearing tumours require gene therapy, uncovering LIMK2-SPOP relationship provides a powerful opportunity to retain WT-SPOP by inhibiting LIMK2, thereby halting disease progression.
Similar content being viewed by others
Background
Prostate cancer (PCa) remains the most prevalent type of urological cancer and ranks second in terms of cancer-related fatality in men in the US with over 191,930 estimated new cases and 33,330 projected deaths in 2020.1 Early stage prostate tumours can be effectively treated by androgen-deprivation therapy;2 nevertheless, almost all tumours eventually progress to castration-resistant PCa (CRPC), which is lethal.3,4 Thus there is an urgent need to uncover the molecular mechanisms of CRPC pathogenesis to identify effective drug targets.
LIM kinase-2 (LIMK2) is a serine/threonine and, at times, a tyrosine kinase. It contains two LIM motifs at the N-terminus, trailed by PDZ and kinase domains.5 LIMK2 regulates actin dynamics in normal cells by controlling the binding of ADF/cofilin to actin.6 LIMK2 is upregulated in several cancers and regulates multiple aspects of cancer development, including cell survival, cell proliferation and metastasis.7,8,9,10 Previously, we identified Aurora-A (AURKA) as a crucial regulator of LIMK2. AURKA and LIMK2 positively regulate each other, establishing a feedback loop, which promotes oncogenesis.11 A recent study from our laboratory further identified LIMK2 as a prospective clinical target for the treatment and prevention of CRPC.12 However, the molecular mechanisms by which it promotes CRPC pathogenesis remains largely unknown. To date, cofilin,6 membrane type 1 matrix metalloproteinase,13 TWIST112 and phosphatase and tensin homologue14 are the only known substrates of LIMK2. Identification of other LIMK2 substrates is expected to not only uncover the molecular mechanisms of LIMK2-induced malignancy but is also expected to provide additional therapeutic targets for disease intervention.
The present study centres on one such LIMK2 target, i.e. Speckle-type POZ protein (SPOP), which we identified utilising our chemical genetic technique.15 SPOP, an E3 ubiquitin ligase adaptor, plays a pivotal role in protein ubiquitination and regulates many cellular events, including regulation of cell cycle, proliferation and apoptosis.16 SPOP can both promote and suppress tumorigenesis depending on its downstream targets in different cell types. It acts as a tumour promoter in renal cell carcinoma and tumour suppressor in prostate, endometrial and other solid tumours.17 In PCa, SPOP ubiquitylates many oncogenic substrates, including androgen receptor (AR),18 steroid receptor coactivator 3 (SRC-3),19 MacroH2A,20 Ci/Gli,21 ERG22 SENP7,23 DEK and TRIM24.24 Not surprisingly, SPOP is the most commonly mutated (15%) gene in PCa.25 These mutations render SPOP ineffective in binding its substrates, which in turn closely correlate with poor prognosis in PCa patients.18,26,27 Importantly, the remaining 85% of PCa cases, which retain wild-type SPOP (WT-SPOP) still progress to CRPC stage, indicating the existence of other critical regulatory mechanisms for downregulating SPOP. SPOP level is indeed reduced in up to 93.5% of prostate tumours bearing WT-SPOP, confirming that SPOP downregulation is an essential step in PCa progression, although no mechanism has been identified to date.27,28 Thus, while numerous downstream targets of SPOP are known in various cancers including PCa, regulation of WT-SPOP in any cancer remains unknown. In this study, we discovered the first posttranslational regulatory mechanism showing that WT-SPOP is degraded by LIMK2, which is critical for stabilising AR, ARv7 and c-Myc and promoting aggressive oncogenic phenotypes in CRPC.
Methods
Cell culture
HEK-293T, Phoenix cells and the two CRPC cell lines, 22Rv1 and C4-2, were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). The HEK293T and Phoenix cells were grown in Dulbecco’s modified Eagle’s medium, while 22Rv1 and C4-2 cells were maintained in RPMI-1640. Culture media in both the cases were supplemented with 10% foetal bovine serum (FBS) and penicillin–streptomycin and cultures were grown at 37 °C with 5% CO2 in a humidified incubator.
LIMK2 and SPOP short hairpin RNAs (shRNAs)
LIMK2 shRNAs were designed and cloned in pLKO.1 vector as reported before.11 Human SPOP shRNA was cloned in pLKO.1 vector. The primer sequences are included in Supplementary Table 1. Lentivirus was produced as reported before.29
Expression plasmids, expression and purification of LIMK2 and SPOP
Haemagglutinin (HA)-tagged LIMK2 was cloned in VIP3 vector at BamHI and Xho1 sites. HA-tagged-SPOP was cloned in VIP3 and TAT-HA vectors at BamHI and Xho1 sites. Various SPOP mutants were created by site-directed mutagenesis and were expressed in Escherichia coli. LIMK2 kinase was cloned, expressed and purified from SF9 insect cells.11 LIMK2 and SPOP retrovirus were generated for infecting PCa cells as described previously.30
In vitro kinase assays
6x-His-LIMK2 LIMK2 was expressed and purified from insect cells using Ni-NTA beads. The kinase was incubated in kinase buffer (50 mM Tris, 10 mM MgCl2) with 100 μM of ATP for 2 h at 30°C to reduce background phosphorylation. Thereafter, the beads were washed with kinase buffer to eliminate extra ATP. For phosphorylation experiments, 2 μg of 6x-His-tagged purified protein (mutant or WT-SPOP) was mixed with LIMK2 and 1 μCi of [γ-32P] ATP for 30 min. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) dye was added, and the samples were boiled for 5 min. Proteins were resolved by SDS-PAGE and exposed for autoradiography.
LIMK2 inhibitor treatment
LIMK2 inhibitor, N-benzyl-N-ethyl-4-(N-phenylsulfamoyl)benzamide, was synthesised as published31 and used at 10 μM concentration.
Immunofluorescence
Immunofluorescence was conducted as before.32 PCa cells were plated on coverslips in a 24-well plate. After 12–16 h, the cells were treated with respective lentivirus (30 h) or LIMK2 inhibitor (10 μM for 12 h). The cells were fixed with 4% formaldehyde, permeabilised using 0.1% Triton X-100 and blocked with phosphate-buffered saline (PBS)/0.1% triton X-100/2% bovine serum albumin solution. The cells were incubated with SPOP or LIMK2 antibodies overnight at 4 °C. Cells were treated with fluorescein isothiocyanate-conjugated secondary antibody for 2 h in dark. Cells was counterstained with 4,6-diamidino-2-phenylindole (Sigma, MO, USA) for 5 min (dilution of 1:50,000). Images were captured using Nikon Eclipse E600 microscope (Nikon Instruments, Melville, NY).
Real-time quantitative PCR (qPCR)
Trizol reagent was used to purify RNA from control and retro/lentivirus-treated cells. RNA was transcribed using the rt-qPCR kit (Promega) according to manufacturer’s instructions. Real-time quantitative reverse transcriptase PCR (rt-qPCR) was performed using 2× SYBR Green master mix (Biorad). The primers used in the study are presented in Supplementary Table 2. Each rt-qPCR experiment was carried out three independent times in triplicate.
Cycloheximide chase assay
CRPC cells were seeded in 6-well plates 12 h prior to infection with the respective retro/lentiviruses. Thirty hours post infection, cycloheximide (10 µM) was added at respective chase times (e.g. 3, 6 h or 2, 4 h) prior to lysis. For LIMK2- CRISPR cells, cells with stable LIMK2 knockout were used. They were seeded at an equal number as the control cells. Cell lysates were probed for the respective protein expressions using western blot analysis.
Ubiquitylation assay
Briefly, PCa cells were co-infected with 6x-His-Ubiquitin and LIMK2 or SPOP retro/lentivirus for 30 h. Thereafter MG132 (10 μM) was added for an additional 12 h to stabilise ubiquitylated proteins. Cells were then harvested and incubated with either specific antibodies or pre-washed Ni-NTA beads for 4 h. Beads were then washed followed by SDS-PAGE and immunoblot analysis. When specific antibodies were used, then ubiquitylated proteins were visualised using 6x-His antibody. For proteins isolated using Ni-NTA beads, substrate-specific antibodies were used for detecting ubiquitylation.
Migration assay
Migration assay was done using Boyden chambers as reported previously.33 For migration assay, 104 cells (harvested using limited trypsin digestion) were suspended into 300 µl of media (without FBS) and added to upper compartment of Boyden Chamber. Fresh culture medium containing 10% FBS was added to the lower compartment. After 4 h, the migrated cells were stained with 0.1% crystal violet and 10 photographs were randomly taken for each sample using a phase-contrast microscope at a magnification of ×200.
MTT (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay
The MTT assay was conducted as before.34 In brief, PCa cells were seeded in a 96-well plate (2000 cells/100 μl/well). After incubation for the indicated time periods (24, 48 and 72 h), 20 µl MTT solution (5 mg/ml) was added to each well. After 4 h at 37 °C, the media was removed and 200 μl dimethyl sulfoxide was added to each well, and absorbance was taken at 570 nm using an enzyme-linked immunosorbent assay plate reader.
Clonogenic assay
Clonogenic assay was conducted as described earlier.35 In all, 10% FBS/RPMI mixed with 0.5% agarose gel was added to 6-well plates and incubated for 1 h. PCa cells (5 × 103) were re-suspended in 10% FBS/RPMI with 0.3% agarose gel media and seeded on the top of base agar. Top agar layer was covered with 500 µl of culture medium containing 10% FBS. The plates were then incubated at 37 °C for 2–3 weeks with fresh media added weekly. Thereafter, colonies were washed with PBS, fixed and stained with crystal violet (0.1%). Colonies were counted using a light phase-contrast microscope.
C4-2 xenografts in nude mice
Male NCRNU-M athymic nude mice (4–6 weeks old) were obtained from Taconic Laboratories. These were kept in Purdue pathogen-free animal facility, with three mice in each cage. Briefly, 1 × 106 cells were resuspended in PBS (100 μl) with 50% matrigel and injected subcutaneously into right and left shoulders. Tumour volume was measured every alternate day, and the mice were euthanised 32 days following injection by CO2 inhalation. Tumour tissues were harvested and flash frozen in liquid nitrogen for further studies.
Statistical analysis
Data are presented as mean +/− SEM of three or more experiments. Statistical analysis was calculated using GraphPad Prism (version 6.07). Statistical significance of difference was determined by the one-way analysis of variance followed by Bonferroni’s post hoc test. P < 0.05 was considered statistically significant.
Results
LIMK2 directly phosphorylates SPOP
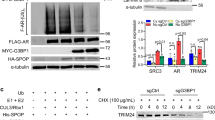
SPOP was identified as a substrate of LIMK2 kinase using our chemical genetic approach. This method uses a modified kinase, which specially uses an orthogonal ATP analogue and transfers the radioactive tag (γ-32P) to its substrates.11,15,36,37,38,39,40,41,42 To confirm the finding, we used 6x-His-SPOP protein and performed a kinase assay with LIMK2. SPOP was directly phosphorylated by LIMK2 in vitro (Fig. 1a).
a LIMK2 directly phosphorylates WT-SPOP protein in vitro. The first lane is LIMK2 alone, the middle lane is SPOP with LIMK2 and the third lane is SPOP alone. The kinase reaction was carried out in 25 µl volume containing 1× kinase buffer and 1 µCi [γ-32P] ATP. b LIMK2 does not interact with SPOP in CRPC cells. C4-2 and 22Rv1 cell lysates were used for immunoprecipitation with IgG antibody or LIMK2 antibody, followed by immunoblotting with SPOP antibody. c SPOP and LIMK2 do not interact with each other. PCa cell lysates were used for immunoprecipitation with IgG or SPOP antibody, followed by immunoblotting with LIMK2 antibody. d Immunofluorescence analysis to detect SPOP localisation in response to LIMK2 inhibitor in C4-2 cells. e Immunofluorescence analysis to establish the location of SPOP in scrambled or LIMK2 shRNA-treated C4-2 cells. f Subcellular localisation of SPOP in scrambled or LIMK2 shRNA-treated C4-2 cells as determined using cellular fractionation. g Immunofluorescence analysis to detect SPOP localisation in response to LIMK2 inhibitor in 22Rv1 cells. h Immunofluorescence analysis to establish the location of SPOP in scrambled or LIMK2 shRNA-treated 22Rv1 cells.
We examined whether LIMK2 and SPOP interact with each other in two CRPC cell lines, C4-2 and 22Rv1. LIMK2 immune complex was isolated and SPOP levels analysed, which showed no SPOP protein in the pull-down immune complex (Fig. 1b). Conversely, pull-down of the SPOP revealed that no LIMK2 was present in the immune complex (Fig. 1c). These results indicate that SPOP and LIMK2 associate transiently in C4-2 and 22Rv1 cells.
Subcellular locations of LIMK2 and SPOP
Since LIMK2 and SPOP proteins did not stably interact with each other in CRPC cells, we next examined their subcellular localisation. SPOP was mainly nuclear with some cytoplasmic distribution (Fig. 1d). LIMK2 inhibition or knockdown using LIMK2 inhibitor or LIMK2-shRNA, respectively, increased SPOP’s cytoplasmic localisation, indicating that LIMK2 promotes SPOP’s nuclear residence (Fig. 1d, e). Interestingly, both LIMK2 inhibition and ablation also increased SPOP’s fluorescence intensity, suggesting that LIMK2 might also impact SPOP’s levels (Fig. 1d, e, raw data acquired using same gain/exposure are shown as Supplementary Fig. 1A, B, respectively). These findings were validated using subcellular fractionation, which too revealed that LIMK2 regulates SPOP’s subcellular localisation (Fig. 1f). In addition, we also investigated SPOP and LIMK2 localisation in 22Rv1 cells, which showed similar residence as in C4-2 cells. Both LIMK2 inhibition and its knockdown impacted SPOP’s cellular residence measurably, confirming that LIMK2 regulates SPOP’s localisation (Fig. 1g, h and Supplementary Fig. 1C, D).
LIMK2 negatively regulates SPOP’s levels and stability
As immunofluorescence data indicated that LIMK2 inhibition/knockdown increases SPOP signal and to determine the consequences of SPOP phosphorylation by LIMK2, we investigated whether LIMK2 regulated SPOP’s expression and stability. Ectopic overexpression of LIMK2 decreased SPOP levels, while LIMK2 knockdown using corresponding shRNA increased SPOP levels in C4-2 cells (Fig. 2a). The changes in SPOP protein levels upon LIMK2 knockdown or overexpression from three independent experiments are depicted in Fig. 2b. Analogous results were observed in 22Rv1 cells (Fig. 2c, d), signifying that LIMK2 negatively regulates SPOP protein levels in CRPC cells.
a LIMK2 knockdown elevates SPOP protein level while LIMK2 overexpression reduces it. b Bar graph shows change in SPOP level with LIMK2 overexpression and knockdown. The data presented are mean ± SEM obtained from three experiments conducted independently. *P < 0.05 and #P < 0.05 compared to C4-2 control cells for LIMK2 and SPOP proteins, respectively. c 22Rv1 cells showing change in SPOP protein level with LIMK2 overexpression and knockdown. d Histogram shows change in SPOP protein level. The data are represented as mean ± SEM obtained from three experiments conducted independently. *P < 0.05 and #P < 0.05 compared to C4-2 control cells for LIMK2 and SPOP proteins, respectively. e, f rt-qPCR analysis depicting mRNA expression levels of SPOP in C4-2 cells infected with LIMK2 and LIMK2 shRNA retro/lentivirus. g, h rt-qPCR was used to quantify SPOP mRNA levels with LIMK2 overexpression and knockdown in 22Rv1 cells. i LIMK2 augments SPOP degradation in PCa cells. C4-2, LIMK2-C4-2 and LIMK2-CRISPR cells were treated with 10 μM cycloheximide for 3 and 6 h, and SPOP level was analysed. j Graphical representation of SPOP degradation rate in cells treated as in i. The results of densitometric scanning are presented graphically with LIMK2 signal normalised to tubulin signal. The data are presented as mean ± SEM acquired from three experiments independently. *P < 0.05 vs 0 h time point of each cell type. k Pattern of LIMK2 degradation in C4-2 and LIMK2-C4-2 cells at the time points where SPOP was chased using cycloheximide. l Quantification of the protein levels (relative to actin) as obtained from k (and two other independent experiments) upon normalisation with respect to the 0 h in each cell type. The data are presented as mean ± SEM acquired from three experiments independently. *P < 0.05, **P < 0.01, ***P < 0.001 vs 0 h time point of each cell type. m LIMK2 increases SPOP degradation by promoting its ubiquitylation. C4-2 cells were co-infected with 6x-His-ubiquitin (6x-His-Ub) along with LIMK2 for 30 h followed by MG132 treatment. Immunoprecipitated SPOP proteins were analysed for ubiquitylation using 6x-His antibody. The experiment was performed three times independently and representative data are included.
To investigate any relationship between LIMK2 and SPOP at the transcriptional stage, we overexpressed LIMK2 in C4-2 and 22Rv1 cells and measured SPOP mRNA levels. As shown in Fig. 2e, while LIMK2 mRNA levels were increased threefold, no change in SPOP mRNA levels was observed. Simultaneously, we knocked down LIMK2 and checked for SPOP mRNA levels. While LIMK2 mRNA level decreased, we did not observe any change in SPOP mRNA levels, suggesting that LIMK2 does not regulate SPOP mRNA expression (Fig. 2f). We obtained similar results in 22Rv1 cells, where neither LIMK2 overexpression nor knockdown had any impact on SPOP mRNA levels, indicating that LIMK2 does not regulate SPOP at this stage (Fig. 2g, h, respectively).
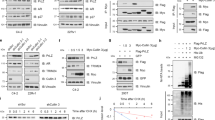
Therefore, SPOP protein stability was analysed using C4-2, LIMK2-knockout cells (generated using CRISPR) and LIMK2-C4-2 cells. These cells were treated with cycloheximide, which showed that LIMK2 knockout stabilised SPOP, while LIMK2 overexpression augmented SPOP degradation (Fig. 2i, j). As a control, LIMK2 levels were analysed in C4-2 and LIMK2-C4-2 cells (Fig. 2k, l). To confirm whether the degradation of SPOP was ubiquitin-dependent, 6x-His-ubiquitin was expressed in C4-2 and LIMK2-C4-2 cells, and SPOP’s stability was examined. LIMK2 overexpression facilitated SPOP ubiquitylation, thereby confirming that LIMK2 degrades SPOP in a ubiquitin-dependent manner (Fig. 2m).
LIMK2 is degraded by SPOP
SPOP acts as an adaptor protein for degrading several of its protein substrates involved in cellular development, physiology and pathology.17 Hence, we checked whether SPOP reciprocally regulates LIMK2 levels in CRPC cells. As indicated, SPOP overexpression indeed decreased LIMK2 levels and its knockdown led to a substantial increase in LIMK2 expression levels in C4-2 cells (Fig. 3a–d). Figure 3b, d depict quantification of LIMK2 levels upon SPOP overexpression and knockdown, respectively, from three separate experiments. Comparable results were observed in 22Rv1 cells, suggesting that SPOP regulates LIMK2 expression levels in CRPC cells (Fig. 3e–h).
a SPOP overexpression decreases LIMK2 levels in C4-2 cells. b Bar graph shows change in LIMK2 level with SPOP overexpression. The data presented as mean ± SEM acquired from three experiments independently. **P < 0.01 vs the level of respective protein in C4-2 control cells. c SPOP ablation increases LIMK2 expression in C4-2 cells. Cells were infected with scrambled shRNA and SPOP-shRNA, and LIMK2 and SPOP levels were analysed. d Bar graph shows change in LIMK2 level with SPOP knockdown. *P < 0.05 and #P < 0.05 compared to C4-2 control cells for SPOP and LIMK2 proteins, respectively. e SPOP overexpression decreases LIMK2 levels in 22Rv1 cells. f Histogram shows change in LIMK2 protein level with SPOP overexpression. The data are represented as mean ± SEM of three experiments. *P < 0.05 and #P < 0.05 compared to C4-2 control cells for SPOP and LIMK2 proteins, respectively. g SPOP depletion increases LIMK2 levels in 22Rv1 cells. h Histogram shows change in LIMK2 protein level with SPOP knockdown. The data are presented as mean ± SEM obtained from three experiments independently. *P < 0.05 and #P < 0.05 compared to C4-2 control cells for SPOP and LIMK2 proteins, respectively. i rt-qPCR analysis representing mRNA levels of LIMK2 in C4-2 cells infected with SPOP retrovirus. j rt-qPCR analysis representing mRNA levels of LIMK2 in C4-2 cells infected with SPOP shRNA. k rt-qPCR was used to quantify LIMK2 mRNA level upon SPOP overexpression in 22Rv1 cells. l qPCR was used to quantify LIMK2 mRNA level upon SPOP knockdown in 22Rv1 cells. m SPOP increases LIMK2 degradation. C4-2, SPOP-C4-2 and SPOP shRNA-C4-2 cells were treated with cycloheximide (10 μM) for 2 and 4 h, and LIMK2 level was analysed. n Graphical depiction of LIMK2 stability. The results of densitometric scanning are presented graphically with LIMK2 signal normalised to tubulin signal. The data are presented as mean ± SEM acquired from three experiments independently. *P < 0.05 vs C4-2 control cells. o Pattern of SPOP degradation in C4-2, SPOP shRNA-C4-2 and SPOP-C4-2 cells measured at the times where LIMK2 was chased using cycloheximide. p Quantification of the protein levels (relative to actin levels) in o (and two other independent experiments) upon normalisation with respect to 0 h of each cell type. The data are presented as mean ± SEM acquired from three experiments independently. *P < 0.05, **P < 0.01 vs 0 h time point of each cell type. q SPOP degrades LIMK2 by enhancing its ubiquitylation. C4-2 cells were co-infected with 6x-His-ubiquitin and SPOP retrovirus for 30 h followed by 12 h MG132 treatment. LIMK2 was isolated and ubiquitylation analysed using 6x-His antibody. Each experiment was performed for a minimum of three times independently and representative data are shown. r Immunofluorescence analysis to establish the location of LIMK2 in scrambled or SPOP shRNA treated C4-2 cells. s Subcellular localisation of LIMK2 in scrambled or SPOP shRNA-treated C4-2 cells as determined by subcellular fractionation. t Immunofluorescence analysis to establish the location of LIMK2 in scrambled or SPOP shRNA-treated 22Rv1cells.
To evaluate whether SPOP-mediated regulation of LIMK2 occurred at the transcriptional or posttranslational stage, SPOP was overexpressed in C4-2 and 22Rv1 cells. While SPOP mRNA levels increased by >2-fold, it did not trigger any change in LIMK2 mRNA abundance, implying that SPOP does not control LIMK2 at the transcriptional stage (Fig. 3i, k). Similarly, SPOP knockdown had no influence on LIMK2 mRNA levels in both C4-2 and 22Rv1 cells (Fig. 3j, l).
We next examined whether SPOP decreases LIMK2 posttranslationally. LIMK2 levels were examined in C4-2, SPOP knocked-down C4-2 and SPOP-overexpressing C4-2 cells exposed to cycloheximide (10 μM) for 2 and 4 h to inhibit protein synthesis. As shown in Fig. 3m, n, overexpression of SPOP significantly augmented the degradation of LIMK2, whereas SPOP downregulation stabilised LIMK2. As a control, SPOP levels were analysed in cycloheximide-treated control C4-2, SPOP-shRNA-C4-2 and SPOP-C4-2 cells as well (Fig. 3o, p). Furthermore, LIMK2 ubiquitylation increased upon SPOP overexpression in C4-2 cells (Fig. 3q). Together, these results suggest that SPOP promotes LIMK2 degradation in CRPC cells.
We also examined whether SPOP regulates LIMK2’s subcellular localisation in C4-2 and 22Rv1 cells (Fig. 3r–t, raw data for Fig. 3r, t acquired using same gain/exposure are shown as Supplementary Fig. 2A, B, respectively). LIMK2 was predominantly localised in cytoplasm with some nuclear localisation. SPOP depletion did not alter LIMK2 localisation within the cells but showed increased LIMK2 fluorescence intensity, confirming the presence of a feedback loop.
LIMK2 phosphorylates SPOP at S59, S171 and S226
Since LIMK2 phosphorylates SPOP, the next step was to determine the phosphorylation sites in SPOP protein. No consensus optimal peptide sequence is known for LIMK2. Following the same strategy as we have previously reported,12 six serine residues that were followed by alanine or glycine were selected as potential phosphorylation sites of LIMK2 on SPOP. These sites were S59, S171, S226, S313, S336 and S358. To further narrow the choices, SPOP was split into three pieces as shown in Fig. 4a. Considering the different SPOP domains, we truncated the protein so that the first piece contained the MATH domain from the first amino acid to lysine 180 (SPOP-1–180). The second piece contained the BTB domain from valine 181 to glutamic acid 300 (SPOP-181–300). The last piece contained the rest of the protein sequence from asparagine 301 to the end (SPOP-301–374). Each of the SPOP fragments was then individually tested for phosphorylation using an in vitro kinase assay with LIMK2. The first (SPOP-1–180) and the second SPOP pieces (SPOP-181–300) showed phosphorylation, while the third fragment (SPOP-301–374) did not (Fig. 4b). Based on the protein fragmentation results shown in (Fig. 4b), we eliminated three potential serine sites, S313, S336 and S358. The remaining three putative sites (S59, S171 and S226) were chosen and corresponding phosphorylation-dead single mutants (S59A, S171A and S226A) were generated using site-directed mutagenesis. All three mutants showed decreased phosphorylation as compared to WT-SPOP (Fig. 4c). Additionally, when all the three sites were mutated (denoted as 3A), the protein was not phosphorylated, confirming that S59, S171 and S226 are the only phosphorylation sites for LIMK2 (Fig. 4d).
a Figure shows protein domains of SPOP. The SPOP protein was fragmented into three pieces: SPOP-1–180 (first fragment), which contains the MATH domain; SPOP-181–300 (second fragment), which contains the BTB domain, and SPOP-301–374 (third fragment), which consisted of the C-terminal region. b LIMK2 phosphorylates the first (SPOP-1–180) and the second fragment (SPOP-181–300) and not the third fragment (SPOP-301–374). c LIMK2 phosphorylates SPOP at S59, S171 and S226 positions. All SPOP proteins were 6x-His-tagged. Kinase reaction was performed for 30 min. The top panel shows autoradiography and the bottom panel shows Coomassie staining. d LIMK2 phosphorylates SPOP at only the aforementioned three sites (S59, S171 and S226), as the corresponding 3A-phospho-resistant mutant did not show any phosphorylation. e Phospho-resistant SPOP is expressed at higher level compared to WT-SPOP in C4-2 cells. C4-2 cells were infected with HA-tagged wild-type SPOP or 3A-SPOP retrovirus for 36 h, and the protein levels of SPOP, LIMK2, HA and actin were analysed using their respective antibodies. f The bar graph shows quantification of WT and mutant SPOP levels obtained from three experiments. Data shown are mean ± SEM of three experiments. *P < 0.05 compared to control cells. g SPOP protein levels in 22Rv1 cells infected with HA-tagged SPOP or 3A-SPOP. h Quantification of SPOP levels obtained from three independent experiments. i LIMK2-mediated phosphorylation of SPOP decreases its stability in C4-2 cells. SPOP levels were analysed in C4-2, SPOP-C4-2 and 3A-SPOP-C4-2 cells treated with cycloheximide for 3 and 6 h. j Graphical depiction of SPOP half-life in C4-2, SPOP-C4-2 and 3A-SPOP-C4-2 cells. k LIMK2-mediated phosphorylation of SPOP decreases its stability in 22Rv1 cells. 22Rv1, SPOP-22Rv1 and 3A-SPOP-22Rv1 cells were treated with cycloheximide for 3 and 6 h, and SPOP levels were analysed. l Graphical depiction of SPOP half-life in 22Rv1, SPOP-22Rv1 and 3A-SPOP-22Rv1 cells. m LIMK2 overexpression augmented the ubiquitylation of WT-SPOP but not of 3A-SPOP in C4-2 cells. WT-SPOP-C4-2 and 3A-SPOP-C4-2 cells were infected with 6x-His-Ubiquitin with or without LIMK2 for 30 h, followed by MG132 treatment for 12 h. SPOP was immunoprecipitated using HA antibody and ubiquitylation analysed. n LIMK2 overexpression augmented the ubiquitylation of WT-SPOP but not of 3A-SPOP in 22Rv1 cells.
LIMK2 decreases SPOP stability via phosphorylation
To determine whether phosphorylation of SPOP by LIMK2 has any effect on SPOP stability, both WT and 3A-SPOP were expressed in C4-2 cells and their protein levels were evaluated. 3A-SPOP was expressed at higher levels compared to WT-SPOP indicating that LIMK2 phosphorylation decreases SPOP stability (Fig. 4e). The increase in 3A-SPOP levels was associated with a concurrent decrease in LIMK2 levels due to the reciprocal loop. The relative protein levels of WT-SPOP and 3A-SPOP obtained from three independent experiments are shown in Fig. 4f. Comparable results were witnessed in 22Rv1 cells, proposing that LIMK2-mediated phospho-regulation of SPOP is a common mechanism in CRPC cells (Fig. 4g, h).
Next, we examined whether phosphorylation affects SPOP protein stability in CRPC cells. We treated C4-2, WT-SPOP and 3A-SPOP-expressing cells with cycloheximide to impede protein synthesis and examined the half-life of SPOP protein. As shown in Fig. 4i, j, phosphorylation decreased SPOP protein stability, as 3A-SPOP was more stable compared to WT-SPOP, followed by endogenous SPOP in C4-2 cells. Comparable results were obtained in 22Rv1 cells, which showed increased stability of 3A-SPOP mutant (Fig. 4k, l). LIMK2 was overexpressed in SPOP-C4-2 and 3A-SPOP-C4-2 cells (or SPOP-22Rv1 and 3A-SPOP-22Rv1 cells) and ubiquitylation of ectopically expressed SPOP was analysed. As shown in Fig. 4m, n, WT-SPOP was highly ubiquitylated as compared to 3A-SPOP, thereby specifying that LIMK2 increases SPOP degradation by phosphorylation.
As our data showed increased cytoplasmic localisation of SPOP upon LIMK2 depletion, we analysed the subcellular localisation of WT-SPOP and 3A-SPOP in both C4-2 and 22Rv1 cells. WT-SPOP showed predominantly nuclear with some cytoplasmic localisation as before in C4-2 and WT-SPOP-C4-2 cells. In contrast, 3A-SPOP showed more cytoplasmic distribution, confirming our earlier finding that LIMK2 contributes to nuclear localisation of SPOP (Supplementary Fig. 3A, B).
SPOP inhibits cell proliferation and invasion of CRPC cells
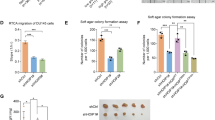
While LIMK2 acts as an oncogene, SPOP’s role in tumorigenesis is context sensitive. Nevertheless, multiple studies have proven that SPOP suppresses tumorigenesis by degrading its oncogenic substrates in PCa.25 Therefore, we evaluated the contribution of LIMK2-SPOP crosstalk in facilitating aggressive phenotypes. Ectopic expression of LIMK2 increased cellular proliferation in C4-2 cells (Fig. 5a). In contrast, overexpression of WT and 3A-SPOP showed decreased cell proliferation as compared to control cells. More importantly, phospho-dead 3A-SPOP maximally inhibited cell proliferation, highlighting the significance of SPOP phosphorylation by LIMK2. Additionally, depletion of LIMK2 further decreased cell proliferation in both SPOP-C4-2 and 3A-SPOP-C4-2 cells. When LIMK2 was overexpressed, a significant increase in cell proliferation of both SPOP-C4-2 and 3A-SPOP-C4-2 was observed (Fig. 5b, c). Comparable results were observed in 22Rv1 cells (Fig. 5d–f).
a WT and 3A-SPOP inhibits cell proliferation in C4-2 cells. C4-2, LIMK2-C4-2, SPOP-C4-2 and 3A-SPOP-C4-2 cells were cultured in 96-well plates for 24, 48 and 72 h followed by MTT assay. b LIMK2 depletion substantially reduces cell growth in SPOP-C4-2 and 3A-SPOP-C4-2 cells. * and # indicate statistically significant difference compared to respective controls; P < 0.05. c LIMK2 overexpression augments cell proliferation in SPOP-C4-2 and 3A-SPOP-C4-2 cells. * and # indicate statistically significant difference compared to respective controls; P < 0.05. d Expression of WT and 3A-SPOP impaired cell proliferation in 22Rv1 cells. 22Rv1, LIMK2-22Rv1, SPOP-22Rv1 and 3A-SPOP-22Rv1 cells were cultured in 96-well plates for 24, 48 and 72 h and subjected to MTT assay. e LIMK2 knockdown decreases cell growth in SPOP-22Rv1 and 3A-SPOP-22Rv1 cells. * and # indicate statistically significant difference compared to respective controls; P < 0.05. f LIMK2 overexpression enhances cell proliferation in SPOP-22Rv1 and 3A-SPOP-22Rv1 cells. * and # indicate statistically significant difference compared to respective controls; P < 0.05. g, h SPOP prevents colony formation in a soft agar assay in C4-2 and 22Rv1 cells. *P < 0.05 compared to vector-expressing control. i Both WT-SPOP and 3A-SPOP inhibit cell migration in C4-2 cells. Representative images of chemotaxis assay. Magnification, ×200. j Chemotaxis assay was performed in C4-2, SPOP-C4-2 and 3A-SPOP-C4-2 cells using Boyden chambers. Histogram shows mean ± SEM of three experiments conducted independently. *P < 0.05 compared to vector control. k, l LIMK2 overexpression increases cell motility in both SPOP-C4-2 and 3A-SPOP-C4-2 cells, although to a higher extent in the former as compared to the latter. m, n LIMK2 depletion inhibits cell motility in SPOP-C4-2 and 3A-SPOP-C4-2 cells with more significant decrease in the latter. o Representative images and p quantitative data of chemotaxis assays measuring migration of 22Rv1, WT-SPOP-22Rv1 and 3A-SPOP-22Rv1 cells. q, r LIMK2 overexpression increases cell motility in both SPOP-22Rv1 and 3A-SPOP-22Rv1 cells. s, t LIMK2 depletion inhibits cell motility in both SPOP-22Rv1 and 3A-SPOP-22Rv1 cells. u 3A-SPOP-expressing cells are more sensitive to enzalutamide (1 μM, treated for 48 h), compared to WT-SPOP-C4-2 cells.
The consequence of SPOP phosphorylation was also observed under clonogenic conditions. SPOP expression significantly decreased colony formation in 22Rv1 and C4-2 cells, compared to control (Fig. 5g, h). 3A-SPOP cells showed more drastic inhibition as compared to WT-SPOP cells, indicating that LIMK2-mediated phosphorylation of SPOP facilitates anchorage-independent growth of CRPC cells.
LIMK2-mediated SPOP phosphorylation promotes cell migration and enzalutamide resistance
We evaluated a potential role of SPOP in cellular migration using a transwell assay. As expected, SPOP overexpression resulted in impaired cell motility (Fig. 5i, j). As before, 3A-SPOP-overexpressing cells showed more drastic inhibition as compared to WT-SPOP cells. Furthermore, LIMK2 knockdown diminished and its overexpression augmented cell migration in both SPOP-C4-2 and 3A-C4-2 cells (Fig. 5k–n), but the effect was more significant in 3A-cells. Comparable results were obtained in 22Rv1 cells (Fig. 5o–t). These results corroborate that LIMK2-mediated phosphorylation of SPOP stimulates cell motility in CRPC cells.
We also investigated whether SPOP phosphorylation by LIMK2 affects enzalutamide resistance. Phospho-resistant SPOP-C4-2 cells were most resistant, whereas WT-SPOP conferred some resistance, compared to parental cells, indicating that SPOP degradation by LIMK2 is one of the mechanisms leading to enzalutamide resistance (Fig. 5u).
LIMK2-mediated phosphorylation of SPOP regulates c-Myc levels
Several oncogenic targets of SPOP are reported in PCa. We focussed on two SPOP substrates, c-Myc and AR, as both are intimately involved with CRPC progression.18,43 Furthermore, c-Myc levels positively correlate with AR in CRPC.44 LIMK2 overexpression increased the levels of both c-Myc and AR (Fig. 6a and Supplementary Fig. 4A). We next analysed c-Myc levels in C4-2, WT and 3A-SPOP-expressing cells, which revealed negative correlation with SPOP levels, indicating that SPOP degradation by phosphorylation is one of the mechanisms by which LIMK2 promotes c-Myc expression (Fig. 6b, c).
a LIMK2 augments c-Myc, ARv7 and AR protein levels in C4-2 cells. b WT-SPOP and 3A-SPOP decrease c-Myc levels. c Bar graph shows change in c-Myc protein level. The data depicted are mean ± SEM of three independent experiments. *P < 0.05 compared to C4-2 control cells. d 3A-SPOP promotes higher ubiquitination and degradation of c-Myc protein as compared to WT-SPOP. e Both WT-SPOP and 3A-SPOP decrease AR protein levels in C4-2 cells. f Histogram showing change in AR protein level. The data depicted are mean ± SEM of three independent experiments. *P < 0.05 compared to C4-2 control cells. g Ectopic expression of WT and 3A-SPOP promote robust ubiquitination and degradation of AR protein, as compared to control C4-2 cells. h Ectopic expression of 3A-SPOP decreases AR and ARv7 protein levels more significantly as compared to WT-SPOP in 22Rv1 cells. i Bar graph shows change in AR and ARv7 protein levels in 22Rv1, SPOP-22Rv1 and 3A-SPOP-22Rv1 cells. The data depicted are mean ± SEM of three experiments. *P < 0.05 compared to 22Rv1 control cells. j 3A-SPOP promotes more robust ubiquitination of AR in 22Rv1 cells as compared to WT-SPOP. k 3A-SPOP promotes more robust ubiquitination of ARv7 in 22Rv1 cells as compared to WT-SPOP. l AR binds SPOP in vector, SPOP and 3A-SPOP-expressing C4-2 cells. SPOP immune complexes were isolated from vector, SPOP and 3A-SPOP-C4-2 cells, followed by AR IB (left panel). Right panel depicts IgG IP in the same cell lines, followed by AR IB. Bottom panel depicts SPOP levels in the input. m WT and 3A-SPOP bind AR with equal efficiency. SPOP and AR association was examined using recombinant SPOP and AR in an in vitro pull-down assay. AR on beads was incubated with 6×-His-SPOP (WT and 3A) and binding was analysed. Right panel shows Coomassie staining of the input. n WT-SPOP inhibits tumour growth in vivo. Tumour growth curve of nude mice inoculated with C4-2 and SPOP-C4-2 cells on right and left shoulders, respectively. The mean value ± SEM were from three animals in each group. o Representative images of tumour-bearing nude mice. Pictures were taken 32 days following injection. p Tumour growth curve of C4-2 and 3A-SPOP-C4-2 cells on subcutaneous tumour growth in athymic nude mice. q Athymic nude mouse injected with C4-2 and 3A-SPOP-C4-2 cells on right and left shoulders, respectively. The images were taken 32 days after inoculation. A representative image is presented. r Expression of EMT markers in tumour tissues obtained from C4-2- and SPOP-C4-2 cell-injected mice. s Proposed model showing the consequences of LIMK2-SPOP signalling in CRPC pathogenesis.
As SPOP degrades c-Myc by ubiquitylation,43 we also investigated the ubiquitylation of c-Myc by overexpressing either WT or 3A-SPOP. Vector-transfected cells were used as a control. While ectopic expression of WT-SPOP significantly increased the ubiquitylation of c-Myc (compared to control cells), 3A-SPOP triggered a higher degree of ubiquitylation, indicating that LIMK2-mediated phosphorylation and subsequent degradation of SPOP is a critical step in promoting CRPC pathogenesis (Fig. 6d).
AR remains one of the most critical targets of SPOP in CRPC.45 Both WT and 3A-SPOP were expressed in C4-2 cells, which resulted in a concomitant decrease in AR levels (Fig. 6e, f). Although 3A-SPOP was expressed at higher levels compared to WT, the decrease in AR levels was almost the same in each case. Similarly, when we analysed potential ubiquitylation of AR in WT and 3A-SPOP-C4-2 cells, they showed similar level of ubiquitylation (Fig. 6g). As C4-2 cells express very low levels of AR, we postulated that even a small increase in SPOP level is sufficient to completely degrade AR in these cells (Fig. 6e).
To test this hypothesis, we used 22Rv1 cells, which express much higher levels of AR. When WT and 3A-SPOP were ectopically expressed in 22Rv1 cells, the decrease in AR level was significantly higher in 3A-expressing cells than in WT (Fig. 6h, i). Similarly, we also observed increased ubiquitylation of AR in 3A-SPOP cells than in WT-expressing cells, thereby validating that LIMK2-mediated phosphorylation and degradation of SPOP is an important contributor to disease progression (Fig. 6j). Interestingly, ectopic expression of both WT and 3A-SPOP decreased ARv7 levels significantly as well, indicating that LIMK2 also regulates ARv7 levels in 22Rv1 cells (Fig. 6h, i). Overexpression of WT and 3A-SPOP in 22Rv1 cells further confirmed that SPOP ubiquitylates ARv7 (Fig. 6k). As observed before, 3A-SPOP triggered enhanced ubiquitylation of ARv7 as compared to WT-SPOP, uncovering a novel mechanism by which LIMK2 upregulates both AR and ARv7. Although ARv7 lacks the SPOP-binding consensus motif present in AR, it was shown to be degraded by SPOP in the presence of full-length AR, possibly via the formation of AR/ARv7 heterodimers in 22Rv1 cells.43
As all known SPOP mutations in PCa affect its binding with substrates, we analysed whether WT and 3A mutant bind differentially to AR. Initially, SPOP was isolated from control, WT and 3A-expressing cells using limiting amount of SPOP antibody (to account for the differences in SPOP expression), which revealed equal levels of AR in all three cell types, indicating that both WT and the mutant bind AR with equal affinity (Fig. 6l). We also confirmed this finding using recombinant WT and 3A-SPOP proteins in a pull-down assay, which too revealed similar binding of AR to both SPOP alleles (Fig. 6m). Together, these results depict that LIMK2-mediated SPOP degradation is a key mechanism that regulates AR stability.
LIMK2-mediated SPOP phosphorylation is crucial for tumorigenesis in vivo
We next evaluated whether LIMK2 regulates SPOP in vivo via phosphorylation. C4-2 and SPOP-C4-2 cells were injected subcutaneously in three nude mice, which resulted in robust tumour formation with C4-2 cells, compared to tumour formed with SPOP-C4-2 cells, confirming the tumour-suppressor role of SPOP in CRPC (Fig. 6n, o and Supplementary Fig. 4B). In another set of experiments, we injected three nude mice with 3A-SPOP-C4-2 and C4-2 cells on left and right shoulders, respectively. As seen in Fig. 6p, q, C4-2 cells formed bigger tumours while the 3A-SPOP-C4-2 cells exhibited no tumour formation. These results indicate that phospho-dead SPOP, which is resistant to LIMK2-mediated degradation possesses higher tumour suppressing potential as compared to WT-SPOP.
LIMK2-mediated SPOP degradation promotes EMT in vivo
Our previous studies have shown that LIMK2 promotes epithelial-to-mesenchymal transition (EMT) in vivo.12 On the other hand, SPOP is known to both promote and inhibit EMT.46 Therefore, we investigated the consequences of SPOP upregulation on various EMT markers in vivo using C4-2 and SPOP-C4-2 xenografts. Furthermore, as our data showed that SPOP downregulates LIMK2 in cells, this regulation was also analysed in vivo. As 3A-SPOP did not form any tumours, it could not be included. We also incorporated E-cadherin, an epithelial marker, as increase in EMT-inducing proteins is accompanied by a concomitant decrease in E-cadherin levels. SPOP expression indeed increased E-cadherin levels with simultaneous decrease in all EMT markers that were analysed, indicating that SPOP downregulates EMT process in CRPC pathogenesis in vivo (Fig. 6r and Supplementary Fig. 4C). Equally importantly, SPOP-expressing xenografts showed reduced levels of LIMK2 confirming the negative regulation of LIMK2 by SPOP in vivo.
Discussion
The most critical problem in patients with PCa is the emergence of CRPC. Although AR signalling inhibitor (ASI)-based therapies (e.g. abiraterone and enzalutamide) are effective initially, resistance develops over time and the disease is no longer curable.3,4 Thus, there is an urgent need for a mechanism-based study to discover novel targets and strategies to treat CRPC patients. We recently identified LIMK2 as a potential drug target in CRPC.12 However, the molecular mechanism of LIMK2-mediated oncogenesis remains unclear as only four direct targets of LIMK2 are known to date.
In this study, we identified SPOP as a LIMK2 substrate. SPOP comprises of two conserved domains, an N-terminal meprin and TRAF homology (MATH) domain (28–166), followed by C-terminal bric-a-brac, tramtrack and broad complex (BTB)/POZ domain (190–297). While the MATH domain binds and recruits substrates, the BTB domain binds the Cullin-3 family of E3 ligases and promotes the ubiquitylation of SPOP-bound substrates.47 Thus SPOP functions as an adaptor protein, which is crucial for the degradation of several proteins involved in normal cellular physiology.
SPOP is the most frequently mutated gene in PCa (~15%). Furthermore, SPOP mutations are rare in other cancers, thereby highlighting a unique mechanism for specifically suppressing SPOP’s antitumorigenic functions in PCa. Most mutations are present on the surface of substrate-binding pocket of MATH domain, which compromises its ability to bind downstream targets thereby preventing the ubiquitylation of several oncogenic targets in PCa. Not surprisingly, SPOP mutants promote enhanced proliferation in vitro and lead to prostate tumorigenesis in vivo.48 Furthermore, numerous downstream substrates of SPOP have been identified in PCa,18,19,20,21,22,23,24,25 uncovering multiple mechanisms by which SPOP inhibits prostate tumorigenesis. As a result, SPOP’s role in PCa has been largely defined by either its mutant alleles or by its downstream targets. However, regulation of WT-SPOP in any cancer including PCa has not yet been reported. This is particularly important as WT-SPOP is retained in the remaining 85% of PCa cases that escape SPOP mutations. As SPOP potently antagonises tumorigenic pathways in PCa, including AR and ARv7 degradation, it points to the existence of other critical regulatory mechanisms that trigger SPOP downregulation during PCa progression.
This study uncovered the first mechanism of WT-SPOP regulation by phosphorylation. LIMK2 directly phosphorylates SPOP at three sites, S59, S171 and S226 (Fig. 4c, d). While S59 is within the MATH domain, S226 falls within the BTB domain. Although many mutations are known in the MATH domain, S59 is not among them. Similarly, S171 and S226 are not known to be mutated in SPOP either. This finding suggests that SPOP mutants may also be subjected to LIMK2-mediated ubiquitination in PCa, further exacerbating the disease. Furthermore, crystal structure studies have revealed that A227 and M228 (following S226) are involved in Cul3 binding, although not as intimately as H231 and M233 residues,49 indicating that S226 phosphorylation by LIMK2 may impact Cul3 binding as well. This in turn should suppress degradation of key oncogenic targets including SRC-3, a potent regulator of AR, promoting PCa.
This study also uncovered that SPOP targets LIMK2 for ubiquitination and proteasomal degradation in a reciprocal loop, thereby suppressing LIMK2-induced cancer-related phenotypes. The phospho-resistant SPOP is more stable and fully inhibits tumorigenesis in vivo. Our previous study revealed a critical role of LIMK2 knockdown in fully reversing CRPC tumorigenesis.12 As AR and ARv7 are key drivers in CRPC, this study uncovered a critical mechanism by which LIMK2 regulates the stability of AR and ARv7 (Fig. 6s). In conclusion, our study shows that SPOP overexpression inhibits cell growth, migratory, invasion ability and EMT in CRPC cells by inhibiting LIMK2 expression. Thus, targeting LIMK2 provides a potent approach to retain functionally active WT-SPOP in cells, which should inhibit not only PCa progression in patients but may also sensitise CRPC to ASIs, such as enzalutamide.
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020).
Coutinho, I., Day, T. K., Tilley, W. D. & Selth, L. A. Androgen receptor signaling in castration-resistant prostate cancer: a lesson in persistence. Endocr. Relat. Cancer 23, T179–T197 (2016).
Watson, P. A., Arora, V. K. & Sawyers, C. L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 15, 701–711 (2015).
Saad, F., Shore, N., Zhang, T., Sharma, S., Cho, H. K. & Jacobs, I. A. Emerging therapeutic targets for patients with advanced prostate cancer. Cancer Treat. Rev. 76, 1–9 (2019).
Scott, R. W. & Olson, M. F. LIM kinases: function, regulation and association with human disease. J. Mol. Med. 85, 555–568 (2007).
Sumi, T., Matsumoto, K., Takai, Y. & Nakamura, T. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by rho-and Cdc42-activated LIM-kinase 2. J. Cell Biol. 147, 1519–1532 (1999).
Prudent, R., Vassal-Stermann, E., Nguyen, C. H., Pillet, C., Martinez, A., Prunier, C. et al. Pharmacological inhibition of LIM kinase stabilizes microtubules and inhibits neoplastic growth. Cancer Res. 72, 4429–4439 (2012).
Zhang, Y., Li, A., Shi, J., Fang, Y., Gu, C., Cai, J. et al. Imbalanced LIMK1 and LIMK2 expression leads to human colorectal cancer progression and metastasis via promoting β-catenin nuclear translocation. Cell Death Dis. 9, 1–0 (2018).
Sun, P., Ren, L., Chen, Q. Q., Tan, X. N., Zhang, T., Li, Y. L. et al. Elevated expression of LIMK2 is an independent prognostic indicator in breast cancer. Int. J. Clin. Exp. Med. 11, 5929–5936 (2018).
Vlecken, D. H. & Bagowski, C. P. LIMK1 and LIMK2 are important for metastatic behavior and tumor cell-induced angiogenesis of pancreatic cancer cells. Zebrafish 6, 433–439 (2009).
Johnson, E. O., Chang, K. H., Ghosh, S., Venkatesh, C., Giger, K., Low, P. S. et al. LIMK2 is a crucial regulator and effector of Aurora-A-kinase-mediated malignancy. J. Cell Sci. 125, 1204–1216 (2012).
Nikhil, K., Chang, L., Viccaro, K., Jacobsen, M., McGuire, C., Satapathy, S. R. et al. Identification of LIMK2 as a therapeutic target in castration resistant prostate cancer. Cancer Lett. 448, 182–196 (2019).
Lagoutte, E., Villeneuve, C., Lafanechère, L., Wells, C. M., Jones, G. E., Chavrier, P. et al. LIMK regulates tumor-cell invasion and matrix degradation through tyrosine phosphorylation of MT1-MMP. Sci. Rep. 6, 1–2 (2016).
Nikhil, K., Kamra, M., Raza, A. & Shah, K. Negative cross talk between LIMK2 and PTEN promotes castration resistant prostate cancer pathogenesis in cells and in vivo. Cancer Lett. 12, S0304–S3835 (2020).
Johnson, E. O., Chang, K. H., Pablo, Y., de., Ghosh, S., Mehta, R., Badve, S. et al. PHLDA1 is a crucial negative regulator and effector of Aurora A kinase in breast cancer. J. Cell Sci. 124, 2711–2722 (2011).
Clark, A. & Burleson, M. SPOP and cancer: a systematic review. Am. J. Cancer Res. 10, 704–726 (2020).
Song, Y., Xu, Y., Pan, C., Yan, L., Wang, Z. W. & Zhu, X. The emerging role of SPOP protein in tumorigenesis and cancer therapy. Mol. Cancer 19, 2 (2020).
An, J., Wang, C., Deng, Y., Yu, L. & Huang, H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 6, 657–669 (2014).
Li, C., Ao, J., Fu, J., Lee, D. F., Xu, J., Lonard, D. et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene 30, 4350–4364 (2011).
Hernández-Muñoz, I., Lund, A. H., Van Der Stoop, P., Boutsma, E., Muijrers, I., Verhoeven, E. et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl Acad. Sci. USA 102, 7635–7640 (2005).
Zhang, Q., Shi, Q., Chen, Y., Yue, T., Li, S., Wang, B. et al. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc. Natl Acad. Sci. USA 106, 21191–21196 (2009).
Gan, W., Dai, X., Lunardi, A., Li, Z., Inuzuka, H., Liu, P. et al. Asara, J.M. SPOP promotes ubiquitination and degradation of the ERG oncoprotein to suppress prostate cancer progression. Mol. Cell 59, 917–930 (2015).
Zhu, H., Ren, S., Bitler, B. G., Aird, K. M., Tu, Z., Skordalakes, E. et al. SPOP E3 ubiquitin ligase adaptor promotes cellular senescence by degrading the SENP7 deSUMOylase. Cell Rep. 13, 1183–1193 (2015).
Theurillat, J. P., Udeshi, N. D., Errington, W. J., Svinkina, T., Baca, S. C., Pop, M. et al. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science 346, 85–89 (2014).
Barbieri, C. E., Baca, S. C., Lawrence, M. S., Demichelis, F., Blattner, M., Theurillat, J. P. et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44, 685–689 (2012).
Wei, X., Fried, J., Li, Y., Hu, L., Gao, M., Zhang, S. et al. Functional roles of speckle-type Poz (SPOP) protein in genomic stability. J. Cancer 9, 3257 (2018).
Garcia-Flores, M., Casanova-Salas, I., Rubio-Briones, J., Calatrava, A., Dominguez-Escrig, J., Rubio, L. et al. Clinico-pathological significance of the molecular alterations of the SPOP gene in prostate cancer. Eur. J. Cancer 50, 2994–3002 (2014).
Hernandez-Llodra, S., Segales, L., Safont, A., Juanpere, N., Lorenzo, M., Fumado, L. et al. SPOP and FOXA1 mutations are associated with PSA recurrence in ERG wt tumors, and SPOP downregulation with ERG-rearranged prostate cancer. Prostate 79, 1156–1165 (2019).
Nikhil, K., Raza, A., Haymour, H. S., Flueckiger, B. V., Chu, J. & Shah, K. Aurora kinase A-YBX1 synergy fuels aggressive oncogenic phenotypes and chemoresistance in castration-resistant prostate cancer. Cancers 12, 660 (2020).
Chang, K. H., Multani, P. S., Sun, K. H., Vincent, F., de Pablo, Y., Ghosh, S. et al. Nuclear envelope dispersion triggered by deregulated Cdk5 precedes neuronal death. Mol. Biol. Cell 22, 1452–1462 (2011).
Goodwin, N. C., Cianchetta, G., Burgoon, H. A., Healy, J., Mabon, R., Strobel, E. D. et al. Discovery of a type III inhibitor of LIM kinase 2 that binds in a DFG-out conformation. ACS Med. Chem. Lett. 6, 53–57 (2014).
Sun, K. H., Lee, H. G., Smith, M. A. & Shah, K. Direct and indirect roles of cyclin-dependent kinase 5 as an upstream regulator in the c-Jun NH2-terminal kinase cascade: relevance to neurotoxic insults in Alzheimer’s disease. Mol. Biol. Cell 20, 4611–4619 (2009).
Shah, K. & Vincent, F. Divergent roles of c-Src in controlling platelet-derived growth factor-dependent signaling in fibroblasts. Mol. Biol. Cell 16, 5418–5432 (2005).
Wang, J., Nikhil, K., Viccaro, K., Lei, C., White, J. & Shah, K. Phosphorylation-dependent regulation of ALDH1A1 by Aurora kinase A: insights on their synergistic relationship in pancreatic cancer. BMC Biol. 15, 1–10 (2017).
Wang, J., Nikhil, K., Viccaro, K., Lei, C., Jacobsen, M., Sandusky, G. et al. Aurora A-Twist1 axis promotes highly aggressive phenotypes in pancreatic carcinoma. J. Cell Sci. 130, 1078–1093 (2017).
Shah, K. & Shokat, K. M. A chemical genetic screen for direct v-Src substrates reveals ordered assembly of a retrograde signaling pathway. Chem. Biol. 9, 35–47 (2002).
Kim, S. & Shah, K. Dissecting yeast Hog1 MAP kinase pathway using a chemical genetic approach. FEBS Lett. 581, 1209–1216 (2007).
Sun, K. H., de Pablo, Y., Vincent, F., Johnson, E. O., Chavers, A. K. & Shah, K. Novel genetic tools reveal Cdk5’s major role in golgi fragmentation in Alzheimer’s disease. Mol. Biol. Cell 19, 3052–3069 (2008).
Sun, K. H., de Pablo, Y., Vincent, F. & Shah, K. Deregulated Cdk5 promotes oxidative stress and mitochondrial dysfunction. J. Neurochem. 107, 265–278 (2008).
Chang, K. H., Vincent, F. & Shah, K. Deregulated Cdk5 triggers aberrant activation of cell cycle kinases and phosphatases inducing neuronal death. J. Cell Sci. 125, 5124–5137 (2012).
Shi, C., Viccaro, K., Lee, H. G. & Shah, K. Cdk5-FOXO3a axis: initially neuroprotective, eventually neurodegenerative in Alzheimer’s disease models. J. Cell Sci. 129, 1815–1830 (2016).
Nikhil, K., Viccaro, K. & Shah, K. Multifaceted regulation of ALDH1A1 by Cdk5 in Alzheimer’s disease. Mol. Neurobiol. 56, 1366–1390 (2018).
Geng, C., Kaochar, S., Li, M., Rajapakshe, K., Fiskus, W., Dong, J. et al. SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene 36, 4767–4777 (2017).
Bai, S., Cao, S., Jin, L., Kobelski, M., Schouest, B., Wang, X. et al. A positive role of c-Myc in regulating androgen receptor and its splice variants in prostate cancer. Oncogene 38, 4977–4989 (2019).
Geng, C., Rajapakshe, K., Shah, S. S., Shou, J., Eedunuri, V. K., Foley, C. et al. Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 74, 5631–5643 (2014).
Wang, Z., Song, Y., Ye, M., Dai, X., Zhu, X. & Wei, W. The diverse roles of SPOP in prostate cancer and kidney cancer. Nat. Rev. Urol. 17, 339–350 (2020).
Zhuang, M., Calabrese, M. F., Liu, J., Waddell, M. B., Nourse, A., Hammel, M. et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell 36, 39–50 (2009).
Blattner, M., Liu, D., Robinson, B. D., Huang, D., Poliakov, A., Gao, D. et al. SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell 31, 436–451 (2017).
Errington, W. J., Khan, M. Q., Bueler, S. A., Rubinstein, J. L., Chakrabartty, A. & Privé, G. G. Adaptor protein self-assembly drives the control of a cullin-RING ubiquitin ligase. Structure 20, 1141–1153 (2012).
Moffat, J., Grueneberg, D. A., Yang, X., Kim, S. Y., Kloepfer, A. M., Hinkle, G. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006).
Acknowledgements
pLKO.1 TRC vector was a gift from David Root (Addgene plasmid # 10878).50
Author information
Authors and Affiliations
Contributions
K.N. performed most of the experiments, followed by M.K. H.S.H. generated SPOP mutants and performed kinase assays. K.S. conceived the idea and supervised the project. K.N. and K.S. wrote the manuscript with input from H.S.H. and M.K.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All experiments were performed in accordance with protocol (protocol # 1111000292; approval date 01/07/2019) approved by the Purdue Animal Care and Use Committee (PACUC) of Purdue University. Male NCRNU-M athymic nude mice (4–6 weeks old) were obtained from Taconic Laboratories. These were kept in Purdue pathogen-free animal facility, with three mice in each cage.
Consent to publish
Not applicable.
Data availability
All data generated or analysed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Funding information
This work was supported by NSF award # 1708823, NIH award 1R01-CA237660 and DOD U.S. Army Medical Research Acquisition Activity, Prostate Cancer Research Program (Award # PC130391).
Additional information
Note This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0).
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nikhil, K., Haymour, H.S., Kamra, M. et al. Phosphorylation-dependent regulation of SPOP by LIMK2 promotes castration-resistant prostate cancer. Br J Cancer 124, 995–1008 (2021). https://doi.org/10.1038/s41416-020-01197-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-020-01197-6
This article is cited by
-
O-GlcNAcylation of SPOP promotes carcinogenesis in hepatocellular carcinoma
Oncogene (2023)
-
Reciprocal deregulation of NKX3.1 and AURKA axis in castration-resistant prostate cancer and NEPC models
Journal of Biomedical Science (2021)