Abstract
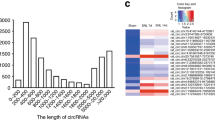
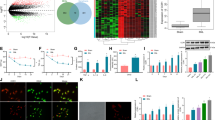
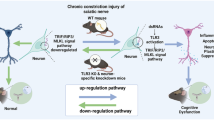
Circular RNAs (ciRNAs) are emerging as new players in the regulation of gene expression. However, how ciRNAs are involved in neuropathic pain is poorly understood. Here, we identify the nervous-tissue-specific ciRNA-Fmn1 and report that changes in ciRNA-Fmn1 expression in spinal cord dorsal horn neurons play a key role in neuropathic pain after nerve injury. ciRNA-Fmn1 was significantly downregulated in ipsilateral dorsal horn neurons after peripheral nerve injury, at least in part because of a decrease in DNA helicase 9 (DHX9), which regulates production of ciRNA-Fmn1 by binding to DNA-tandem repeats. Blocking ciRNA-Fmn1 downregulation reversed nerve-injury-induced reductions in both the binding of ciRNA-Fmn1 to the ubiquitin ligase UBR5 and the level of ubiquitination of albumin (ALB), thereby abrogating the nerve-injury-induced increase of ALB expression in the dorsal horn and attenuating the associated pain hypersensitivities. Conversely, mimicking downregulation of ciRNA-Fmn1 in naïve mice reduced the UBR5-controlled ubiquitination of ALB, leading to increased expression of ALB in the dorsal horn and induction of neuropathic-pain-like behaviors in naïve mice. Thus, ciRNA-Fmn1 downregulation caused by changes in binding of DHX9 to DNA-tandem repeats contributes to the genesis of neuropathic pain by negatively modulating UBR5-controlled ALB expression in the dorsal horn.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
References
Treede RD, Rief W, Barke A, Aziz Q, Bennett MI, Benoliel R, et al. Chronic pain as a symptom or a disease: the IASP Classification of Chronic Pain for the International Classification of Diseases (ICD-11). Pain. 2019;160:19–27.
Cheng L, Duan B, Huang T, Zhang Y, Chen Y, Britz O, et al. Identification of spinal circuits involved in touch-evoked dynamic mechanical pain. Nat Neurosci. 2017;20:804–14.
Zheng Q, Xie W, Lückemeyer DD, Lay M, Wang XW, Dong X, et al. Synchronized cluster firing, a distinct form of sensory neuron activation, drives spontaneous pain. Neuron. 2022;110:209–20.e6.
Pan Z, Du S, Wang K, Guo X, Mao Q, Feng X, et al. Downregulation of a dorsal root ganglion-specifically enriched long noncoding RNA is required for neuropathic pain by negatively regulating RALY-triggered Ehmt2 expression. Adv Sci. 2021;8:e2004515.
Huang Y, Chen SR, Chen H, Luo Y, Pan HL. Calcineurin inhibition causes α2δ-1-mediated tonic activation of synaptic NMDA receptors and pain hypersensitivity. J Neurosci. 2020;40:3707–19.
Araldi D, Bonet IJM, Green PG, Levine JD. Contribution of G protein alpha subunits to analgesia, hyperalgesia and hyperalgesic priming induced by sub-analgesic and analgesic doses of fentanyl and morphine. J Neurosci. 2022;42:1196–210.
Pan Z, Li GF, Sun ML, Xie L, Liu D, Zhang Q, et al. MicroRNA-1224 splicing circularRNA-Filip1l in an Ago2-dependent manner regulates chronic inflammatory pain via targeting Ubr5. J Neurosci. 2019;39:2125–43.
Chen G, Zhang YQ, Qadri YJ, Serhan CN, Ji RR. Microglia in pain: detrimental and protective roles in pathogenesis and resolution of pain. Neuron. 2018;100:1292–311.
Su S, Yudin Y, Kim N, Tao YX, Rohacs T. TRPM3 channels play roles in heat hypersensitivity and spontaneous pain after nerve injury. J Neurosci. 2021;41:2457–74.
Mazzitelli M, Neugebauer V. Amygdala group II mGluRs mediate the inhibitory effects of systemic group II mGluR activation on behavior and spinal neurons in a rat model of arthritis pain. Neuropharmacology. 2019;158:107706.
Chen L, Kong R, Wu C, Wang S, Liu Z, Liu S, et al. Circ-MALAT1 functions as both an mRNA translation brake and a microRNA sponge to promote self-renewal of hepatocellular cancer stem cells. Adv Sci. 2020;7:1900949.
Qu X, Li Z, Chen J, Hou L. The emerging roles of circular RNAs in CNS injuries. J Neurosci Res. 2020;98:1485–97.
Huang S, Li X, Zheng H, Si X, Li B, Wei G, et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–76.
Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–85.
Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci. 2019;22:1903–12.
Lukiw WJ. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet. 2013;4:307.
Doxakis E. Insights into the multifaceted role of circular RNAs: implications for Parkinson’s disease pathogenesis and diagnosis. NPJ Parkinsons Dis. 2022;8:7.
Zurawska A, Mycko MP, Selmaj KW. Circular RNAs as a novel layer of regulatory mechanism in multiple sclerosis. J Neuroimmunol. 2019;334:576971.
Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R, et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci. 2018;38:32–50.
Zhang SB, Lin SY, Liu M, Liu CC, Ding HH, Sun Y, et al. CircAnks1a in the spinal cord regulates hypersensitivity in a rodent model of neuropathic pain. Nat Commun. 2019;10:4119.
Cai W, Zhang Y, Su Z. ciRS-7 targeting miR-135a-5p promotes neuropathic pain in CCI rats via inflammation and autophagy. Gene. 2020;736:144386.
Spires TL, Hannan AJ. Molecular mechanisms mediating pathological plasticity in Huntington’s disease and Alzheimer’s disease. J Neurochem. 2007;100:874–82.
Jimenez-Sanchez M, Licitra F, Underwood BR, Rubinsztein DC. Huntington’s Disease: mechanisms of pathogenesis and therapeutic strategies. Cold Spring Harb Perspect Med. 2017;7:a024240.
Sellier C, Buijsen RAM, He F, Natla S, Jung L, Tropel P, et al. Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron. 2017;93:331–47.
Faruq M, Scaria V, Singh I, Tyagi S, Srivastava AK, Mukerji M. SCA-LSVD: a repeat-oriented locus-specific variation database for genotype to phenotype correlations in spinocerebellar ataxias. Hum Mutat. 2009;30:1037–42.
Dewan R, Chia R, Ding J, Hickman RA, Stein TD, Abramzon Y, et al. Pathogenic Huntingtin repeat expansions in patients with frontotemporal dementia and amyotrophic lateral sclerosis. Neuron. 2021;109:448–60.e4.
Matsuura T, Fang P, Pearson CE, Jayakar P, Ashizawa T, Roa BB, et al. Interruptions in the expanded ATTCT repeat of spinocerebellar ataxia type 10: repeat purity as a disease modifier? Am J Hum Genet. 2006;78:125–9.
Burrell JR, Halliday GM, Kril JJ, Ittner LM, Götz J, Kiernan MC, et al. The frontotemporal dementia-motor neuron disease continuum. Lancet. 2016;388:919–31.
Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–7.
Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, et al. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–80.
Hannan AJ. Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet. 2018;19:286–98.
Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–7.
Piché J, Van Vliet PP, Pucéat M, Andelfinger G. The expanding phenotypes of cohesinopathies: one ring to rule them all! Cell Cycle. 2019;18:2828–48.
Burberry A, Wells MF, Limone F, Couto A, Smith KS, Keaney J, et al. C9orf72 suppresses systemic and neural inflammation induced by gut bacteria. Nature. 2020;582:89–94.
Akta T, Avşar Ilık İ, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–9.
Pan Z, Zhang Q, Liu X, Zhou H, Jin T, Hao LY, et al. Methyltransferase-like 3 contributes to inflammatory pain by targeting TET1 in YTHDF2-dependent manner. Pain. 2021;162:1960–76.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–87.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Pan Z, Zhang M, Ma T, Xue ZY, Li GF, Hao LY, et al. Hydroxymethylation of microRNA-365-3p regulates nociceptive behaviors via Kcnh2. J Neurosci. 2016;36:2769–81.
Pan Z, Xue ZY, Li GF, Sun ML, Zhang M, Hao LY, et al. DNA hydroxymethylation by ten-eleven translocation methylcytosine dioxygenase 1 and 3 regulates nociceptive sensitization in a chronic inflammatory pain model. Anesthesiology. 2017;127:147–63.
Jiang BC, Cao DL, Zhang X, Zhang ZJ, He LN, Li CH, et al. CXCL13 drives spinal astrocyte activation and neuropathic pain via CXCR5. J Clin Invest. 2016;126:745–61.
Hugel S, Schlichter R. Presynaptic P2X receptors facilitate inhibitory GABAergic transmission between cultured rat spinal cord dorsal horn neurons. J Neurosci. 2000;20:2121–30.
Bagshaw ATM. Functional mechanisms of microsatellite DNA in eukaryotic genomes. Genome Biol Evol. 2017;9:2428–43.
Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–73.
Kwok A, Raulf N, Habib N. Developing small activating RNA as a therapeutic: current challenges and promises. Ther Deliv. 2019;10:151–64.
Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18:119.
Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y, et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer. 2019;18:158.
Wang X, Xing L, Yang R, Chen H, Wang M, Jiang R, et al. The circACTN4 interacts with FUBP1 to promote tumorigenesis and progression of breast cancer by regulating the expression of proto-oncogene MYC. Mol Cancer. 2021;20:91.
Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–80.e21.
Gao YJ, Ji RR. c-Fos and pERK, which is a better marker for neuronal activation and central sensitization after noxious stimulation and tissue injury? Open Pain J. 2009;2:11–7.
Gao YJ, Ji RR. Light touch induces ERK activation in superficial dorsal horn neurons after inflammation: involvement of spinal astrocytes and JNK signaling in touch-evoked central sensitization and mechanical allodynia. J Neurochem. 2010;115:505–14.
Lee T, Pelletier J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget. 2016;7:42716–39.
Ding X, Jia X, Wang C, Xu J, Gao SJ, Lu C. A DHX9-lncRNA-MDM2 interaction regulates cell invasion and angiogenesis of cervical cancer. Cell Death Differ. 2019;26:1750–65.
Du WW, Xu J, Yang W, Wu N, Li F, Zhou L, et al. A neuroligin isoform translated by circNlgn contributes to cardiac remodeling. Circ Res. 2021;129:568–82.
Li Y, Chen B, Zhao J, Li Q, Chen S, Guo T, et al. HNRNPL circularizes ARHGAP35 to produce an oncogenic protein. Adv Sci. 2021;8:2001701.
Rozga J, Piątek T, Małkowski P. Human albumin: old, new, and emerging applications. Ann Transpl. 2013;18:205–17.
Mendez CM, McClain CJ, Marsano LS. Albumin therapy in clinical practice. Nutr Clin Pract. 2005;20:314–20.
Bihari S, Bannard-Smith J, Bellomo R. Albumin as a drug: its biological effects beyond volume expansion. Crit Care Resusc. 2020;22:257–65.
Rastogi RP, Singh SP, Häder DP, Sinha RP. Detection of reactive oxygen species (ROS) by the oxidant-sensing probe 2’,7’-dichlorodihydrofluorescein diacetate in the cyanobacterium Anabaena variabilis PCC 7937. Biochem Biophys Res Commun. 2010;397:603–7.
Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9:5068.
Adachi M, Koyama H, Long Z, Sekine M, Furuchi T, Imai K, et al. L-Glutamate in the extracellular space regulates endogenous D-aspartate homeostasis in rat pheochromocytoma MPT1 cells. Arch Biochem Biophys. 2004;424:89–96.
Weissberg I, Wood L, Kamintsky L, Vazquez O, Milikovsky DZ, Alexander A, et al. Albumin induces excitatory synaptogenesis through astrocytic TGF-β/ALK5 signaling in a model of acquired epilepsy following blood-brain barrier dysfunction. Neurobiol Dis. 2015;78:115–25.
Senatorov VV, Jr., Friedman AR, Milikovsky DZ, Ofer J, Saar-Ashkenazy R, Charbash A, et al. Blood-brain barrier dysfunction in aging induces hyperactivation of TGFβ signaling and chronic yet reversible neural dysfunction. Sci Transl Med. 2019;11:eaaw8283.
Schukur L, Zimmermann T, Niewoehner O, Kerr G, Gleim S, Bauer-Probst B, et al. Identification of the HECT E3 ligase UBR5 as a regulator of MYC degradation using a CRISPR/Cas9 screen. Sci Rep. 2020;10:20044.
Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grøfte M, et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709.
Hu X, Pickering EH, Hall SK, Naik S, Liu YC, Soares H, et al. Genome-wide association study identifies multiple novel loci associated with disease progression in s ubjects with mild cognitive impairment. Transl Psychiatry. 2011;1:e54.
Rutz S, Kayagaki N, Phung QT, Eidenschenk C, Noubade R, Wang X, et al. Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature. 2015;518:417–21.
Christensen T, Jensen L, Bouzinova EV, Wiborg O. Molecular profiling of the lateral habenula in a rat model of depression. PLoS One. 2013;8:e80666.
Acknowledgements
The work was supported by grants from the National Natural Science Foundation of China (82171234, 81971041, 81671096 to ZQP; 82171233 to HJW; 82201391 to QHW; 32200818 to LY; 81901132 to ZYX); Jiang Su-Specially Appointed Professor Project, Natural Science Foundation of Jiangsu Province (BK20201460), and Key project of the Natural Science Foundation of Jiangsu Education Department (22KJA320008), and Key Project of Shanghai Chongming District (CKY2019-1).
Author information
Authors and Affiliations
Contributions
ZQP, conceived the project. ZQP and JLC supervised all experiments. XDL, YT, TJ, HLZ, MZ, and LYH performed the experiments; XDL, YT, SYL, YH, QQL, KHY, RNW, HLZ, MZ, QHW, LY, HJW, FQL, WS and ZQP analyzed data; XDL, HJW, and ZQP wrote the draft of paper. YXT discussed the results and edited the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Xd., Jin, T., Tao, Y. et al. DHX9/DNA-tandem repeat-dependent downregulation of ciRNA-Fmn1 in the dorsal horn is required for neuropathic pain. Acta Pharmacol Sin 44, 1748–1767 (2023). https://doi.org/10.1038/s41401-023-01082-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-023-01082-x