Abstract
Acute myeloid leukemia (AML) with CEBPA bZIP in-frame mutations (CEBPAbZIP-inf) is classified within the favorable-risk group by the 2022 European LeukemiaNet (ELN-2022). However, heterogeneous clinical outcomes are still observed in these patients. In this study, we aimed to investigate the mutation profiles and transcriptomic patterns associated with poor outcomes in patients with CEBPAbZIP-inf. One hundred and thirteen CEBPAbZIP-inf patients were identified in a cohort of 887 AML patients homogeneously treated with intensive chemotherapy. Concurrent WT1 or DNMT3A mutations significantly predicted worse survival in AML patients with CEBPAbZIP-inf. RNA-sequencing analysis revealed an enrichment of interferon (IFN) signaling and metabolic pathways in those with a shorter event-free survival (EFS). CEBPAbZIP-inf patients with a shorter EFS had higher expression of IFN-stimulated genes (IRF2, IRF5, OAS2, and IFI35). Genes in mitochondrial complexes I (NDUFA12 and NDUFB6) and V (ATP5PB and ATP5IF1) were overexpressed and were associated with poorer survival, and the results were independently validated in the TARGET AML cohort. In conclusion, concurrent WT1 or DNMT3A mutations and a dysregulated immune and metabolic state were correlated with poor survival in patients with CEBPAbZIP-inf, and upfront allogeneic transplantation may be indicated for better long-term disease control.
Similar content being viewed by others
Introduction
Acute myeloid leukemia (AML) with mutant CCAAT/enhancer binding protein α (CEBPA) accounts for 5.1–18.9% of patients with AML; a higher incidence rate is observed in patients with AML from Asia than in those from Western countries [1,2,3,4,5,6,7]. We and others have showed that CEBPA double mutation (CEBPAdm) predicted better overall survival (OS) whereas CEBPA single mutation (CEBPAsm) did not have this survival advantage [8,9,10,11]. Accordingly, CEBPAdm AML has been recognized as a separate entity in the 2016 World Health Organization (WHO) Classification (WHO-2016) [12] and a favorable-risk category in 2017 European LeukemiaNet (ELN) recommendations. Concomitant WT1 mutations or CSF3R mutations helped predict a poor prognosis in patients with CEBPAdm [7, 13].
Recently, it was discovered that the favorable outcomes of patients with CEBPA mutations are confined to those with in-frame mutations in bZIP (CEBPAbZIP-inf), no matter whether they have CEBPAsm or CEBPAdm [4, 14]. These findings impact the current 5th WHO Classification (WHO-2022) [15], 2022 ELN recommendations (ELN-2022) [16] and International Consensus Classification (ICC) [17], resulting in the changing of the category, “AML with biallelic mutations of CEBPA” into “AML with in-frame bZIP mutations of CEBPA.” However, the 5-year event-free survival (EFS) was less than 50% [4], and the cumulative incidence of relapse was approaching 40% in this category [18]. It remains largely unknown how to explain the heterogeneous outcomes in this so-called “favorable category” at the mutational and transcriptomic level. The identification of dysregulated pathways that are independent of the mutational background, may provide novel options for therapeutic targeting.
In this study, we aimed to investigate the prognostic relevance of mutations and transcriptomics in AML patients with CEBPAbZIP-inf from a large cohort of 887 unselected de novo non-M3 AML patients. Our study showed that patients with poor survival were characterized by enrichment of interferon (IFN) pathways and oxidative phosphorylation. These data provide insights into non-genetic heterogeneity that may guide precise risk stratification and hold therapeutic potential in AML with CEBPAbZIP-inf. The findings were also well-validated in the TARGET AML cohort.
Methods and materials
Subjects
We consecutively enrolled 887 newly diagnosed de novo non-M3 AML patients who received standard chemotherapy at the National Taiwan University Hospital (NTUH). Diagnosis and classification of AML were made according to the 2022 ICC [12] and WHO-2022 [15]. Detailed patient characteristics and treatment regimens were summarized in the Supplementary Data [19]. This study was approved by the Institutional Review Board of the NTUH, and written informed consent was obtained from all participants in accordance with the Declaration of Helsinki (approval number: 201709072RINC and 202109078RINB).
Cytogenetics
Chromosomal analyses were performed as described previously [20]. Karyotypes were classified using Medical Research Council (MRC)-defined risk groups [21].
Mutation analysis
Gene mutations were examined via targeted next-generation sequencing (NGS), using the TruSight myeloid sequencing panel (Illumina, San Diego, CA, USA), which included 15 full exon genes and 39 oncogenic hotspot genes. HiSeq platform (Illumina, San Diego, CA, USA) was used for sequencing with a median reading depth of 12000×. Owing to suboptimal sequencing sensitivity, we verified CEBPA mutations via Sanger sequencing. Analysis of FLT3-ITD was performed via polymerase chain reaction (PCR), followed by fluorescence capillary electrophoresis and that of KMT2A-PTD, by PCR followed by Sanger sequencing [22].
RNA-sequencing (RNA-seq) analysis
Bone marrow (BM) mononuclear cells from 136 AML patients (36 with CEBPAbZIP-inf, 11 with CEBPAnonbZIP-inf, 89 with wild type of CEBPA and normal karyotype, CEBPAwt) were performed with RNA-seq. The purified RNA was used to prepare the sequencing libraries using the TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer’s recommendations. The qualified libraries were then sequenced on Illumina NovaSeq 6000 with 150 bp paired-end mode. Trimming of adaptor sequences and removal of reads of low quality were performed using cutadapt (v2.3). Quantified reads were aligned to the human genome (GRCh38.p12) by STAR (v2.6.1 a) [23], and then gene-level read counts were generated based on the annotations of Gencode (v28) [24]. The read counts cross all samples were normalized using the trimmed mean of the M values method as implemented in the calcNormFactors function of edgeR package [25], and gene expression in terms of log2(CPM + 1) (counts per million reads) was computed for further analysis. We used limma to assess the differentially expressed genes (DEGs) between conditions [26]. The linear model was fit to the normalized expression data using a design matrix contrasting patients with short and long EFS. Log2 fold changes (logFC) were computed, along with adjusted P values (false discovery rate, FDR) corrected for multiple hypothesis testing using the Benjamini–Hochberg (BH) method.
We used Gene Set Enrichment Analysis (GSEA) software to investigate systematic enrichments of CEBPA mutation-governed expressional profiles with gene sets curated in the Molecular Signatures Database (MSigDB) [27]. The statistical significance of the degree of enrichment was assessed by a 1000-time random permutation test [28].
External datasets
The gene expression data of GSE15210, derived from Affymetrix HG-U133 oligonucleotide microarrays (n = 61, 7 with CEBPAdm, 8 with CEBPAsm, and 46 with CEBPAwt), and the TCGA AML RNA-seq data generated by the Illumina HiSeq 2000 platform (n = 173) were downloaded for validation purposes [10, 29]. To evaluate the prognostic significance of mitochondria complexes in an independent AML patient cohort, RNA-seq, gene mutation and clinical data of the TARGET AML cohort (n = 493) were downloaded from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/) [30]. Data from patients in the AMLSG cohort, the UK-NCRI trials, the TCGA cohort and NTUH Yunlin Branch cohort with available molecular annotations were further used to validate the prognostic relevance of concurrent mutations in CEBPAbZIP-inf [29, 31, 32].
Statistical analysis
The discrete variables were compared using the chi-square tests, but if the expected values of contingency tables were smaller than 5, Fisher’s exact test was used. Mann–Whitney U tests were used to compare continuous variables and medians of distributions. OS was measured from the date of first diagnosis to the date of last follow-up or death from any cause. EFS was measured from the date of diagnosis until treatment failure, relapse from first complete remission (CR1) or death from any cause, whichever occurred first [33]. Multivariate Cox proportional hazard regression analysis was used to investigate independent prognostic factors for OS and EFS. A P value < 0.05 was considered statistically significant. Statistical analyses were performed in the SPSS 23 (SPSS Inc., Chicago, IL, USA) and R software 4.3.1 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Patient characteristics and clinical outcomes
A total of 142 patients (16%) were identified as having CEBPA mutations, including 113 (12.7%) with CEBPAbZIP-inf and 29 (3.3%) patients with CEBPAnonbZIP-inf. The majority of CEBPAbZIP-inf were CEBPAdm (96 of 113, 85.0%) (Supplementary Table 1). The CEBPAnonbZIP-inf consisted of 23 CEBPAsm, 4 CEBPAdm outside the bZIP core region, and 2 CEBPAdm with nonsense/out-of-frame bZIP mutations. Patients with CEBPAbZIP-inf had significantly younger age, higher white blood cell (WBC) count and peripheral blood blast percentage at diagnosis compared with those with CEBPAwt. CEBPAbZIP-inf also differed from CEBPAnonbZIP-inf in terms of age, hemoglobin level, and peripheral blood blast percentage (Table 1).
The CR1 rate was significantly higher for patients with CEBPAbZIP-inf than those with CEBPAnonbZIP-inf (92.9% vs. 72.4%, P = 0.005) and CEBPAwt (92.9% vs. 73.2%, P < 0.0001). In contrast, the relapse rate was lower for patients with CEBPAbZIP-inf than those with CEBPAnonbZIP-inf (37.1% vs. 57.1%, P = 0.088) and CEBPAwt (37.1% vs. 54.5%, P = 0.002). After a median follow-up time of 7.1 years, patients with CEBPAbZIP-inf had significantly better OS and EFS than those with CEBPAnonbZIP-inf (OS, median, not reached (NR) vs. 42.1 months, P = 0.023; EFS, median, 138.2 vs. 10.9 months, P = 0.002, respectively) and CEBPAwt (OS, median, NR vs. 23.4 months, P < 0.0001; EFS, median, 138.2 vs. 10.4 months, P < 0.0001, respectively) (Fig. 1). Intriguingly, according to the WHO-2022 [15], patients with CEBPA mutations had significantly better EFS and a trend towards better OS than those with CEBPAothers (EFS, median, 138.2 vs. 10.4 months, P = 0.014; OS, median, NR vs. 23.4 months, P = 0.149, respectively) (Supplementary Fig. 1).
Frequency of concurrent mutations and its prognostic relevance in CEBPA bZIP-inf
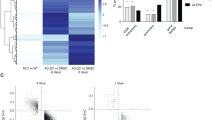
Approximately two-thirds (76 of 113, 67.3%) of patients with CEBPAbZIP-inf had at least one concurrent mutation, and the most frequently mutated genes were GATA2 (29.1%), WT1 (14.5%), NRAS (12.7%), FLT3-ITD (11.8%), and TET2 (10.9%). Patients with CEBPAbZIP-inf had significantly higher frequencies of mutations in GATA2 and WT1, but lower frequencies of mutations in PTPN11, RUNX1, DNMT3A, IDH1, IDH2, SRSF2, NPM1, TP53, and KMT2A-PTD than patients with CEBPAwt. On the other hand, CEBPAbZIP-inf was associated with lower frequencies of mutations in TET2, IDH1, IDH2, SRSF2, STAG2, and NPM1 compared with CEBPAnonbZIP-inf (Fig. 2A and Supplementary Table 2). The results supported the different concerted mutations in biological cooperation among groups with different CEBPA mutational statuses.
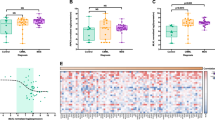
A Frequencies of concurrent mutations in patients with different CEBPA mutation statuses. Mutations with a frequency of at least 1% in one subgroup were shown. *P < 0.05 between CEBPAbZIP-inf and CEBPAwt, **P < 0.05 between CEBPAbZIP-inf and CEBPAnonbZIP-inf. B, C Concurrent WT1 or DNMT3A mutations were associated with significantly shorter OS and a trend towards shorter EFS in patients with CEBPAbZIP-inf. D Allogeneic hematopoietic stem cell transplantation (Allo-HSCT) in first complete remission was associated with significantly longer EFS and a trend towards longer OS than post-remission chemotherapy (PR-CT) in CEBPAbZIP-inf patients with WT1 or DNMT3A mutations.
Next, we studied the prognostic impact of concurrent mutations with the aim of risk-stratifying the patients with CEBPAbZIP-inf more precisely. Concurrent WT1 or DNMT3A mutations predicted significantly worse OS (WT1 mutation, median, 28.2 months vs. NR, P = 0.020 and DNMT3A mutation, median, 6.1 months vs. NR, P = 0.013) and a trend towards worse EFS (WT1 mutation, median, 10.6 vs. 138.2 months, P = 0.064 and DNMT3A mutation, 3.3 vs. 138.2 months, P = 0.083) in patients with CEBPAbZIP-inf (Fig. 2B, C). Importantly, the outcome of CEBPAbZIP-inf with concurrent WT1 or DNMT3A mutations was similar to the ELN-2022 intermediate-risk group in terms of OS (P = 0.251) and EFS (P = 0.220). However, GATA2, TET2, ELN-2022 defined adverse gene mutations, and FLT3-ITD did not influence CR1, relapse rate, EFS and OS (Supplementary Fig. 2). Allogeneic hematopoietic stem cell transplantation (allo-HSCT) in CR1 was associated with significantly longer EFS and a trend towards longer OS compared with post-remission chemotherapy (PR-CT) only in CEBPAbZIP-inf patients with DNMT3A or WT1 mutations (EFS, median, NR vs. 8.9 months, P = 0.025; OS, median, NR vs. 14.0 months, P = 0.066, respectively) (Fig. 2D). For CEBPAbZIP-inf patients without DNMT3A or WT1 co-mutations, allo-HSCT in CR1 was associated with similar OS (P = 0.413) and EFS (P = 0.168) compared with PR-CT (Supplementary Fig. 3 and Supplementary Table 3). Given the limited number of cohorts with available CEBPAbZIP-inf status, we validated our findings with datasets comprising patients with CEBPAdm. This decision was supported by evidences indicating that the majority (94.1% in the current study and 90% in the study by Taube F et al.) [4] of CEBPAdm can be classified as CEBPAbZIP-inf. Similarly, concurrent DNMT3A mutation was associated with significantly poorer OS (median, 16.3 months vs. not reached, P = 0.029) compared with DNMT3A wild-type cases in the validation cohort (Supplementary Fig. 4 and Supplementary Table 4).
Characteristics and clinical outcomes of patients with CEBPA bZIP-inf with relapse
There were 39 patients with CEBPAbZIP-inf who experienced a relapse. Twenty-nine (82.9%) of 35 patients who underwent re-induction chemotherapy achieved second complete remission (CR2). Among the 6 patients refractory to re-induction chemotherapy, two patients were found to have GATA2 mutations, two patients had WT1 mutations, two patients had cytogenetic evolution, and one patient had central nervous system involvement. Notably, the presence of WT1 mutation was associated with a lower probability of attaining CR2 (50% vs. 92.6%, P = 0.031), and a trend towards inferior OS from the time of relapse (median, 2.8 vs. 26.5 months, P = 0.144).
The median OS from the time of relapse was 23.4 months. Twenty-one patients received subsequent allo-HSCT, including 14 patients (66.7%) in CR2. Notably, patients with allo-HSCT after relapse had significantly better OS compared with those without allo-HSCT (median, 124 vs. 13.1 months, P = 0.001). Furthermore, allo-HSCT in CR2 was associated with significantly better OS compared with allo-HSCT in other disease statuses (median, not reached vs. 19.9 months, P = 0.044) (Supplementary Fig. 5 and Supplementary Table 5).
Comparison of transcriptional signatures between patients with CEBPA bZIP-inf and those with CEBPA nonbZIP-inf or CEBPA wt
Gene expression profiles of the 36 patients with CEBPAbZIP-inf were compared with those of the 89 patients with CEBPAwt and normal karyotype. The principle component analysis (PCA) showed obvious separation of clustering for transcriptomes of CEBPAbZIP-inf and CEBPAwt AML (Fig. 3A). CEBPAbZIP-inf was associated with 327 upregulated and 849 downregulated genes based on a |logFC|>1 cutoff and a P value < 0.01, with HOXA9 (|logFC| = 5.91), and HOXA5 (|logFC| = 4.97) among the highest ranked downregulated genes (Fig. 3B, C, Supplementary Table 6). By using the MsigDB Hallmark gene sets, we found that CEBPAbZIP-inf transcriptome was enriched for MYC targets (normalized enrichment score (NES) 2.92, FDR 0), oxidative phosphorylation (NES 2.10, FDR 0), and E2F targets (NES 1.80, FDR 3.42E-04). The oncogenic gene sets revealed positive enrichment of the MYC target pathway (NES 2.12, FDR 0) and negative enrichment of genes upregulated upon knockdown of HOXA9 (NES -2.18, FDR 0) (Fig. 3D, E). As previously reported, both MYC targets and E2F targets are characterized by a proliferation signature, and dysregulation of these pathways has been witnessed in a wide variety of cancers, including myeloid neoplasms [34,35,36].
A PCA showing a separation of clustering of transcriptomes between AML with CEBPAbZIP-inf and CEBPAwt. B Volcano plot showing DEGs with genes downregulated (logFC<0) and upregulated (logFC>0), respectively, in patients with CEBPAbZIP-inf compared with CEBPAwt. Multiple genes in the HOX family were downregulated in CEBPAbZIP-inf. C Heat map of normalized gene expression values of the top upregulated and downregulated DEGs. The DEGs were listed in Supplementary Table 6. D, E Enriched and depleted MSigDB oncogenic and Hallmark gene sets between CEBPAbZIP-inf and CEBPAwt.
For validation purposes, targets of MYC and E2F were shown to be significantly enriched in the TCGA cohort when we compared the patients with CEBPAbZIP-inf (n = 7) to CEBPAwt with intermediate-risk cytogenetics (n = 14) (Supplementary Fig. 6A). Since there is no other public RNA-seq cohort for patients with CEBPAbZIP-inf, we performed validation in a public dataset of patients with CEBPAdm. In GSE15210, a total of 83 genes were differentially expressed between CEBPAdm and CEBPAwt patients [10]. Fifty-one (83.6%) out of 61 downregulated genes and 22 (100%) of 22 upregulated genes were also significantly differentially expressed in our RNA-seq data when comparing the transcriptomes between patients with CEBPAdm and CEBPAwt. The major downregulated genes belonged to the homeobox gene family (HOXA9, HOXA10, HOXA5, HOXA3, HOXA6, etc). The results confirmed the concordance of our RNA-seq data with others (Supplementary Fig. 6B).
Association of dysregulated immune and metabolic pathways with short EFS in patients with CEBPA bZIP-inf
Among the 39 patients with CEBPAbZIP-inf who had relapsed, only 8 (20.5%) had WT1 or DNMT3A mutations. Since concurrent mutations could only partly explain the poor clinical outcomes in CEBPAbZIP-inf, we hypothesized that, in addition to mutations, transcriptomic variation underpins a large proportion of functional variation. To elucidate why some patients with CEBPAbZIP-inf still had poor clinical outcomes at the transcriptomic level, we compared the RNA-seq data between CEBPAbZIP-inf patients with short EFS (n = 16, EFS < 2 years) and long EFS (n = 20). There were 138 DEGs with |logFC|>0.585 (absolute fold change >1.5 or <0.67) and P value < 0.05, which could separate the patients well on unsupervised clustering and PCA (Fig. 4A–C and Supplementary Table 7). This finding highlighted the possibility that some genes with lower fold changes were also important determinants of clinical outcomes. The MsigDB Hallmark gene sets showed significant enrichment of immune and metabolic pathways in CEBPAbZIP-inf with short EFS. The enriched immune pathways include IFN alpha and gamma response (NES 1.89, FDR 0.003; NES 1.52, FDR 0.027, respectively). The Reactome gene sets and Gene Ontology on biological process revealed an enrichment of IFN alpha signaling and type I IFN response (Fig. 4D, E and Supplementary Fig. 7A). STING1, IRF2, IRF5, OAS2, IFI35, and FADD were significantly overexpressed in patients with short EFS (Fig. 4F). Higher expression of STING1, IRF5, OAS2, IFI35, and FADD were associated with a trend towards shorter EFS in CEBPAbZIP-inf (Supplementary Fig. 8). To validate the results in an independent cohort, RNA-seq data from 20 CEBPA-mutated patients in the TARGET AML cohort were analyzed. Similarly, gene sets related to response to IFN were significantly enriched in those with shorter EFS (Supplementary Fig. 7B).
A Heat map of normalized gene expression values of the top upregulated and downregulated DEGs in patients with short EFS. The DEGs were listed in Supplementary Table 7. B PCA showing separate clustering of transcriptomes between CEBPAbZIP-inf AML with short and long EFS based on the top DEGs. C Volcano plot showing DEGs with genes downregulated (logFC<0) and upregulated (logFC>0), respectively, in CEBPAbZIP-inf patients with short EFS compared with long EFS. Important genes in the metabolic and immune pathways are highlighted. D Enriched and depleted MSigDB Hallmark and KEGG gene sets between patients with short and long EFS. E GSEA enrichment plots showing overrepresentation of IFN response and metabolic gene set signatures within the CEBPAbZIP-inf transcriptome. F, G Scatter plots illustrating the log2-transformed, CPM-normalized expression values in samples associated with short vs. long EFS for genes involved in IFN response (F) and oxidative phosphorylation (G). *P value < 0.05. H Kaplan–Meier plots for EFS for patients with low expression (blue lines) and high expression (red lines) of NDUFA12, NDUFB6, ATP5PB, ATP5IF1 at diagnosis. The mean expression value for the respective gene in all CEBPAbZIP-inf samples was used to discretize between low and high expression. I High expression of ATP5F1C was associated with significantly shorter OS and EFS in patients with CEBPAbZIP-inf.
The enriched metabolic pathways included oxidative phosphorylation (NES 2.50, FDR 0), fatty acid metabolism (NES 1.82, FDR 0.002), and adipogenesis (NES 1.72, FDR 0.005). The KEGG gene sets also revealed perturbed pathways involving oxidative phosphorylation (NES 2.32, FDR 0) and the proteasome (NES 2.15, FDR 0) (Fig. 4D, E). Molecules in mitochondrial complex I (NDUFB10, NDUFC1, NDUFV2, NDUFAF1, NDUFB5, NDUFA12 and NDUFB6) and complex V (ATP5PB, ATP5F1C and ATP5IF1) were significantly overexpressed in patients with short EFS (Fig. 4G). Higher expression of NDUFA12 (P = 0.029), NDUFB6 (P = 0.047), ATP5PB (P = 0.049), and ATP5IF1 (P = 0.011) was associated with significantly shorter EFS in CEBPAbZIP-inf, and higher expression of ATP5F1C was associated with both significantly shorter OS and EFS (P = 0.022, and P = 0.018, respectively) (Fig. 4H, I). Next, we investigated whether there are different clinic-biological features between those with high or low expression of ATP5F1C, but found no discernible difference with regard to karyotypic changes or mutations (Supplementary Table 8). The prognostic impact of mitochondrial complexes was validated in the TARGET AML cohort, which demonstrated that higher expression of NDUFA12 (P = 0.076) and NDUFB6 (P = 0.013) also correlated with shorter EFS in CEBPA-mutated patients (Supplementary Fig. 9).
We additionally studied the genetic and transcriptomic heterogeneity within the CEBPAnonbZIP-inf group (n = 11). Concurrent GATA2 mutation was associated with a trend towards longer EFS, while TET2 mutation was associated with a trend towards shorter OS, albeit the small number of patients. There were 822 DEGs between CEBPAnonbZIP-inf patients with short and long EFS, of which 24 coincided with the DEGs that differentiated the outcomes in patients with CEBPAbZIP-inf. The detailed enriched pathways on GSEA were shown in Supplementary Fig. 10.
To address the question whether enrichment of interferon signaling and metabolic pathways were associated with shorter EFS in other AML subtypes, we analyzed RNA-seq data from 60 AML patients with NPM1 mutation and compared the transcriptomes between patients with short EFS (EFS < 2 years, n = 34) and those with long EFS (n = 26). Those with short EFS did not have a significant enrichment of metabolic pathways, such as oxidative phosphorylation (NES −1.06, FDR 0.35) and fatty acid metabolism (NES 0.67, FDR 0.97). However, we observed a negative enrichment of interferon signaling (NES -3.34, FDR 0) in those with short EFS (Supplementary Fig. 11). Although enriched interferon signaling pathways have previously been associated with unfavorable prognostic implications in the overall AML cohort [37], our findings suggested that the enriched pathways are context-dependent and differ across various AML subtypes.
Discussion
The current study leverages a large cohort of AML to study the genomic and transcriptomic pattern that contribute to poor survival in AML with CEBPAbZIP-inf, a new disease category in the ELN-2022 and 2022 ICC. We identified WT1 and DNMT3A mutations, dysregulated immune and metabolic signatures that highly correlated with poor survival. We have generated a publicly available database for patients with CEBPAbZIP-inf that contains patient demographics, clinical survival data, RNA-seq, and NGS data, which serves as a resource for the AML research community.
The reports on the prognostic impact of concurrent mutations in CEBPAbZIP-inf have been re limited to GATA2, WT1, and TET2 [4, 18]. Mutant GATA2 is associated with improved survival, while WT1 and TET2 mutations are associated with a lower probability of survival [4]. In our study, concurrent WT1 or DNMT3A mutations were associated with significantly poorer survival. We are the first to report the prognostic relevance of DNMT3A mutations in CEBPAbZIP-inf, and the result has been validated in external databases. However, due to the small number of DNMT3A co-mutant patients in both our cohort and the external databases, further investigation in a larger independent cohort is still warranted to validate these findings. Recently, it was discovered that CEBPA and DNMT3A cooperate to induce leukemogenesis. Mutant CEBPA interacts with the long splice isoform DNMT3A and increase the aberrant methylation especially on PRC2 target genes [38]. We additionally found that CEBPAbZIP-inf patients with WT1 or DNMT3A mutations had improved survival if receiving allo-HSCT in CR1. In addition, WT1-mutated patients had a lower probability of attaining CR2 and displayed a trend towards inferior OS following relapse. These results highlight the importance of utilizing a personalized treatment approach in determining the appropriate timing of allo-HSCT within the ELN-2022 favorable category.
Since WT1 or DNMT3A mutations are presented in only 20.5% patients with CEBPAbZIP-inf who had relapsed, it’s apparent that non-genetic drivers underpin a large proportion of functional variation in leukemia. Indeed, the RNA-seq revealed enrichment of IFN and metabolic pathways in those with short EFS. Cumulative evidences have supported the role of immune microenvironment in AML, and the activation of IFN-related pathways has been shown to have negative prognostic implications in AML [37, 39]. Chronic activation of the IFN signaling pathway actually creates an immune-suppressed environment and mediates resistance to various cancer treatments [40]. In our study, enriched IFN-related pathways also correlated with a poorer survival. Patients with short EFS and long EFS expressed comparable amounts of leukemia-associated antigens CD34, CD123, and CD117 (Supplementary Fig. 12). This observation suggests that bulk BM RNA-seq could capture elements of the tumor immune microenvironment in addition to features of the leukemic cell compartment.
Another notable discovery is the dysregulated metabolism in those with short EFS. The results corroborate the findings that aberrant metabolic states are hallmarks of various types of cancer [41]. Oxidative phosphorylation, involving mitochondria complexes I-V, has been reported to play a role in leukemia stem cell maintenance in myeloid leukemia [42, 43]. Genes in the mitochondrial complexes I and V were most prominently dysregulated and also correlated with survival in the TARGET AML cohort. It’s noteworthy that many genes in the mitochondrial complexes had fold change less than 2, suggesting biologically relevant functions occur even at very low fold changes in RNA levels [44].
The use of bulk BM aspirates is a potential limitation of our analysis. Single-cell RNA-seq or mass cytometry will further elucidate which cell component makes the major contribution. The number of patients with CEBPA mutations was not adequate in the TARGET AML cohort, which potentially decreased the reliability of the external validation. Besides, we did not perform mechanistic studies to elucidate the detailed molecular pathways in CEBPAbZIP-inf. Further studies are needed to investigate whether targeting of these pathways could sensitize the leukemic cells and ultimately reduce relapse in these patients.
In conclusion, our results highlight the importance of the combination of genetic and transcriptomic approaches to fully elucidate the heterogeneities within a WHO-2022/ICC-defined entity. We identified WT1 mutations, DNMT3A mutations, dysregulated interferon and oxidative phosphorylation as negative prognosticators in patients with CEBPAbZIP-inf. We provide a cohort with complete patient demographics, clinical survival, NGS, and RNA-seq data that will nourish further research in the AML community.
Data availability
RNA-seq data can be accessed at NCBI GEO under GSE253086. Clinical and mutation data and computer codes with nonidentifiable patient information are accessible on reasonable request. Evaluation of such requests will be conducted by the corresponding author (H.-A.H.) to determine their appropriateness.
References
Preudhomme C, Sagot C, Boissel N, Cayuela JM, Tigaud I, de Botton S, et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood. 2002;100:2717–23.
Lin LI, Chen CY, Lin DT, Tsay W, Tang JL, Yeh YC, et al. Characterization of CEBPA mutations in acute myeloid leukemia: most patients with CEBPA mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res. 2005;11:1372–9.
Frohling S, Schlenk RF, Stolze I, Bihlmayr J, Benner A, Kreitmeier S, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22:624–33.
Taube F, Georgi JA, Kramer M, Stasik S, Middeke JM, Röllig C, et al. CEBPA mutations in 4708 patients with acute myeloid leukemia: differential impact of bZIP and TAD mutations on outcome. Blood. 2022;139:87–103.
Su L, Tan Y, Lin H, Liu X, Yu L, Yang Y, et al. Mutational spectrum of acute myeloid leukemia patients with double CEBPA mutations based on next-generation sequencing and its prognostic significance. Oncotarget. 2018;9:24970–9.
Konstandin NP, Pastore F, Herold T, Dufour A, Rothenberg-Thurley M, Hinrichsen T, et al. Genetic heterogeneity of cytogenetically normal AML with mutations of CEBPA. Blood Adv. 2018;2:2724–31.
Tien FM, Hou HA, Tang JL, Kuo YY, Chen CY, Tsai CH, et al. Concomitant WT1 mutations predict poor prognosis in acute myeloid leukemia patients with double mutant CEBPA. Haematologica. 2018;103:e510–e3.
Pabst T, Eyholzer M, Fos J, Mueller BU. Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favourable prognosis. Br J cancer. 2009;100:1343–6.
Schmidt L, Heyes E, Grebien F. Gain-of-function effects of N-terminal CEBPA mutations in acute myeloid leukemia. BioEssays. 2020;42:e1900178.
Dufour A, Schneider F, Metzeler KH, Hoster E, Schneider S, Zellmeier E, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol. 2010;28:570–7.
Wouters BJ, Löwenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113:3088–91.
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405.
Wei H, Zhou C, Liu B, Lin D, Li Y, Wei S, et al. The prognostic factors in acute myeloid leukaemia with double-mutated CCAAT/enhancer-binding protein alpha (CEBPAdm). Br J Haematol. 2022;197:442–51.
Tarlock K, Lamble AJ, Wang YC, Gerbing RB, Ries RE, Loken MR, et al. CEBPA-bZip mutations are associated with favorable prognosis in de novo AML: a report from the Children’s Oncology Group. Blood. 2021;138:1137–47.
Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–19.
Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–77.
Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. International Consensus Classification of myeloid neoplasms and acute leukemia: integrating morphological, clinical, and genomic data. Blood. 2022;140:1200–22.
Wakita S, Sakaguchi M, Oh I, Kako S, Toya T, Najima Y, et al. Prognostic impact of CEBPA bZIP domain mutation in acute myeloid leukemia. Blood Adv. 2022;6:238–47.
Lo MY, Tsai XC, Lin CC, Tien FM, Kuo YY, Lee WH, et al. Validation of the prognostic significance of the 2022 European LeukemiaNet risk stratification system in intensive chemotherapy treated aged 18 to 65 years patients with de novo acute myeloid leukemia. Am J Hematol. 2023;98:760–9.
Hou HA, Tsai CH, Lin CC, Chou WC, Kuo YY, Liu CY, et al. Incorporation of mutations in five genes in the revised International Prognostic Scoring System can improve risk stratification in the patients with myelodysplastic syndrome. Blood Cancer J. 2018;8:39.
Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood. 1998;92:2322–33.
Lee WH, Lin CC, Tsai CH, Tseng MH, Kuo YY, Liu MC, et al. Effect of mutation allele frequency on the risk stratification of myelodysplastic syndrome patients. Am J Hematol. 2022;97:1589–98.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–73.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Tien FM, Hou HA, Tsai CH, Tang JL, Chiu YC, Chen CY, et al. GATA2 zinc finger 1 mutations are associated with distinct clinico-biological features and outcomes different from GATA2 zinc finger 2 mutations in adult acute myeloid leukemia. Blood Cancer J. 2018;8:87.
Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74.
Bolouri H, Farrar JE, Triche T Jr., Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24:103–12.
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21.
Tazi Y, Arango-Ossa JE, Zhou Y, Bernard E, Thomas I, Gilkes A, et al. Unified classification and risk-stratification in acute myeloid leukemia. Nat Commun. 2022;13:4622.
Hou HA, Lin CC, Chou WC, Liu CY, Chen CY, Tang JL, et al. Integration of cytogenetic and molecular alterations in risk stratification of 318 patients with de novo non-M3 acute myeloid leukemia. Leukemia. 2014;28:50–8.
Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, Hansen AS, Gouw AM, Felsher DW. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol. 2022;19:23–36.
Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–97.
Ahmadi SE, Rahimi S, Zarandi B, Chegeni R, Safa M. MYC: a multipurpose oncogene with prognostic and therapeutic implications in blood malignancies. J Hematol Oncol. 2021;14:121.
Vadakekolathu J, Minden MD, Hood T, Church SE, Reeder S, Altmann H, et al. Immune landscapes predict chemotherapy resistance and immunotherapy response in acute myeloid leukemia. Sci Transl Med. 2020;12:eaaz0463.
Chen X, Zhou W, Song RH, Liu S, Wang S, Chen Y, et al. Tumor suppressor CEBPA interacts with and inhibits DNMT3A activity. Sci Adv. 2022;8:eabl5220.
Mulherkar N, Scadden DT. What is the role of the bone marrow microenvironment in AML? Best Pract Res Clin Haematol. 2021;34:101328.
Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci USA. 2008;105:18490–5.
Kim J, DeBerardinis RJ. Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metab. 2019;30:434–46.
de Beauchamp L, Himonas E, Helgason GV. Mitochondrial metabolism as a potential therapeutic target in myeloid leukaemia. Leukemia. 2022;36:1–12.
Saito K, Zhang Q, Yang H, Yamatani K, Ai T, Ruvolo V, et al. Exogenous mitochondrial transfer and endogenous mitochondrial fission facilitate AML resistance to OxPhos inhibition. Blood Adv. 2021;5:4233–55.
St Laurent G, Shtokalo D, Tackett MR, Yang Z, Vyatkin Y, Milos PM, et al. On the importance of small changes in RNA expression. Methods. 2013;63:18–24.
Acknowledgements
We would like to acknowledge the service provided by the DNA Sequencing Core of the First Core Laboratory, National Taiwan University College of Medicine. This work was partially sponsored by grants from the Ministry of Science and Technology (Taiwan) (MOST 104–2314-B-002–128-MY4, 106-2314-B-002-226-MY3, 108-2628-B-002-015, 109-2314-B-002-213, 111-2314-B-002-279 and 112-2314-B-002-197) and the Ministry of Health and Welfare (Taiwan) (MOHW 107-TDU-B-211-114009 and 111-TDU-B-221-114001).
Author information
Authors and Affiliations
Contributions
F.-M.T. was responsible for study design, literature collection, data management and interpretation, bioinformatics analysis, statistical analysis and manuscript writing; Y.-C.Y. and X.-C.T. were responsible for bioinformatics analysis; Y.-Y.K., M.-H.T. and L.-I.L. were responsible for mutation analysis and interpretation; M.-Y.L., C.-Y.C., W.-H.L., C.-C.L., Y.-S.W., B.-S.K., M.-Y., J.-L.T. and W.-C.C. contributed patient samples and clinical data; Y.-L.P., M.-H.T. and M.-C.L. performed the gene mutation and chromosomal studies. H.-F.T. coordinated the study and wrote and revised the manuscript. H.-A.H. planned, designed, conceived and coordinated the study, analyzes the statistics and wrote and revised the manuscript. All authors provided final approval of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tien, FM., Yao, CY., Tsai, X.CH. et al. Dysregulated immune and metabolic pathways are associated with poor survival in adult acute myeloid leukemia with CEBPA bZIP in-frame mutations. Blood Cancer J. 14, 15 (2024). https://doi.org/10.1038/s41408-023-00975-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41408-023-00975-8
This article is cited by
-
CEBPA mutations in acute myeloid leukemia: implications in risk stratification and treatment
International Journal of Hematology (2024)