Abstract
Background
Prostate cancer (PCa) is among the most commonly diagnosed malignancies in men. Although 5-year survival in patients with localised disease reaches nearly 100%, metastatic disease still remains incurable. Therefore, there is a need for markers indicating metastatic dissemination.
Methods
EGFR overexpression (EGFRover) was tracked in 1039 primary tumours, circulating tumour cells from 39 d’Amico high-risk patients and metastatic samples from 21 castration-resistant PCa cases. EGFR status was compared to clinical parameters and multiple molecular factors were assessed using immunohistochemistry and gene ontology analysis. The functional aspect of EGFR was evaluated by plating PC-3 cells on soft and rigid matrices.
Results
EGFRover was found in 14% of primary tumours, where it was associated with shorter metastasis-free survival and was an independent indicator of worse overall survival. EGFRover correlated with a pro-migratory and pro-metastatic phenotype of tumour cells as well as rich collagen fibre content. All circulating tumour cells (detected in 13% of cases) were positive for EGFR, independent of their EMT-related phenotype. EGFRover was more prevalent in castration-resistant bone metastases (29% of patients) and supported growth of human PCa cells on rigid matrices mimicking bone stiffness.
Conclusions
EGFRover is a stable, EMT-independent marker of PCa disseminating to rigid organs, preferentially bones.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) is the second most frequent malignancy in men worldwide.1 Although 5-year survival in patients with localised PCa is nearly 100%, metastatic disease still remains incurable.2,3 Therefore, there is an urgent need for markers that could help to detect initial stages of tumour dissemination, probability of recurrence and predict preferred sites of metastasis in order to personalise patients’ treatment.
Epithelial–mesenchymal transition (EMT) and plasticity are involved in metastatic progression of PCa.4,5,6 In addition, significant roles for the epidermal growth factor receptor (EGFR) have been suggested in prostate tumorigenesis and progression.7,8 EGFR expression was previously shown to be associated with high grade, advanced stage and high risk for prostate-specific antigen (PSA) recurrence9 and bone metastases.10 In addition, EGFR was also shown to control bone development.11 Indeed, as one of the regulators of EMT, (de)differentiation, proliferation and angiogenesis, EGFR might initiate and/or promote tumour dissemination and metastasis and thus may be considered as a surrogate marker of high metastatic potential.10 However, there is a lack of a complex study evaluating EGFR expression in PCa in the context of tumour characteristics and at various stages of PCa dissemination.
Thus, in the current study, the expression of EGFR protein was assessed in the dissemination cascade—throughout the disease process from primary tumours to disseminated circulating tumour cells (CTCs) and metastatic samples obtained from castrate-resistant PCa (CRPC) patients at the time of death. It was also compared to clinical parameters and multiple molecular factors (including EMT-related proteins, collagen fibre content, vascular and lymphatic vessels numbers) to evaluate its feasibility as a stable marker in the PCa dissemination process.
Methods
PCa patients of cohort I to study primary tumours
One-thousand two hundred PCa patients were included in this study (Supplementary Table 1) based on their signed informed consent, after the approval of the local Ethics Committee (i.e. Ethik Kommission der Aerztekammer Westfalen-Lippe und der Medizinischen Fakultaet der Westfaelischen Wilhelms-Universitaet Muenster, Germany, no. 2007–467–f–S). The patients underwent radical prostatectomy at the Department of Urology in the Prostate Centre University Clinic Münster (Germany) between 1993 and 2004. The variable clinico-pathological and molecular parameters were documented as described.12,13 Time to biochemical recurrence was defined as time between prostatectomy and the time point of first serum PSA increase >0.1 ng/mL followed by another value >0.1 ng/mL after surgery. Metastasis-free survival was defined as the time between prostatectomy and occurrence of clinically defined metastases. Overall survival was defined as the time between the prostatectomy and patient death. Last follow-up was completed in June 2019. The median follow-up was 76 months (range 0.1–273 months).
PCa patients of cohort II to study CTCs
Fifty-nine d’Amico high-risk PCa patients treated in the Martini-Clinic at the University Medical Center Hamburg-Eppendorf (Hamburg, Germany) between 2012 and 2013 were enrolled in this study after informed consent based on the approval of the local ethical review board number PV3779 as presented in Supplementary Table 2. Blood samples (at mean volume of 7.5 mL, range 5–12 mL; first 2 mL of collected blood discarded to avoid contamination by skin cells) were collected into EDTA tubes. Last follow-up was completed in September 2015. The median observation time was 13 months (range 1–25 months).
PCa patients of cohort III to study metastases
Visceral and bone metastases were obtained from 21 PCa patients who died of metastatic CRPC and who signed written informed consent for a rapid autopsy performed within 6 h of death, under the aegis of the Prostate Cancer Donor Program at the University of Washington and approved by the Institutional Review Board of the University of Washington (Supplementary Table 3).
Tissue microarrays (TMAs)
TMAs with primary or metastatic PCa samples were prepared as previously described.13,14
The Cancer Genome Atlas (TCGA) prostate adenocarcinoma (PRAD) data set
RNA-seq data (RNASeqV2, RSEM normalised) covering normalised counts of sequences aligning to 20,531 genes were obtained for 497 PRAD patients from TCGA portal (data status of 28 January 2016). The methods of biospecimen procurement, RNA isolation and RNA sequencing were previously described by TCGA Research Network.15
Isolation of CTCs
Peripheral blood was processed within 24 h of collection. Peripheral blood mononuclear cell (PBMC) fraction, preferably containing CTCs, was enriched using Ficoll density gradient centrifugation, resuspended in 5 mL of 1× phosphate-buffered saline (PBS) and centrifuged to prepare microscopic slides, each containing 500,000 cells. The slides were left overnight to air-dry at room temperature and used within 24 h for further CTC analysis.
Immunohistochemical (IHC) detection and evaluation of EGFR
To detect EGFR, TMA sections were deparaffinised and treated with Proteinase K Ready-to-Use (Dako) for 6 min and Perioxidase-Blocking Solution (Dako) for 5 min. TMAs were incubated overnight at 4 °C with mouse monoclonal anti-EGFR in vitro diagnostic antibody (E30, Dako) diluted 1:20, envisioned by EnVision Kit, Rabbit/Mouse (Dako) and counterstained with haematoxylin (Merck, Germany). The intensity (negative, weak, moderate or strong), subcellular localisation of the staining (membranous, cytoplasmic, nuclear) and the percentage of positive tumour cells were documented. Two tumour samples (TMA tissue cores) from each patient were assessed individually by two independent observers, experienced in IHC analysis. The EGFR intensity (negative, weak, moderate, strong) was evaluated according to the analogical recommendations for HER2 testing in breast cancer proposed by American Society of Clinical Oncology.16 To evaluate overall score corresponding to one patient, maximal intensity of EGFR was chosen from two analysed tumour samples. If one tissue core was uninformative, the overall score corresponded to the remaining one.
IHC detection and evaluation of other proteins
IHC for vimentin, epithelial cell adhesion molecule (EpCAM) and keratins K8/18 and K19 was performed, evaluated and categorised as negative vs. positive staining as described.13,14 The number of vascular or lymphatic vessels was examined in each tumour sample as the number of vessels with visible lumen, positive for CD34 and podoplanin staining, respectively.12
EGFR/EpCAM/pan-keratin/CD45 immunocytochemical staining on CTCs
Immunocytochemical staining identifying CTCs was performed for each patient on 3 slides containing 500,000 PBMCs each. The slides were fixed for 10 min with the Fixation Solution B for Epithelial Cell Detection Kit (Micromet AG; 135 μL diluted in 10 mL of 1× PBS), incubated for 5 min with Peroxidase-Blocking Solution (Dako) and subsequently for 20 min with AB blocking serum (Bio-Rad Medical Diagnostics) diluted 1:10 in 1× PBS. CD45 was detected by incubation with a mouse antibody (NCL-LCA-RP, Novocastra, diluted 1:100, 45 min) and secondary rabbit polyclonal anti-mouse antibody labelled with horseradish peroxidase (Dako, diluted 1:100, 30 min) followed by addition of DAB substrate (Dako, diluted 1:50, 10 min). EGFR was detected by incubation with rabbit polyclonal antibody (sc-03, SantaCruz, diluted 1:100, overnight at +4 °C) and secondary anti-rabbit antibody labelled with Alexa 555 (Life Technologies, diluted 1:200, 45 min). EpCAM was detected by incubation with mouse antibody (NCL-ESA, Novocastra, diluted 1:100, 45 min) and secondary anti-mouse Alexa 350–conjugated antibody (Life Technologies, diluted 1:200, 45 min). Subsequently, cells were incubated for 45 min with anti-pan-keratin antibody AE1/AE3 (eBioscience, diluted 1:700) and C11 (Cell Signalling Technology, diluted 1:300) both directly labelled with Alexa 488. Nuclei of the cells were counterstained with Red-Dot (Biotum, diluted 1:200, 30 min) and covered with coverslips with one drop of Moviol (Sigma Aldrich). Three slides per patient were screened and evaluated under the fluorescence microscope (Axioplan2) in five fluorescent channels and brightfield for putative CTCs under magnification ×400 and ×600. A cell was classified as a CTC based on its cellular and nuclear morphology (inclusion criteria: intact and non-apoptotic cell morphology, intact non-leucocyte-like nucleus, non-granulocyte-like morphology, cell diameter of minimum 5 µm) and absence of CD45 staining. Keratins, EGFR and EpCAM expression was evaluated in such cells and documented. If no CTCs were found, a subsequent three slides were stained and analysed to confirm CTC status in such patients.
Collagen fibre content evaluation
Collagen fibres were visualised with polarised light microscopy17 using Olympus BX63 microscope equipped with a camera (Leica DFC450C) and ×10 objective (UPlanSApo, NA 0.4). The percentage content of collagen fibres was evaluated and divided into 3 groups—low (<25%), moderate (25–50%) and high (>50%)—and correlated to other molecular markers.
Gene ontology analysis
The patients from TCGA PRAD data set were categorised into four subgroups according to the normalised mRNA expression of EGFR and α-1 type I collagen (COL1A1). For both genes, patients were dichotomised according to the third quartile (Q3) cut-off. Further, phenotypes EGFRpositiveCOL1A1positive (n = 31, 6%), EGFRnegativeCOL1A1positive (n = 93, 19%), EGFRpositiveCOL1A1negative (n = 93, 19%) and EGFRnegativeCOL1A1negative (n = 280, 56%) were defined in those patients. Differences in gene expression between the 4 subgroups were estimated with Kruskal–Wallis test with Benjamini–Hochberg correction; p values < 0.05 and false discovery rate values < 0.05 were considered statistically significant. Low-expression genes (median count in each group = 0 and median count in all samples < 100) were excluded, leaving 11,075 transcripts for further analysis. For the EGFRpositiveCOL1A1positive group, transcripts with the lowest/highest expression in comparison to the three remaining groups were selected based on expression medians. Selected genes were associated with functional annotations using the Functional Annotation Tool by DAVID Bioinformatics Resources 6.8.18,19 EASE Score, a modified Fisher’s exact p value, was used to assess gene enrichment. Multiple testing was corrected using Benjamini correction. The data were analysed using the R statistical environment (3.6.1).20
PCa cell culture and fluorescence-activated cell sorting (FACS) analysis
PCa cell line PC-3 was obtained from ATCC (American Type Culture Collection) and cultured in Ham’s F12 (Biowest), supplemented with 10% foetal bovine serum (Euroclone) and 2 mM L-Glutamine (Euroclone). For FACS analysis aiming to separate EGFRhigh and EGFRlow subpopulations, the cells were trypsinised with 0.05% trypsin (Life Technologies) for 5–10 min and incubated with 5 µg/mL anti-EGFR antibody (Thermo Fisher, clone 30H45L48) for 30 min on ice. After washing in 1×PBS, the cells were incubated for 30 min on ice with secondary anti-rabbit antibody conjugated with Alexa-488 (Jackson Immuno Research, Cat. 211-542-171). Then the samples were sorted using FACSAria using 488 nm laser (BD Bioscience) and analysed with the BD FACSDiva 8.0.1 software (BD Bioscience, IFOM license).
Cell plating and their outgrowth measurement on matrices with different rigidity
Two distinct populations of high or low EGFR PC-3 cells sorted by FACS were plated on hydrogels characterised by different rigidity (i.e. 0.2, 2, 8 and 25 kPa) bound to 6-well glass bottom plates (Cell Guidance Systems), previously coated with 0.01 mg/mL rat tail collagen type I (Corning), and incubated for 48 h under standard culture conditions followed by fixation for 10 min in 4% paraformaldehyde/1×PBS. The cells were permeabilised in 0.1%Triton-X/1×PBS, incubated in blocking solution (1% bovine serum albumin in 1×PBS) and primary anti-Ki-67 antibody (Abcam, ab16667). Afterwards, a secondary anti-rabbit antibody conjugated with Alexa-488 (Jackson ImmunoResearch) was added for 45 min. Nuclei were stained with NucBlue dye (Hoechst 33342, Thermo Fisher). The samples were kept in 1×PBS at 4 °C until microscopic analysis. The images were acquired with Olympus 1×81 microscope, equipped with ×10 objective and cellSens software. At least 50 cells from 7 different fields of view were imaged. The single nuclei and Ki-67-positive cells were counted. The cells from clusters were analysed only when the nuclei were separable. The percentage of Ki-67-positive cells was evaluated and analysed using the Prism software.
Statistics
Statistical analysis was performed using SPSS version 25.0 licensed for the University of Gdańsk. Chi-square or Fisher’s exact tests were used in order to compare EGFR expression to molecular factors and clinico-pathological parameters. Mann–Whitney test was used to compare continuous variables. Associations between EGFR expression and time-to-biochemical recurrence or time-to-death were evaluated using log-rank test and presented as Kaplan–Meier plots. To estimate hazard risk, Cox hazard potential regression analysis (95% confidence interval (CI)) was performed. Cases with missing values were excluded from the study or subanalyses. Uninformative or technically damaged samples were excluded from the analyses. All results were considered statistically significant if p < 0.05. The study was conducted according to REMARK study recommendations,21 in accordance with the Helsinki Declaration of 1975 and STROBE checklist.22
Results
EGFR overexpression is associated with poor prognosis in PCa patients
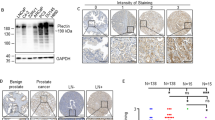
Membranous and membranous/cytoplasmic expression of EGFR with different intensity was evaluated in 1841 primary PCa samples from 1033 patients. EGFR staining intensity was divided into two groups—negative/weak/moderate (EGFRneg-to-mod) and strong (EGFRover; Fig. 1a)—present in 890 (86.2%) and 143 (13.8%) patients, respectively. Interestingly, in primary tumours EGFRover was not associated with EGFR gene dosage (and specifically EGFR gains) assessed by fluorescent in situ hybridisation (n = 277, Supplementary Fig. 1a). It also did not correlate to the shorter (≤18) length of CA repeats in intron I of EGFR gene23 in the small subset of primary prostate carcinomas (n = 26, data not shown). In addition, there was no correlation between EGFR expression and clinico-pathological parameters (i.e. age, T status, N status, preoperative PSA and Gleason score) in the whole cohort of patients (data not shown).
At landmark time points of 5 months, among patients with EGFRover, metastasis-free survival was significantly shorter than in the EGFRneg-to-mod group [(n = 776, Kaplan–Meier log-rank analysis, p = 0.005), Fig. 1b]. This result was even more pronounced in patients who did not reach a PSA concentration <0.1 ng/mL [(n = 59, Kaplan–Meier log-rank analysis, p = 0.018), Supplementary Fig. 1b] or d’Amico high-risk group [(n = 517, Kaplan–Meier log-rank analysis, p = 0.001), Supplementary Fig. 1c]. In addition, EGFRover was a marker of poor overall survival in both univariate (Cox, p = 0.034, hazard ratio (HR) = 1.202, 95% CI = 1.014–1425, Fig. 1c) and multivariate analysis (Cox, p = 0.022, HR = 1.221, 95% CI = 1.029–1.449, Fig. 1c).
EGFR overexpression correlates with pro-migratory and pro-metastatic phenotype of tumour cells
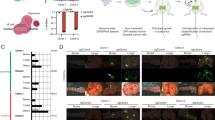
EGFRover correlated with the expression of mesenchymal cell marker vimentin (n = 415, Chi2 = 7.632, p = 0.006, Fig. 2a), intermediate epithelial–mesenchymal phenotype24 characterised by phenotype K8/18/19(+)vim(+) and K8/18/19(−)vim(−), (n = 393, Chi2 = 9.002, p = 0.029, Fig. 2b) and the loss of expression of EpCAM (n = 501, Chi2 = 8.645, p = 0.003, Fig. 2c). Moreover, EGFRover occurred more frequently in the tumours with a higher number of intratumoural lymphatic vessels and blood vessels assessed using podoplanin and CD34 staining, respectively (n = 472, Chi2 = 11.541, p = 0.009, Fig. 2d and Supplementary Fig. 1d, e).
a Vimentin expression in EGFRneg-to-mod and EGFRover cases, n = 415 tumours. b Expression of EMT-related markers in EGFRneg-to-mod and EGFRover cases, n = 383 tumours. c EpCAM expression in EGFRneg-to-mod and EGFRover cases, n = 501 tumours. d Prevalence of blood and lymphatic vessels in EGFRneg-to-mod and EGFRover cases, n = 472 tumours. e Representative images of collagen content quantification (left panel), collagen content distribution in EGFRneg-to-mod and EGFRover cases, n = 120 patients. f GO BP terms enriched in genes upregulated in EGFRpositiveCOL1A1positive tumours; top 20 terms with the lowest p value plotted against fold enrichment and ordered according to −log10(FDR); dot size represents the number of genes associated with the term, dot colour represents −log10(FDR); analysed with Functional Annotation Tool by DAVID Bioinformatics Resources 6.81. g GO BP terms enriched in genes downregulated in EGFRpositiveCOL1A1positive tumours; top 20 terms with the lowest p value plotted against fold enrichment and ordered according to −log10(FDR); dot size represents the number of genes associated with the term, dot colour represents −log10(FDR); analysed with Functional Annotation Tool by DAVID Bioinformatics Resources 6.81.
In primary tumours from 120 PCa patients, percentage content of the collagen fibre content was quantified. A high collagen fibre content (>50%) was associated with the EGFRover phenotype of tumour cells (n = 120, Chi2 = 7.114, p = 0.029, Fig. 2e).
With the gene ontology analysis, multiple genes involved in cell migration, adhesion and proliferation, as well as angiogenesis regulation, were significantly upregulated in tumours expressing EGFR and COL1A1 (Fig. 2f and Supplementary Data 1). On the contrary, multiple genes involved in translation, transcription and mitochondrial metabolism were downregulated in this subgroup of tumours (Fig. 2g and Supplementary Data 2).
EGFR is an EMT-independent marker of CTCs in d’Amico high-risk PCa patients
Single CTCs (n = 11) and CTC clusters (n = 2) were isolated from 5 (12.8%) of 39 analysed d’Amico high-risk patients. The CTC yield varied between 1 and 5 CTCs/1,500,000 PBMCs per patient. All detected CTCs showed strong EGFR and were negative for CD45 (Fig. 3a–c and Supplementary Fig. 2a). Other common CTC markers including pan-keratins and EpCAM were also evaluated. Only three patients had CTCs positive for EpCAM, including all cells from both clusters (Fig. 3a, c). Pan-keratin status of the detected CTCs was heterogonous, varying from negative (five cells) through weak (six cells) to moderate expression—five cells in two clusters (Fig. 3a–c). CTCs’ yield and phenotype were not associated with any tumour characteristics (Supplementary Table 4). Despite the short observation time after surgery (≤25 months) and limited follow-up cohort, patients with EGFRover CTCs had significantly shorter time to biochemical recurrence and time to metastasis than patients negative for CTCs (n = 25, Kaplan–Meier log-rank analysis, p = 0.002 and p < 0.001, respectively, Supplementary Fig. 2b, c). In addition, 3 of those 5 patients with EGFRover CTCs developed distant metastases to lymph nodes (patient with CTCs negative for EpCAM) or bones (patient with weakly positive CTCs for EpCAM), whereas none of the patients developed metastasis in the cohort negative for CTCs (n = 23, Fisher’s exact test, p = 0.005).
a Representative CTC cluster composed of two cells positive for EGFR, pan-keratin and EpCAM and negative for CD45; scale bar 10 μm. b Representative single CTC positive for EGFR and pan-keratin and negative for EpCAM and CD45. c Characteristics of CTCs detected in d’Amico high-risk patients. Number of CTCs expressing EGFR, EpCAM and pan-keratin/total detected CTCs.
EGFR overexpression is most frequent in CRPC metastases to bones
Membranous and membranous/cytoplasmic EGFR expression was also evaluated in 75 tissue cores from castration-resistant metastases of 21 patients (Fig. 4a, b). EGFR expression was significantly more frequent in castration-resistant bone metastases, when compared to its distribution in primary tumours (n = 39 vs. n = 1841, Chi2 = 11.543, p = 0.009, Fig. 4c, d). Interestingly, also EpCAM strong intensity reached 76% in bone metastases from CRPC patients (Fig. 4d). In bone metastases, the percentage of EGFR-positive tumour cells frequently reached 100% per tumour sample (Fig. 4e) and its mean value was significantly higher than in the cohort of EGFR-positive primary tumours (92%, n = 13 vs. 58%, n = 1258; two-tailed Mann–Whitney test, p < 0.0001). There was no correlation between EGFRover and EpCAM, K8/18/19 and vimentin expression in castration-resistant bone and/or visceral metastases (data not shown). In addition, there was a borderline correlation between higher prevalence of collagen fibres (>50%) and the EGFRover phenotype of tumour cells in metastases (n = 71, Chi2 = 5.934, p = 0.051, Fig. 4f).
a Representative immunohistochemical staining of EGFRover in bone metastasis. b Representative immunohistochemical staining EGFRover in visceral metastases. c EGFR staining intensity distribution in primary tumours (n = 1841), visceral metastases (n = 36) and bone metastases (n = 39). d EGFR and EpCAM staining intensity distribution in bone and visceral metastases. e Percentage of EGFR-positive cells in bone and visceral metastases. f Collagen content distribution in EGFRneg-to-mod and EGFRover metastases (n = 71). g Representative images of EGFRlow and EGFRhigh PC-3 cell growth on soft (0.2 kPa) and rigid (25 kPa) matrices. Quantification of PC-3 cells proliferation using Ki-67 marker on soft (0.2 kPa, n = 70 cells) and rigid (25 kPa, n = 91 cells) matrices.
High expression of EGFR improves PC-3 proliferation on rigid matrices
On collagen-coated rigid matrices (25 kPa), mimicking bone tissues rigidity, PC-3 cells sorted according to their high EGFR expression, adhered and grew more efficiently than sub-lines with low EGFR expression (two-tailed Mann–Whitney test, p < 0.0001, Fig. 4g and Supplementary Fig. 3a, b). On the other hand, such increase in adhesion and growth was not observed on soft substrate (0.2 kPa). Interestingly, PC-3 cells plated on matrices with different rigidity (0.2 and 25 kPa) revealed no significant change in the epithelial/mesenchymal phenotype characterised by K19/vimentin ratio assessed by real-time PCR (data not shown).
Discussion
Our data indicate that EGFRover might be a candidate biomarker of stable marker of PCa dissemination cascade.
EGFR overexpression, found both in primary tumour and CTCs, was an indicator of poor prognosis. In primary tumours, it was associated with shorter metastasis-free survival, which has been previously shown to be related with a significant risk of death from PCa.25 Our observation is similar to the ones from other groups,9 performed, however, mainly in CRPC.26,27
The expression of EGFR was reported to be associated with EMT,28,29 which promotes pro-migratory and pro-survival behaviour of tumour cells, generating their aggressive phenotype.4 Based on our data, EGFR overexpression correlated with pro-migratory and pro-metastatic phenotype of PCa tumour cells. In EGFRover tumours, we also found a higher collagen fibre content that can influence cell migration, invasiveness and proliferation and indicate worse prognosis,30,31 Gene ontology analysis further confirmed that cancers with high expression of both EGFR and gene-encoding type I collagen have upregulated genes involved in cell migration and adhesion. Tumours with strong EGFR expression had a larger number of blood and/or lymphatic vessels that could facilitate haematogenous and lymphatic dissemination of cancer cells.32
Of note, EMT is a phenomenon hindering detection of CTCs in bloodstream.33,34,35 Our results suggest that EGFR seems to be a stable signature of PCa progression, which might serve as surrogate marker of CTCs undergoing EMT. Importantly, based on our collected data, in a d’Amico high-risk patient cohort, inclusion of EGFR can improve both CTC detection and stratification of patients. However, this result needs further confirmation in a larger cohort of patients as the absolute number of CTC-positive patients and actual number of CTCs in the present study are too low to allow for strong conclusions.
In our study, EGFRover was enriched in bone metastases, suggesting that organ-specific factors such as its stiffness or tumour microenvironment might result in regulation of harbouring and/or nesting of EGFR-positive tumour cells. PCa cells that disseminate show an exquisite tropism for the bone.36 In an autopsy study, 90% of the men who had died with haematogenous metastases of PCa were diagnosed with bone metastases.37 However, the possible molecular mechanisms involved in governing bone metastases tropism is still poorly understood.36 Bone is among tissues characterised by elastic moduli with the greatest stiffness.38 It was also reported that EGFR can be involved in rigidity sensing after associating with nascent adhesions under rigidity-dependent tension.39 Moreover, human squamous cell carcinoma cells, in response to matrix stiffening, increased EGFR expression and invasiveness.40 In our study, EGFR positively correlated with collagen fibre content that can indicate tumour stiffness.40,41 Together with our data showing improved proliferation of EGFR-overexpressing PCa cells on rigid matrices, it can be speculated that EGFR can promote growth of cells on rigid substratum and bone metastases.
To sum up, the data collected within this study suggest that EGFR is a marker of PCa dissemination, independent of EMT.
References
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A. & Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Wu, J. N., Fish, K. M., Evans, C. P., Devere White, R. W. & Dallera, M. A. No improvement noted in overall or cause-specific survival for men presenting with metastatic prostate cancer over a 20-year period. Cancer 15, 818–823 (2014).
Crawford, E. D., Petrylak, D. & Sartor, O. Navigating the evolving therapeutic landscape in advanced prostate cancer. Urol. Oncol. 35, 1–13 (2017).
Montanari, M., Rossetti, S., Cavaliere, C., D’Aniello, C., Malzone, M. G., Vanacore, D. et al. Epithelial-mesenchymal transition in prostate cancer: an overview. Oncotarget 23, 35376–35389 (2017).
Wang, L., Song, G., Tan, W., Qi, M., Zhang, L., Chan, J. et al. miR-573 inhibits prostate cancer metastasis by regulating epithelial-mesenchymal transition. Oncotarget 3, 35978–35990 (2015).
Stylianou, N., Lehman, M. L., Wang, C., Fard, A. T., Rockstroh, A., Fazli, L. et al. A molecular portrait of epithelial–mesenchymal plasticity in prostate cancer associated with clinical outcome. Oncogene 38, 913–934 (2019).
DeHaan, A. M., Wolters, N. M., Keller, E. T. & Ignatoski, K. M. W. EGFR ligand switch in late stage prostate cancer contributes to changes in cell signaling and bone remodeling. Prostate 69, 528–537 (2009).
Traish, A. M. & Morgentaler, A. Epidermal growth factor receptor expression escapes androgen regulation in prostate cancer: a potential molecular switch for tumour growth. Br. J. Cancer 101, 1949–1956 (2009).
Schlomm, T., Kirstein, P., Iwers, L., Daniel, B., Steuber, T., Walz, J. et al. Clinical significance of epidermal growth factor receptor protein overexpression and gene copy number gains in prostate cancer. Clin. Cancer Res. 13, 6579–6584 (2007).
Day, K. C., Hiles, G. L., Kozminsky, M., Dawsey, S. J., Paul, A., Broses, L. J. et al. HER2 and EGFR overexpression support metastatic progression of prostate cancer to bone. Cancer Res. 77, 74–85 (2017).
Linder, M., Hecking, M., Glitzner, E., Zwerina, K., Holcmann, M., Bakiri, L. et al. EGFR controls bone development by negatively regulating mTOR-signaling during osteoblast differentiation. Cell Death Differ. 25, 1094–1106 (2018).
Smentoch, J., Szade, J., Żaczek, A. J., Eltze, E., Semjonow, A., Brandt, B. et al. Low numbers of vascular vessels correlate to progression in hormone-naïve prostate carcinomas undergoing radical prostatectomy. Cancers (Basel) 11, 1356 (2019).
Omari, A., Nastały, P., Stoupiec, S., Bałabas, A., Dąbrowska, M., Bielińska, B. et al. Somatic aberrations of BRCA1 gene are associated with ALDH1, EGFR, and tumor progression in prostate cancer. Int. J. Cancer 144, 607–614 (2019).
Nastały, P., Filipska, M., Morrissey, C., Eltze, E., Semjonow, A., Brandt, B. et al. ALDH1-positive intratumoral stromal cells indicate differentiated epithelial-like phenotype and good prognosis in prostate cancer. Transl. Res. 203, 49–56 (2019).
Abeshouse, A., Ahn, J., Akbani, R., Ally, A., Amin, S., Andry, C. D. et al. The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015).
Wolff, A. C., Hammond, M. E. H., Hicks, D. G., Dowsett, M., McShane, L. M., Allison, K. H. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast. J. Clin. Oncol. 131, 18–43 (2013).
Changoor, A., Tran-Khanh, N., Méthot, S., Garon, M., Hurtig, M. B., Shive, M. S. et al. A polarized light microscopy method for accurate and reliable grading of collagen organization in cartilage repair. Osteoarthr. Cartil. 19, 126–135 (2011).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2014).
McShane, L. M., Altman, D. G., Sauerbrei, W., Taube, S. E., Gion, M. & Clark, G. M. REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res. Treat. 100, 229–235 (2006).
von Elm, E., Altman, D. G., Egger, M., Pocock, S. J., Gøtzsche, P. C. & Vandenbroucke, J. P. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: Guidelines for reporting observational studies. Int. J. Surg. 12, 1495–1499 (2014).
Buerger, H., Gebhardt, F., Schmidt, H., Beckmann, A., Hutmacher, K., Simon, R. et al. Length and loss of heterozygosity of an intron 1 polymorphic sequence of egfr is related to cytogenetic alterations and epithelial growth factor receptor expression. Cancer Res. 60, 854–857 (2000).
Yang, J., Antin, P., Berx, G., Blanpain, C., Brabletz, T., Bronner, M. et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020).
Xie, W., Regan, M. M., Buyse, M., Halabi, S., Kantoff, P., Sartor, O. et al. Metastasis-free survival is a strong surrogate of overall survival in localized prostate cancer. J. Clin. Oncol. 20, 3097–3104 (2017).
Josefsson, A., Linder, A., Flondell Site, D., Canesin, G., Stiehm, A., Anand, A. et al. Circulating tumor cells as a marker for progression-free survival in metastatic castration-naïve prostate cancer. Prostate 77, 849–858 (2017).
Okegawa, T., Itaya, N., Hara, H., Tambo, M. & Nutahara, K. Epidermal growth factor receptor status in circulating tumor cells as a predictive biomarker of sensitivity in castration-resistant prostate cancer patients treated with docetaxel chemotherapy. Int. J. Mol. Sci. 17, 2008 (2016).
Lo, H. W., Hsu, S. C., Xia, W., Cao, X., Shih, J. Y., Wei, Y. et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 67, 9066–9076 (2007).
Zuo, J. H., Zhu, W., Li, M. Y., Li, X. H., Yi, H., Zeng, G. Q. et al. Activation of EGFR promotes squamous carcinoma SCC10A cell migration and invasion via inducing EMT-like phenotype change and MMP-9-mediated degradation of E-cadherin. J. Cell Biochem. 112, 2508–2517 (2011).
Wei, S. C., Fattet, L., Tsai, J. H., Guo, Y., Pai, V. H., Majeski, H. E. et al. Matrix stiffness drives epithelial–mesenchymal transition and tumour metastasis through a TWIST1–G3BP2 mechanotransduction pathway. Nat. Cell Biol. 17, 678–688 (2015).
Xu, S., Xu, H., Wang, W., Li, S., Li, H., Li, T. et al. The role of collagen in cancer: from bench to bedside. J. Transl. Med. 17, 309 (2019).
Dhakal, H. P., Naume, B., Synnestvedt, M., Borgen, E., Kaaresen, R., Schlichting, E. et al. Vascularization in primary breast carcinomas: its prognostic significance and relationship with tumor cell dissemination. Clin. Cancer Res. 14, 2341–2350 (2008).
Bednarz-knoll, N., Alix-panabières, C. & Pantel, K. Clinical relevance and biology of circulating tumor cells. Breast Cancer Res. 13, 228 (2011).
Hille, C. & Pantel, K. Circulating tumour cells in prostate cancer. Nat. Rev. Urol. 15, 265–266 (2018).
Pantel, K., Hille, C. & Scher, H. I. Circulating tumor cells in prostate cancer: from discovery to clinical utility. Clin. Chem. 65, 87–99 (2019).
Wong, S. K., Mohamad, N. V., Giaze, T. R., Chin, K. Y., Mohamed, N. & Ima-Nirwana, S. Prostate cancer and bone metastases: the underlying mechanisms. Int. J. Mol. Sci. 20, 2581 (2019).
Bubendorf, L., Schöpfer, A., Wagner, U., Sauter, G., Moch, H., Willi, N. et al. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum. Pathol. 31, 578–583 (2000).
Butcher, D. T., Alliston, T. & Weaver, V. M. A tense situation: forcing tumour progression. Nat. Rev. Cancer 9, 108–122 (2009).
Saxena, M., Liu, S., Yang, B., Hajal, C., Changede, R., Hu, J. et al. EGFR and HER2 activate rigidity sensing only on rigid matrices. Nat. Mater. 16, 775–781 (2017).
Grasset, E. M., Bertero, T., Bozec, A., Friard, J., Bourget, I., Pisano, S. et al. Matrix stiffening and EGFR cooperate to promote the collective invasion of cancer cells. Cancer Res. 78, 5229–5242 (2018).
Riegler, J., Labyed, Y., Rosenzweig, S., Javinal, V., Castiglioni, A., Dominguez, C. X. et al. Tumor elastography and its association with collagen and the tumor microenvironment. Clin. Cancer Res. 24, 4455–4467 (2018).
Acknowledgements
We thank all patients who agreed to donate the blood for this study. We thank the patients and their families who were willing to participate in the Prostate Cancer Donor Program, the investigators Dr. Robert Vessella, Dr. Celestia Higano, Dr. Bruce Montgomery, Dr. Evan Yu, Dr. Peter Nelson, Dr. Paul Lange, Dr. Martine Roudier, and Dr. Lawrence True for their contributions to the University of Washington Medical Center Prostate Cancer Donor Rapid Autopsy Program. We express our gratitude to Mrs. Małgorzata Stoupiec for her excellent technical assistance. The authors thank IFOM Imaging Facility and IFOM Cell Biology Unit.
Author information
Authors and Affiliations
Contributions
Conceptualisation—P.N., B. Brandt and N.B.-K.; methodology—S.S., M.P., P.N., J.S., P.M., A.J.Ż. and N.B.-K.; formal analysis—S.S., M.P., P.N., J.S. and N.B.-K.; investigation—S.S., M.P., P.N., J.S., E.E. and N.B.-K.; resources—M.A., B. Beyer, P.T., T.S., C.M., A.J.Ż., M.G., E.E., A.S., P.M., K.P., B. Brandt and N.B.-K.; data curation—S.S., M.P., J.S., P.N. and NBK; original draft preparation—S.S., M.P., P.N. and N.B.-K.; draft review and editing—all authors; visualisation—P.N., S.S., M.P. and N.B.-K.; supervision, project administration and funding acquisition—N.B.-K.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Primary tumour samples were collected from patients according to a standard protocol approved by the University Clinic Münster (Germany) ethics committee and upon signing an informed consent. Blood samples were collected from patients according to a standard protocol approved by the University Medical Center Hamburg-Eppendorf (Hamburg, Germany) ethics committee and upon signing an informed consent. Metastatic castration-resistant prostate cancer samples were collected from patients according to a standard protocol approved by the University of Washington ethics committee and upon signing an informed consent. This study was performed in accordance with the tenets of the Declaration of Helsinki. The cell line employed was purchased from ATCC, Rockville, MD, USA.
Consent to publish
All informed consents were obtained. This report does not contain any individual person’s information.
Data availability
The data that support the findings of this work are available from the corresponding author upon request.
Competing interests
Professor Dr. med. Klaus Pantel is an Editorial Board Member of British Journal of Cancer.
Funding information
This research was funded by National Science Centre, with the grant numbers 2017/26/D/NZ5/01088 (to N.B.-K.) and DEC-2017/01/X/NZ4/02120 (to N.B.-K.) and ERC Advanced Investigator Grant INJURMET (K.P.). The rapid autopsy program was funded in part by the Pacific Northwest Prostate Cancer SPORE number P50CA97186 (to C.M.). This work was supported by fellowships from the Italian Association for Cancer Research number 19416 (to P.N.) and Fondazione Umberto Veronesi Post-doctoral fellowship (to P.N.).
Additional information
Note This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0).
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nastały, P., Stoupiec, S., Popęda, M. et al. EGFR as a stable marker of prostate cancer dissemination to bones. Br J Cancer 123, 1767–1774 (2020). https://doi.org/10.1038/s41416-020-01052-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-020-01052-8
This article is cited by
-
sBioSITe enables sensitive identification of the cell surface proteome through direct enrichment of biotinylated peptides
Clinical Proteomics (2023)
-
Microfluidic-based prostate cancer model for investigating the secretion of prostate-specific antigen and microRNAs in vitro
Scientific Reports (2023)
-
CRISPR/Cas9-mediated deletion of Interleukin-30 suppresses IGF1 and CXCL5 and boosts SOCS3 reducing prostate cancer growth and mortality
Journal of Hematology & Oncology (2022)
-
SBSN drives bladder cancer metastasis via EGFR/SRC/STAT3 signalling
British Journal of Cancer (2022)