Abstract
CKLF (chemokine-like factor)-MARVEL transmembrane domain containing protein 6 (CMTM6) is a novel regulator to maintain the stability of PD-L1. CMTM6 can colocalize and interact with PD-L1 on the recycling endosomes and cell membrane, preventing PD-L1 from lysosome-mediated degradation and proteasome-mediated degradation thus increasing the half-life of PD-L1 on the cell membrane. The difficulties in obtaining stable full-length PD-L1 and CMTM6 proteins hinder the research on their structures, function as well as related drug development. Using lauryl maltose neopentyl glycol (LMNG) as the optimized detergent and a cell membrane mimetic strategy, we assembled a stable membrane-bound full-length CMTM6-PD-L1 complex with amphipol A8-35. When the PD-1/PD-L1-CMTM6 interactions were analyzed, we found that CMTM6 greatly enhanced the binding and delayed the dissociation of PD-1/PD-L1, thus affecting immunosuppressive signaling and anti-apoptotic signaling. We then used the CMTM6-PD-L1 complex as immunogens to generate immune repertoires in camels, and identified a functional anti-CMTM6 nanobody, called 1A5. We demonstrated that the anti-CMTM6 nanobody greatly decreased T-cell immunosuppression and promoted apoptotic susceptibility of tumor cells in vitro, and mainly relied on the cytotoxic effect of CD8+ T-cells to exert tumor growth inhibitory effects in CT26 tumor-bearing mice. In conclusion, the stable membrane-bound full-length CMTM6-PD-L1 complex has been successfully used in studying PD-1/PD-L1-CMTM6 interactions and CMTM6-targeting drug development, suggesting CMTM6 as a novel tumor immunotherapy target.
Similar content being viewed by others
Introduction
Targeting programmed death ligand 1 (PD-L1) expression and regulatory pathway-related molecules is a promising approach to improve the efficacy and response rate of anti-PD-L1 therapy. CKLF (chemokine-like factor)-MARVEL transmembrane domain containing protein 6 (CMTM6), a type 3 membrane protein characterized by a MARVEL domain with four transmembrane helices [1, 2], was identified as a novel regulator to maintain the stability of membranal PD-L1 [3, 4] through haploid genetic screening and CRISPR-Cas9 knockout library screening. CMTM6 can colocalize and interact with PD-L1 on the recycling endosomes and cell membrane, preventing PD-L1 from lysosome-mediated degradation and proteasome-mediated degradation to increase the half-life of PD-L1 on the cell membrane [3, 4].
CMTM6 has recently been reported to be associated with the progression of multiple malignancies and patient prognosis [5,6,7,8,9,10,11]. And CMTM6 was reported to be involved in DNA methylation of hepatocellular carcinoma, maintain cancer genome stability, drive cisplatin resistance and induce M2 macrophages polarization [12,13,14,15]. In addition, the level of tumor CMTM6 has been reported to be associated with immunotherapy effects [7, 8, 16]. These works indicate that CMTM6 is a promising novel target for tumor therapy.
Difficulties in obtaining full-length PD-L1 and CMTM6 membrane protein hinder the investigation of PD-1/PD-L1/CMTM6 interaction, the structure analysis of intact PD-L1 or CMTM6, the understanding of PD-L1 and CMTM6 intracellular signaling, and the development of CMTM6-targeting drugs. Cell membrane mimics and nanobodies are commonly applied to assist in the research of membrane protein interactions. Providing a membrane-like environment based on the reconstitution of membrane proteins in lipid bilayers represents a straightforward approach to better understanding the biological function of these proteins. The membrane mimetic system has been effectively utilized to substitute detergents after the solubilization step and preserve the native state of membrane proteins in detergent-free solutions [17,18,19]. Nanobodies are frequently used in protein crystallography and cell biology due to their small size, high affinity, target specificity, thermal stability, and reversible folding; conformation-specific nanobodies targeting membrane proteins can manipulate cells directly on the cell surface and are exquisite tools for basic science and drug discovery [20, 21].
In this study, we aim to overcome the instability of transmembrane PD-L1 and CMTM6 and provide stable CMTM6-PD-L1 complexes for the mechanism exploration, structure analysis, and drug development of CMTM6.
Materials and methods
Cell lines
HEK293F was purchased from Thermo Fisher and grown in FreeStyle™ 293 expression medium (12338018, Thermo Fisher Scientific, MA, USA). HEK293S, Jurkat, RKO, BxPC-3, and CT26 cell lines were purchased from the American Type Culture Collection (ATCC, MD, USA). HEK293S cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; MA0212, Meilunbio, Dalian, China) supplemented with 1% penicillin/streptomycin (15140122, Invitrogen, MA, USA) and 10% heat-inactivated fetal bovine serum (FBS; 10100147, Life Technologies, MA, USA). Jurkat, CT26, and BxPC-3 cells were cultured in RPMI-1640 medium (MA0215, Meilunbio, Dalian, China) containing 1% penicillin/streptomycin and 10% FBS. RKO cells were cultured in Minimum Essential Medium (MEM; MA0217, Meilunbio, Dalian, China) supplemented with 1% penicillin/streptomycin and 10% FBS. The CT26/hPD-L1 cell line was provided by GemPharmatech (Nanjing, China).
Animals
Female BALB/c mice (6–8 weeks old) were purchased from Shanghai Sipp-BK Laboratory Animal Co. Ltd. All mice were maintained under specific pathogen-free (SPF) conditions in the animal facility of the Shanghai Institute of Materia Medica, Chinese Academy of Sciences. BALB/c-hPD-1/hPD-L1 mice were purchased from GemPharmatech Co., Ltd. and housed in SPF condition (GemPharmatech, Nanjing, China). Animal care and experiments were performed in accordance with GemPharmatech and the Shanghai Institute of Materia Medica, using protocols approved by the Institutional Laboratory Animal Care and Use Committee (IACUC).
Protein expression and purification
The cDNA of full-length human PD-L1 (NCBI reference sequence NM_014143.3) and human CMTM6 (NCBI reference sequence NM_026036.3) were subcloned separately into the CMV vector. A C-terminal Flag tag was fused to CMTM6. PD-L1 and CMTM6 were co-expressed in HEK293S cells (2.0 × 106 cells/mL), then were infected with two separate recombinant baculoviruses, each containing the gene for one of them at a 1:1.5 multiplicity of infection for 48 h at 30 °C with rotation at 135 rpm.
For purification of the CMTM6-PD-L1 complex, collected cell pellets were thawed and resuspended in a buffer containing 20 mM of HEPES, 150 mM of NaCl, 10% (v/v) of glycerol and a protease inhibitor cocktail. The suspension was pressurized with 500 p.s.i N2 for 30 min in an EmulsiflexC3 homogenizer (AVESTIN, Ottawa, Canada). After depressurization, the lysate was centrifuged to remove nuclei and unlysed cells, and then ultracentrifuged at 180,000 × g for 1 h. The pelleted membranes were solubilized in a buffer consisting of 20 mM of HEPES, 150 mM of NaCl, 10% (v/v) of glycerol, 1% (w/v) of LMNG, and 0.2% (w/v) of CHS for 2 h with constant tiring. The solubilized material was then clarified by ultra-centrifugation for 1 h at 180,000 × g. The clarified solubilized material was loaded onto the 3 mL M2 flag column pre-equilibrated in buffer A: 20 mM of HEPES, 150 mM of NaCl, 10% (v/v) of glycerol, 0.05% (w/v) of LMNG, and 0.01% (w/v) of CHS at a flow rate of 0.5 mL/min. The column was washed with 100 mL of buffer A before elution in buffer A supplemented with 0.2 mg/mL of flag peptide. Elution fractions were pooled, protein complex purity was analyzed by SDS-PAGE, then was concentrated and subjected to size exclusion chromatography (SEC; Superdex 200 Increase 10/300 column; GE Healthcare, MA, USA). The peak fractions were pooled and concentrated, and biotinylated A8-35 (BAM01, Anatrace, OH, USA) was added at a 3:1 molar ratio (A8-35:CMTM6-PD-L1 complex) and incubated for 3 h at 4 °C, then biobeads were added and incubated for 6 h at 4 °C. The mixture was concentrated, and protein monodispersity was assayed by SEC.
Llama immunization and library construction
One adult camelid (Camelus bactrianus; approved and under the supervision of the Shanghai Institute of Materia Medica, Chinese Academy of Sciences) was immunized subcutaneously seven times with HEK293S cells (~108) coinfected with human PD-L1 and human CMTM6, and peripheral blood lymphocytes (200 mL) were isolated 3–5 days after the last immunization. The nanobody phage library was generated with at least 108 transformants as previously described.
Selection of nanobodies by phage display
Before each round of panning, rescue and amplify nanobody-displaying phage particles by adding helper phage. The phage library grown at 37 °C with shaking (200 rpm) to an OD600 of between 0.4 and 0.5 was infected with ~20-fold helper phage for 30 min at 37 °C without shaking. Centrifuge the infected cells at 2800 × g for 10 min at room temperature (RT), and discard the supernatant carefully to remove traces of glucose. Resuspend the cell pellet in 50 mL of 2YT supplemented with 100 µg/mL of ampicillin and 25 µg/mL of kanamycin into a 250 mL baffled Erlenmeyer flask. Incubate overnight at 37 °C and 200 rpm for the amplification of nanobody-displaying phage. Pellet the phage particles by mixing 10% (w/v) PEG3000/1.25 M NaCl solution, then resuspend the precipitated phage particles in ice-cold PBS and centrifuge several times to completely remove bacterial contaminants.
The nanobody selection was performed on MaxiSorp plates (436110, Nunc, MA, USA) coated with 10 µg/mL of neutravidin overnight at 4 °C. The coated wells were blocked with 2% (v/v) milk/PBS for 2 h at RT, biotin-A8-35-CMTM6-PD-L1 was added and immobilized on the plates with shaking, and then phages were incubated for 30 min at 4 °C with shaking. Wells were then washed 10 times with PBS, and bound phages were finally eluted with a 0.25 mg/mL of trypsin solution for 30 min at RT with shaking. To allow recovery of the phage that eluted from each well, infect exponentially grown 3 mL of TG1 cells (200123, Agilent, CA, USA) in a 50 mL Falcon tube with 50 µL of eluted phage. To achieve target-specific phage enrichment, consecutive rounds of selection may be needed.
Spread the overnight-grown phage infected TG1 cells over culture plates and incubate overnight at 37 °C. Individual TG1 colonies from plates were picked and grown in a 96-well round-bottom culture plate filled with 1 mL of 2YT. After 3 h growth, induce nanobody expression by adding 1 mM IPTG (I6758, Sigma, MO, USA). Pellet the cell, then thaw and freeze several times to release the nanobodies from the periplasm. Then we could detect nanobody binding by ELISAs.
Nanobody expression and purification
The purified plasmids were transformed into the Top10F’ competent (C303003, Invitrogen, MA, USA) and coated on LB plates containing 100 µg/mL of ampicillin at 37 °C overnight. A single clone was selected and grown overnight, and then inoculated into 1 L medium at 37 °C and 220 rpm until the OD value reached 0.6–0.8. Nanobody expression was induced with 1 mM IPTG and the clone was grown overnight at 220 rpm and 28 °C. On the second day, the bacteria were collected by centrifugation at 4500 rpm at RT for 15 min, followed by ultrasonication, and then purified by a pernickel affinity chromatography column.
Biolayer interferometry binding assays
The binding kinetics of nanobody 1A5 with the CMTM6-PD-L1 complex was analyzed by biolayer interferometry using an Octet-Red96 device (Pall ForteBio, CA, USA). Purified nanobody 1A5 (20 µg/mL) buffered in 20 mM of HEPES, 150 mM of NaCl was immobilized onto activated Ni-NTA biosensors and incubated with twofold serial dilutions of biotin-A8-35-CMTM6-PD-L1 in running buffer. Fitting curves were constructed using ForteBio Data Analysis software. The affinity measurement of 1A5 and PD-L1 ECD, PD-1 and CMTM6-PD-L1 complex, PD-1 and PD-L1 ECD was also applied to the procedure. For the competition assay, biotin-A8-35-CMTM6-PD-L1 was bound to streptavidin sensors. The association of the nanobody 1A5 was measured for 300 s at 37 °C, and then PD-1 was added in the presence of 1A5, and vice versa.
Construction of CMTM6 knockout RKO cells and PD-L1 knockout RKO cells
CMTM6/PD-L1 knockout RKO cells can be achieved by using lentiviral vectors [3, 4]. Briefly, hCMTM6 sgRNA (5′-CCGGGTCCTCCTCCGTAGTG-3′) and hPD-L1 sgRNA (5′-ACTGCTTGTCCAGATGACTT-3′) were subcloned into the LentiCRISPRv2 lentiviral expression vector (VT8107, Youbio, Changsha, China) and transfected into HEK293T cells with the package plasmids. Then the virosomes were collected and infected into RKO cells. CMTM6 knockout RKO cells and PD-L1 knockout RKO cells were obtained after puromycin resistance selection. PD-L1 knockout RKO cells (3 × 105 cells) were incubated with the Hu CD274 PE MIH1 (557924, BD Biosciences, CA, USA) and PD-L1 signals were detected by flow cytometry (ACEA NovoCyte, CA, USA). CMTM6 knockout RKO cells were incubated with the Anti-CMTM6 antibody (ab264067, Abcam, MA, USA) and Goat Anti-Rabbit IgG H&L (Alexa Fluor 488; ab150077, Abcam, MA, USA) and CMTM6 signals were detected by flow cytometry.
Jurkat effector cells reporter assay
Jurkat-NFAT-Luciferase cells were constructed by stable transfection of pLV-NFAT-RE-Luci by lentivirus. Jurkat-NFAT-Luciferase cells were stimulated with 50 ng/mL of phorbol-12-myristate-13-acetate (PMA) and 1 µg/mL of ionomycin for 18 h. Activation markers and PD-1 were detected by flow cytometry. In 24-well plates, each well was added with different RKO cells (300 μL, 3 × 105 cells/mL; 100 ng/mL human IFN-γ induced for 24 h). After 24 h, Jurkat-NFAT-Luciferase cells (300 μL, 3 × 105 cells/mL) were added into the wells. After co-culture of cells for 24 h, cells were collected and 120 μL of cell lysate (11401ES60, Yeasen, Shanghai, China) was added. The lysate, after sufficient lysis, was centrifuged at 10,000–15,000 rpm for 3–5 min. Firefly luciferase assay reagent (100 μL) was added to the supernatant. The solution was mixed 2–3 times (no vortex mixing) before measuring the RLU (relative light unit) in an automatic microplate reader SpectraMax (Molecular Devices, CA, USA).
Apoptosis assay
The apoptosis assay was modified from the published literature [22]. To test the blocking activity of CMTM6 deficiency, 10 μg/mL of PD-1-Fc or IgG was coated on 24 well-plates for 18 h at 4 °C. Cas9 control RKO cells or CMTM6 knockout RKO cells or PD-L1 knockout RKO cells (500 μL, 5 × 105 cells/mL) were added into the 24 well-plates and cultured with staurosporine (STP; 1 μM, Meilunbio, Dalian, China) for 18 h. Cells (5 × 105) were harvested and stained with Annexin V-FITC/PI (40302ES20, Yeasen, Shanghai, China) at 25 °C for 15 min. After being washed, Annexin V-FITC and PI signals of cells were detected by flow cytometry. To test the blocking activity of 1A5, different proteins were coated on 24 well-plates for 18 h at 4 °C (10 μg/mL of IgG, 10 μg/mL of IgG and 10 μg/mL of PD-1-Fc, 10 μg/mL of 1A5 and 10 μg/mL of PD-1-Fc, 10 μg/mL of KN035 and 10 μg/mL of PD-1-Fc).
Detection of 1A5 binding to CMTM6 expressed HEK293S cells
HEK293S cells were infected with human or mouse CMTM6 recombinant baculovirus. HEK293S/hCMTM6 or HEK293S/mCMTM6 cells (2.5 × 105) and 2 μg/mL of 1A5 were incubated at 4 °C for 20 min. After being washed, cells were stained with HA-Tag Rabbit mAb (PE Conjugate; 14904 S, CST, MA, USA) solution for 20 min at 4 °C. After being washed, data were obtained by flow cytometry.
In vivo treatment
To evaluate the antitumor activity of 1A5, CT26 tumor cells (1 × 106 cells) were subcutaneously inoculated into each female BALB/c mouse. Mice were randomly grouped into 2 groups of 10 mice each, including the vehicle group (PBS) and the 1A5 group. 1A5 was administered subcutaneously at a dose of 100 μg/each every 2 days for a total of five doses. Tumors were measured every day. Tumor volume was calculated as tumor volume (mm3) = tumor length × width × width/2, and tumor growth curves were plotted. To evaluate the activity of 1A5 on CMTM6 knockout tumors, CMTM6 knockout CT26 cells (sgRNA sequence: 5′-CCTGGCCGCCTACTTCGTCC-3′) were constructed [3, 4]. CMTM6 knockout CT26 tumor cells (1 × 106 cells) were subcutaneously inoculated into each female BALB/c mouse. Mice were randomly grouped into 2 groups of 8 mice each, including the vehicle group (PBS) and the 1A5 group. 1A5 was administered subcutaneously at a dose of 100 μg/each every 2 days for a total of seven doses. To evaluate whether the antitumor activity of 1A5 depends on CD8+ T cells, 6–8-week-old BALB/c mice were randomly divided into 3 groups, with 5 animals in each group. Mice were inoculated subcutaneously with CT26 cells (1 × 106; induced by 100 ng/mL IFN-γ for 24 h). The corresponding groups were the vehicle group, 1A5 group, and 1A5 + anti-CD8 group. CD8+ T cells were deleted by intraperitoneal injection of anti-CD8 antibodies (100763, Biolegend, CA, China) on day 3 (200 μg/each), day 4 (100 μg/each) and day 5 (100 μg/each). To evaluate the antitumor activity of 1A5 in humanized mice, six- to eight-week-old dual-humanized BALB/c mice were inoculated with CT26/hPD-L1 tumor cells (5 × 105 cells, 100 μL) subcutaneously on the right side of the back of the mice above the thigh. When the average tumor volume reached 80–120 mm3, the mice were randomly divided into two groups (hIgG1 group and 1A5-Fc group) according to the tumor volume, with six mice in each group. hIgG1 was administered at a dose of 130 μg/each, while 1A5-Fc was administered at a dose of 200 μg/each. The dose was performed every 3 days for a total of six doses, during which tumor growth was observed and recorded.
Immunophenotype analysis of tumor-bearing mice
Tumor tissues (180 mg) were minced and digested by hyaluronidase (20426ES60, Yeasen, Shanghai, China) and collagenase IV (40510ES60, Yeasen, Shanghai, China) into single cell suspensions and then were subjected to cell extraction by lymphocyte separation medium (7211011, Dakewe, Shenzhen, China) for T cell sorting and to remove erythrocytes and debris from the sample suspension by sterile erythrocyte lysis solution (40401ES60, Yeasen, Shanghai, China). Samples were blocked with anti-CD16/CD32 (FcγRIII/FcγRII, 2.4G2, 553141, BD Biosciences CA, USA), incubated with surface marker antibodies for 20 min at 4 °C and then permeabilized with BD Cytofix/Cytoperm (554714) buffer before intracellular labeling antibodies were added for 30 min at 4 °C. Flow cytometry analysis was performed using ACEA NovoCyte and data processing was done through NovoExpress software. Antibody staining of tumor tissue cells for flow cytometry analysis was performed following the antibody manufacturer’s recommendations. The antibodies used in immunotyping include PE-Cyanine7 Rat Anti-Mouse F4/80 (25-4801-82, eBioscience, MA, USA), APC Rat anti-Mouse CD86 (558703, BD Biosciences, CA, USA), Ms CD206 Alexa 647 MR5D3 (53-2061-82, eBioscience, MA, USA), FITC Hamster Anti-Mouse CD3e (561806, BD Biosciences, CA, USA), PerCP-Cy™ 5.5 Rat Anti-Mouse CD4 (550954, BD Biosciences, CA, USA), APC Rat Anti-Mouse CD8a (553035, BD Biosciences, CA, USA), PE-Cy™ 7 Hamster Anti-Mouse CD11c (558079, BD Biosciences, CA, USA), BB515 Rat Anti-Mouse I-A/I-E (565254, BD Biosciences, CA, USA), PE Hamster Anti-Mouse CD80 (557369, BD Biosciences, CA, USA).
RNA-seq
About 45 mg of fresh tumor tissues were obtained and total RNA was extracted for transcriptome library construction. The transcriptome library sequencing was done by BGI (Shenzhen, China). Based on the RNA-seq raw data, we analyzed differential genes, immune-related signature gene levels [23], T-cell infiltration levels, and apoptosis pathway gene expression. Differential expression was evaluated with DESeq. For a given gene to be considered significantly differentially expressed, we required a fold change of 1.5:1 or greater and a false discovery rate (FDR) corrected P value of 0.05 or less. Genes in the heatmap were clustered according to the log2(fold-change). Immune signature scores are defined as the mean log2(fold-change) among all genes in each gene signature during immune-related signature gene level analysis. Immune cell infiltration within tumor tissues was estimated by ImmuCellAI [24].
Statistical methods and software
Statistical analysis was performed using GraphPad Prism 8 Software. A Student’s t test was used for comparison between the two groups. Multiple comparisons were performed using one-way ANOVA followed by Tukey’s multiple comparisons test. Detailed statistical methods and sample sizes in the experiments are described in each figure legend. No statistical methods were used to predetermine the sample size. All statistical tests were two-sided, and P values < 0.05 were considered to be significant. ns not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
Results
Acquisition of stable membranal CMTM6-PD-L1 complex
The previously hard-to-obtain stable full-length proteins of PD-L1 and CMTM6 have hindered the comprehensive investigation of their functions, which has also led to the fact that no full PD-L1 protein structure or CMTM6 structure has been reported to date. We hypothesized that the construction of the CMTM6-PD-L1 complex would contribute to obtaining stable PD-L1 and CMTM6 full-length proteins. First, the genes for PD-L1FL, CMTM6FL, and CMTM61-163 (C-terminal truncation) were cloned and expressed by the baculovirus system. The commonly used lauryl maltose neopentyl glycol (LMNG) was applied to solubilize the infected HEK293S cells. Size exclusion chromatography (SEC) results showed that both PD-L1FL and CMTM6FL alone existed mainly in aggregates with poor monomeric protein peak shapes, indicating that both full-length monomers were unstable and challenging to prepare (Fig. 1a).
a SEC elution profiles of solubilized PD-L1FL, CMTM6FL, CMTM61-163-PD-L1FL complex, and CMTM6FL-PD-L1FL complex. b The left image shows the glycosylation sites of human PD-L1. The right pictures show the SDS-PAGE results of CMTM6-PD-L1 complexes before and after deglycosylation. c Purification and identification of deglycosylated CMTM6-PD-L1 complex by SEC and SDS-PAGE. d Detergent optimization and bulk purification of the CMTM6-PD-L1 complex. The SEC elution profiles are shown. e Assembly and identification of CMTM6-PD-L1-amphipol A8-35. f Purification and identification of PD-1-PD-L1-CMTM6 complex. g Representative BLI sensorgrams of PD-L1 ECD or CMTM6-PD-L1 complex binding to immobilized PD-1.
Notably, when PD-L1FL and CMTM6FL were co-expressed and purified, the monomer peaks were symmetrical and homogeneous, implying the CMTM6FL-PD-L1FL complex was stable (Fig. 1a). In addition, the identification result of CMTM61-163-PD-L1FL was inferior to that of CMTM6FL-PD-L1FL (Fig. 1a), suggesting the intact CMTM6 structure is essential for stabilizing PD-L1FL. These results found that CMTM6 was also effective in maintaining membrane PD-L1 stability in a non-cellular environment and also reaffirmed that CMTM6 is a PD-L1-interacting protein. The purified CMTM6-PD-L1 complex was identified by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and it was found that PD-L1 had three uniformly distributed protein bands, which was due to glycosylation modification confirmed by treatment with the deglycosylase Endo HF (Fig. 1b). To avoid the protein heterogeneity caused by glycosylation, PD-L1 deglycosylated mutants (PD-L1DG) and CMTM6 were co-expressed and purified, and it was found that PD-L1 deglycosylation did not affect the stability and homogeneity of the PD-L1-CMTM6 complex (Fig. 1c), which may suggest that glycosylation is not involved in the interaction between CMTM6 and PD-L1.
Then, we assessed the oligomeric or monomeric state, stability, and overall homogeneity of CMTM6-PD-L1 complex in several detergents, including n-dodecyl-β-d-maltoside (DDM), lauryl maltose neopentyl glycol (LMNG), and decyl maltose neopentyl glycol (DMNG), to find a detergent that solubilized the complex efficiently from the lipid environment while maintaining their native three-dimensional fold and function (Fig. 1d). LMNG was found to be superior and was applied to purified complexes in large quantities. Specifically, PD-L1 and CMTM6 gene co-transfected cells were resuspended in buffer containing 0.05% LMNG, crushed and ultracentrifuged. The precipitate was lysed with 1% LMNG, and CMTM6 and PD-L1 proteins were purified by affinity chromatography. The persistent presence of detergents in solution may alter the structure and biological function of the membrane protein. To provide a cell membrane-like environment for the purified CMTM6-PD-L1 complex, we reconstituted the CMTM6-PD-L1 complex in amphipol A8-35 as a membrane mimetic to preserve the correct folding and stability of membrane proteins (Fig. 1e). Amphipol A8-35 is an amphiphilic polymer that does not form micelles but instead binds tightly around the hydrophobic transmembrane segments of proteins. LMNG-purified complexes were exchanged for amphipol A8-35 by dialysis in the presence of bio-beads SM-2. And SEC results revealed that the CMTM6-PD-L1 complex was homogenously prepared by A8-35 (Fig. 1e). The biotin-A8-35-CMTM6-PD-L1 complex was used in the following studies.
Evaluation of PD-1/PD-L1-CMTM6 interaction
To identify that the function of the obtained CMTM6-PD-L1 complex was not affected and apply the complex to the PD-1/PD-L1-CMTM6 interaction study, we co-incubated PD-1 extracellular domain (ECD) with A8-35-CMTM6-PD-L1, and subsequently collected samples of the complex peak by SEC, which were identified by SDS-PAGE to contain PD-1, PD-L1, and CMTM6, indicating that the PD-1-PD-L1-CMTM6 tricomplex was successfully assembled and the packing of A8-35 did not affect the complex function (Fig. 1f). Then we measured the binding affinity of PD-1 to PD-L1 and the CMTM6-PD-L1 complex using biolayer interferometry technology (BLI). PD-1 showed comparable binding affinity to PD-L1, consistent with results published [25, 26]. Notably, we observed that the binding affinity of the CMTM6-PD-L1 complex to PD-1 was two orders of magnitude higher than that of PD-L1 (2.0 × 10−9 M and 1.86 × 10−7 M, respectively), which was largely attributed to greatly slow disassociation rate between PD-1 and the CMTM6-PD-L1 complex (>6 min) (Fig. 1g). The dissociation time of PD-1 and PD-L1 interactions is only tens of seconds. These findings suggested that CMTM6 might augment the binding of PD-1 to PD-L1 and delay their dissociation, which is a novel mechanism for CMTM6 to stabilize PD-L1. We hypothesized that CMTM6 could also modulate the signal of the PD-1/PD-L1 axis by enhancing their interaction.
Generation of potent and selective CMTM6 binding nanobodies
The preservation of the natural structure of the antigen molecules is the prerequisite to obtain antibody drugs. So, in addition to using the complex to study protein interactions, we intended to utilize the generated stable CMTM6-PD-L1 membrane protein complex for CMTM6-targeting nanobody development (Fig. 2a). The camels were immunized with the A8-35-CMTM6-PD-L1 complexes. The high-quality VHHs phage library, with a capacity of 6.3 × 107 colony forming units (CFU), was generated and candidate nanobodies were screened and enriched for two rounds by A8-35-CMTM6-PD-L1 based ELISAs (Supplementary Fig. S1). After the exclusion of the PD-L1-targeting nanobodies by PD-L1-based ELISAs, a CMTM6-targeting nanobody with the best properties, called 1A5, was selected and expressed for subsequent experiments. The SEC profile exhibited a single symmetrical peak, indicating the high solubility and homogeneity of the nanobodies, and the SDS-PAGE result showed a single band of ~15 kDa, as expected for the size of 1A5 (Fig. 2b). We applied the BLI technique to determine the affinity of 1A5 to CMTM6-PD-L1 complex at 16.7 nM (Fig. 2c). The undetectable affinity of 1A5 to PD-L1 suggested that 1A5 mainly interacted with CMTM6 protein (Fig. 2c). At the cellular level, 1A5 could effectively bind to HEK293S cells overexpressing human or murine CMTM6 (Fig. 2d). The application of the stable membrane CMTM6-PD-L1 complex makes 1A5 the first reported antibody targeting the extracellular loops of CMTM6. It is also proved that stable membrane-bound complexes are beneficial for solving the problems of antigen acquisition and antibody development for structurally specific membrane proteins.
a Camel immunization and anti-CMTM6 nanobodies screening. b The SEC profiles and SDS-PAGE were used to characterize the nanobody 1A5. c Binding kinetics of nanobody 1A5 to PD-L1 ECD or CMTM6-PD-L1 complex, as measured by BLI using OctetRED96. d Flow cytometry histograms show the binding of 1A5 to CMTM6 on HEK293 cells expressing human or murine CMTM6. e The interaction between ECL1 and ECL2 of human or murine CMTM6 structure by AlphaFold. f The binding of 1A5 to ECL1 and ECL2 mutants of human CMTM6 by flow cytometry.
We attempted to map the binding site between the nanobody 1A5 and CMTM6. The predicted structures of human and mouse CMTM6 by AlphaFold2 show that extracellular domains 1 (ECL1) and 2 (ECL2) are structurally highly conserved and distributed with multiple polar amino acids (Fig. 2e). Further analysis shows that C62 and C63 on ECL1 form disulfide bonds directly, and T63 on ECL1 forms hydrogen bonds with D129 on ECL2 (Fig. 2e). The interaction of these four amino acids may stabilize the conformation of ECL1 and ECL2. So, we considered six residues (C62, T63, C65, D129, R130, T131) and measured the change in binding ability of 1A5 at these mutations by flow cytometry. Results showed that the binding of 1A5 to the C62G mutant was enhanced (Fig. 2f). We postulated that the C62G mutation destroyed the disulfide bond, affected CMTM6 conformation, and thus changed the binding of 1A5 to CMTM6. Taken together, these modeling and mutation data provide a further understanding of the 1A5-CMTM6 interaction and CMTM6 structure.
CMTM6 nanobody interfered with PD-L1 signaling in vitro
In PD-1/PD-L1 interactions, PD-L1 can transmit immunosuppressive signals to PD-1 [27] and receive anti-apoptotic signals from PD-1 [22]. To investigate the bioactivity of 1A5 at a cellular level, we constructed the T-cell activation model and the tumor cell apoptosis model, respectively.
Reversed PD-L1 repression can be measured by the level of nuclear factor of activated T-cell (NFAT) activation, thus we constructed Jurkat-NFAT-luciferase cells to co-culture with RKO colorectal cancer cells [induced by interferon-γ (IFN-γ) to express high levels of PD-L1] to perform a reporter assay (Supplementary Fig. S2). Compared to co-culturing with Cas9 control RKO cells, co-culturing with CMTM6-deficient tumor cells greatly diminished T-cell immunosuppression (Fig. 3a and Supplementary Fig. S3). And the inhibition effect was blocked by 1A5 in a concentration-dependent manner. A high concentration of 1A5 could significantly block the immunosuppression (Fig. 3a). The activation level of Jurkat was elevated by 3.67-fold when 20 μg/mL of 1A5 was incubated. RKO cells were not directly subjected to apoptosis in the absence of CMTM6 or PD-L1 (Supplementary Fig. S4).
a Effect of CMTM6 knockout or PD-L1 knockout or 1A5 on blocking the inhibitory signal from RKO cells to Jurkat-NFAT-Luciferase cells. The data (n = 5) are shown in relative luciferase units (RLU). b Annexin V-PI staining of Cas9 control/PD-L1-knockout/CMTM6-knockout RKO cells or WT RKO cells treated with STP after pretreatment of PD-1-Fc or IgG or 1A5. Bar graphs represent the percentage of apoptotic cells (n = 3). c The CT26 tumor growth in mice treated with 1A5 or PBS and tumor weights are shown (n = 10). d BALB/c mice were subcutaneously inoculated with CMTM6 knockout CT26 cells and were treated with 1A5 or PBS. The tumor growth in mice is shown (n = 8). e PD-1/PD-L1 dual-humanized BALB/c mice (n = 6) were subcutaneously inoculated with CT26/hPD-L1 cells and were treated with hIgG1 or 1A5. The tumor growth in mice is shown. The data are presented as the mean ± SEM. **P < 0.01; ***P < 0.001; ns, not significant by one-way ANOVA with multiple comparison tests or unpaired t-test.
Apoptosis assays showed that the death of RKO cells induced with staurosporine (STP) was lower by 2.7-fold after pretreatment of PD-1, and the death of PD-L1-knockout RKO cells and CMTM6-knockout RKO cells was unaffected by PD-1 pretreatment. And 1A5 also abolished the anti-apoptotic effects of PD-1, demonstrating 1A5 can interfere with anti-apoptotic signal transmission from T-cells to tumor cells, leading to susceptibility to death (Fig. 3b). These results suggested that CMTM6 may act with PD-L1 to transmit and receive signals between T cells and tumor cells by modulating the interaction between PD-1 and PD-L1 and that the anti-CMTM6 nanobody may also affect the PD-1/PD-L1 bidirectional signaling.
Several possibilities were speculated for how 1A5 intervened in PD-1/PD-L1 signaling. Flow cytometry assays showed that 1A5 could not block the PD-1/PD-L1 binding, nor did it affect the membranal PD-L1 level of tumor cells (Supplementary Fig. S5). Thus, we speculated that 1A5 may affect PD-L1 signaling by altering the structure of the PD-1/PD-L1/CMTM6 tricomplex.
Therapeutic efficacy of CMTM6 nanobody in syngeneic tumor models
Since 1A5 could ramp up tumor cell susceptibility to apoptosis and reverse T-cell suppression, its antitumor activity was evaluated further. BALB/c mice bearing CT26 cells were treated subcutaneously with 100 μg of 1A5 every 2 days after injection of the tumor cells. 1A5 suppressed colon tumor growth in mice with an inhibition rate of 34.8% (Fig. 3c). Similar findings have been observed in CT26/hPD-L1 tumors (Fig. 3e). In contrast, 1A5 had no impact on the growth rate of the CMTM6-deficient CT26 tumors (Fig. 3d). Taken together, these studies indicated that 1A5 had efficacious and CMTM6-dependent anti-colon cancer activity in vivo.
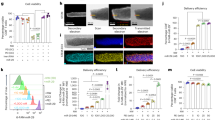
Immunophenotyping was performed to explore whether 1A5 could affect the tumor-infiltrating immune cells. In CT26 tumor-bearing mice, 1A5 promoted the infiltration of intratumoral CD8+ T-cells by 22.85% (Fig. 4a), decreased infiltration of M2 macrophages by 15.29%, and marginally increased infiltration of dendritic cells (Supplementary Fig. S6). Then, using anti-CD8a antibody-based immune cell depletion studies in CT26 tumor-bearing mice, the efficacy of 1A5 was abrogated by the depletion of CD8+ T-cells (Fig. 4b). To examine the gene expression alterations induced by 1A5 treatment, we analyzed CT26 tumor tissues by RNA sequencing. Compared to the vehicle group, 1A5 resulted in upregulated differential genes predominantly enriched in immune system-related pathways, especially cytotoxic effect, apoptotic process, and antigen-presenting process (Fig. 4c, d and Supplementary Fig. S7). The gene expression levels of effector molecules of intra-tumoral cytotoxic cells were analyzed, such as IFN-γ, granzyme A, granzyme B, perforin, tumor necrosis factor alpha (TNF-α), factor Related Apoptosis Ligand (FASL), lymphotoxin A, and lymphotoxin B and found that it was 1A5 increased the expression of cytotoxic cell effector molecules (Fig. 4d). Additional assessment of immune infiltration suggested that administration of 1A5 amplified the infiltration of antitumor immune cells, particularly CD8+ T-cell subsets (Fig. 4e and Supplementary Fig. S7). Thus, the above data identified CD8+ T-cells as critical effector cells for the antitumor activity of 1A5 and indicated that CMTM6 could be exploited as a tumor immunotherapy target.
a CD8+ T-cells in tumors treated with PBS or 1A5 were quantified by flow cytometry (n = 3). b The CT26 tumor volumes and weights in mice treated with PBS, 1A5 or anti-murine CD8 antibody and 1A5 are shown (n = 5). c RNA-sequencing analysis of CT26 tumor tissues from BALB/c mice treated with 1A5 or PBS (n = 3). The bubble plots show the GO and KEGG pathway enrichment analysis of differential genes upregulated by 1A5 treatment compared to PBS treatment. d Heatmap shows the upregulated gene expression pattern of effector molecules of intratumoral cytotoxic cells after treatment [defined as the log2(fold change)]. The signature score defined as the mean log2(fold change) among all genes in the signature is shown as scatter plots. e Cytotoxic cell infiltration analysis by the ImmuCellAI. The data are presented as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ns not significant by one-way ANOVA with multiple comparison tests or unpaired t-test.
Discussion
After PD-1/PD-L1 antibody development, another prominent research subject is targeting PD-L1 expression and regulatory pathway-related molecules [28]. In recent years, diverse interacting proteins of PD-L1 have been found to regulate the cell membrane level and subcellular localization of PD-L1 [29]. Huntingtin-interacting protein 1-related protein (HIP1R) acts on the intracellular domain of PD-L1 for lysosomal destruction [30]. Trafficking protein particle complex subunit 4 (TRAPPC4) recycles endocytic PD-L1 to the cell surface [31]. In 2017, two papers pointed out that CMTM6 is a key regulator of PD-L1 expression [3, 4]. CMTM6 hinders STUB1-mediated polyubiquitination of PD-L1 and colocalizes with PD-L1 in recycling endosomes to suppress lysosome-mediated degradation. Nonetheless, there has been a lack of progress on the interaction pattern of CMTM6/PD-L1, the expression profile of CMTM6 in tumors, and the role of CMTM6 in tumor immunity, which may be attributed to the unique protein structure of CMTM6 and the dearth of antibody tools.
The typical low abundance of membrane proteins in their natural hosts makes it necessary to overexpress PD-L1 and CMTM6 either in heterologous systems or through in vitro translation/cell-free expression. Heterologous expression of proteins, in turn, leads to multiple obstacles due to the unpredictability of compatibility between PD-L1 and CMTM6 for expression in a given host. The highly hydrophobic and/or amphipathic nature of membrane proteins also leads to challenges in producing a homogeneous, stable, and pure sample for structural studies. The recent introduction of amphipol as a membrane mimetic appears promising for the study of membrane proteins in a native-like environment.
CMTM6 is a potential antitumor target. In this work, we further explore whether CMTM6 plays a role in the PD-1/PD-L1 interaction and whether targeting CMTM6 is an applicable approach for oncotherapy. We have found that CMTM6 could delay the dissociation of PD-1/PD-L1 to stabilize their interaction, which may be one of the reasons that CMTM6 deficiency and CMTM6 nanobody could interfere with T-cell immunosuppression and tumor cell anti-apoptotic signaling. Consistently, in vivo, anti-CMTM6 nanobody inhibited CT26 tumor growth mainly by depending on CD8+ T-cells to remodel the tumor immune microenvironment and promote intratumoral apoptotic pathway expression, which is similar to the mechanism of action of anti-PD-L1 antibodies. Of course, our current understanding of CMTM6 and the nanobody-mediated regulation of the PD-1/PD-L1 axis is rudimentary, and additional research is required; affinity maturation modification and combination strategies must be applied to improve the outcomes of CMTM6 nanobody treatment.
In addition, whether CMTM6 regulates structural changes or oligomerization of PD-L1 and thus affects PD-L1 signaling still needs to be investigated. It was found that PD-L1 appeared in the form of monomers and dimers by analyzing the structure of the extracellular domain of PD-L1 [32, 33]. And some small compounds can cover the PD-1/PD-L1 binding interface through PD-L1 dimerization, thereby blocking the PD-1/PD-L1 interaction [34, 35]. These observations raise the question of how the full-length PD-L1 resides on the membrane, and whether the existing form will be changed by the interaction with CMTM6. There are many examples of membrane proteins affecting the dimerization of their interacting proteins. For instance, fibroblast growth factor receptor (FGFR) is unable to recognize FGF factors when it exists as a monomer but is able to recognize and bind FGF factors when it forms a coreceptor with the chaperone molecule Klotho [36]. In this work, we co-expressed and purified the full-length CMTM6-PD-L1 complex and found that there are also some non-monomeric peaks of potential protein multimers. However, given the aggregation associated with protein purification, whether CMTM6 can promote PD-L1 multimerization still needs to be determined by more sophisticated methods, such as three-dimensional structure resolution and fluorescence resonance energy transfer (FRET).
In summary, through the membrane mimetic system, we overcame the full-length instability of PD-L1 and CMTM6. For the first time, we succeeded in constructing a stable membranal CMTM6-PD-L1 complex, which can be used for studying PD-1/PD-L1-CMTM6 interactions, PD-L1/CMTM6 membrane protein intact structures, and drug development. CMTM6, as a chaperonin molecule, can strengthen the PD-1/PD-L1 interaction, stabilize PD-L1 and regulate PD-L1 signaling. The functional CMTM6-targeting nanobody we developed can elicit effective antitumor cytotoxic responses, which also suggests for the first time that CMTM6 could be a novel target for antibody-mediated tumor immunotherapy.
References
Han W, Ding P, Xu M, Wang L, Rui M, Shi S, et al. Identification of eight genes encoding chemokine-like factor superfamily members 1–8 (CKLFSF1–8) by in silico cloning and experimental validation. Genomics. 2003;81:609–17.
Jin C, Ding P, Wang Y, Ma D. Regulation of EGF receptor signaling by the MARVEL domain-containing protein CKLFSF8. FEBS Lett. 2005;579:6375–82.
Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–5.
Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–10.
Guan X, Zhang C, Zhao J, Sun G, Song Q, Jia W. CMTM6 overexpression is associated with molecular and clinical characteristics of malignancy and predicts poor prognosis in gliomas. EBioMedicine. 2018;35:233–43.
Chen L, Yang QC, Li YC, Yang LL, Liu JF, Li H, et al. Targeting CMTM6 suppresses stem cell-like properties and enhances antitumor immunity in head and neck squamous cell carcinoma. Cancer Immunol Res. 2020;8:179–91.
Martinez-Morilla S, Zugazagoitia J, Wong PF, Kluger HM, Rimm DL. Quantitative analysis of CMTM6 expression in tumor microenvironment in metastatic melanoma and association with outcome on immunotherapy. Oncoimmunology. 2020;10:1864909.
Wu X, Lan X, Hu W, Zhang W, Lai X, Xu S, et al. CMTM6 expression in M2 macrophages is a potential predictor of PD-1/PD-L1 inhibitor response in colorectal cancer. Cancer Immunol Immunother. 2021;70:3235–48.
Li X, Chen L, Gu C, Sun Q, Li J. CMTM6 significantly relates to PD-L1 and predicts the prognosis of gastric cancer patients. PeerJ. 2020;8:e9536.
Zhao Y, Zhang M, Pu H, Guo S, Zhang S, Wang Y. Prognostic Implications of Pan-Cancer CMTM6 Expression and Its Relationship with the Immune Microenvironment. Front Oncol. 2020;10:585961.
Zheng Y, Wang C, Song A, Jiang F, Zhou J, Li G, et al. CMTM6 promotes cell proliferation and invasion in oral squamous cell carcinoma by interacting with NRP1. Am J Cancer Res. 2020;10:1691–709.
Wang H, Fan Y, Chen W, Lv Z, Wu S, Xuan Y, et al. Loss of CMTM6 promotes DNA damage-induced cellular senescence and antitumor immunity. Oncoimmunology. 2022;11:2011673.
Mohapatra P, Shriwas O, Mohanty S, Ghosh A, Smita S, Kaushik SR, et al. CMTM6 drives cisplatin resistance by regulating Wnt signaling through the ENO-1/AKT/GSK3beta axis. JCI Insight. 2021;6:e143643.
Pang X, Wang SS, Zhang M, Jiang J, Fan HY, Wu JS, et al. OSCC cell-secreted exosomal CMTM6 induced M2-like macrophages polarization via ERK1/2 signaling pathway. Cancer Immunol Immunother. 2021;70:1015–29.
Huang Y, Zhu Y, Yang J, Pan Q, Zhao J, Song M, et al. CMTM6 inhibits tumor growth and reverses chemoresistance by preventing ubiquitination of p21 in hepatocellular carcinoma. Cell Death Dis. 2022;13:251.
Wang Z, Peng Z, Liu Q, Guo Z, Menatola M, Su J, et al. Co-expression with membrane CMTM6/4 on tumor epithelium enhances the prediction value of PD-L1 on Anti-PD-1/L1 therapeutic efficacy in gastric adenocarcinoma. Cancers (Basel). 2021;13:5175.
Althoff T, Mills DJ, Popot JL, Kühlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–64.
Kevany BM, Tsybovsky Y, Campuzano ID, Schnier PD, Engel A, Palczewski K. Structural and functional analysis of the native peripherin-ROM1 complex isolated from photoreceptor cells. J Biol Chem. 2013;288:36272–84.
Tsybovsky Y, Orban T, Molday RS, Taylor D, Palczewski K. Molecular organization and ATP-induced conformational changes of ABCA4, the photoreceptor-specific ABC transporter. Structure. 2013;21:854–60.
Yang EY, Shah K. Nanobodies: Next generation of cancer diagnostics and therapeutics. Front Oncol. 2020;10:1182.
Ahuja S, Rougé L, Swem DL, Sudhamsu J, Wu P, Russell SJ, et al. Structural analysis of bacterial ABC transporter inhibition by an antibody fragment. Structure. 2015;23:713–23.
Azuma T, Yao S, Zhu G, Flies AS, Flies SJ, Chen L. B7-H1 is a ubiquitous antiapoptotic receptor on cancer cells. Blood. 2008;111:3635–43.
Lan Y, Zhang D, Xu C, Hance KW, Marelli B, Qi J, et al. Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci Transl Med. 2018;10:eaan5488.
Miao YR, Zhang Q, Lei Q, Luo M, Xie GY, Wang H, et al. ImmuCellAI: A unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh). 2020;7:1902880.
Maute RL, Gordon SR, Mayer AT, McCracken MN, Natarajan A, Ring NG, et al. Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc Natl Acad Sci USA. 2015;112:E6506–E6514.
Cheng X, Veverka V, Radhakrishnan A, Waters LC, Muskett FW, Morgan SH, et al. Structure and interactions of the human programmed cell death 1 receptor. J Biol Chem. 2013;288:11771–85.
Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–34.
Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52:17–35.
Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms controlling PD-L1 expression in cancer. Mol Cell. 2019;76:359–70.
Wang H, Yao H, Li C, Shi H, Lan J, Li Z, et al. HIP1R targets PD-L1 to lysosomal degradation to alter T cell-mediated cytotoxicity. Nat Chem Biol. 2019;15:42–50.
Ren Y, Qian Y, Ai L, Xie Y, Gao Y, Zhuang Z, et al. TRAPPC4 regulates the intracellular trafficking of PD-L1 and antitumor immunity. Nat Commun. 2021;12:5405.
Chen Y, Liu P, Gao F, Cheng H, Qi J, Gao GF. A dimeric structure of PD-L1: functional units or evolutionary relics? Protein Cell. 2010;1:153–60.
Zak KM, Grudnik P, Magiera K, Dömling A, Dubin G, Holak TA. Structural biology of the immune checkpoint receptor PD-1 and its ligands PD-L1/PD-L2. Structure. 2017;25:1163–74.
Zak KM, Grudnik P, Guzik K, Zieba BJ, Musielak B, Dömling A, et al. Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1). Oncotarget. 2016;7:30323–35.
Guo Y, Jin Y, Wang B, Liu B. Molecular mechanism of small-molecule inhibitors in blocking the PD-1/PD-L1 pathway through PD-L1 dimerization. Int J Mol Sci. 2021;22:4766.
Urakawa I, Yamazaki Y, Shimada T, Iijima K, Hasegawa H, Okawa K, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–4.
Acknowledgements
This work is supported by the National Natural Science Foundation of China (No. 31670743), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA12040326), Science and Technology Commission of Shanghai Municipality (No. 3918JC141540001), Joint Research Fund for Overseas, Hong Kong and Macao Scholars (No. 81628013), Natural Science Foundation of Shanghai (16ZR1442900), National Science Foundation for Young Scholar projects (118180359901) and The grand from the Shanghai Institute of Materia Medica, Chinese Academy of Sciences (CASIMM0120164013, SIMM1606YZZ-06, SIMM1601KF-06, 55201631121116101, 55201631121108000, 5112345601, 2015123456005, CASIMM0120202003). We would like to thank colleagues from the Shanghai Institute of Materia Medica, Chinese Academy of Sciences, who provided advice and technical support.
Author information
Authors and Affiliations
Contributions
YG, LKG, XMJ, and YRL designed the experiments and analyzed the data. XMJ and YRL performed the experiments and prepared the paper. XLY and RQC assisted in performing the experiments and preparing the paper. All authors approved the final draft of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jia, Xm., Long, Yr., Yu, Xl. et al. Construction of stable membranal CMTM6-PD-L1 full-length complex to evaluate the PD-1/PD-L1-CMTM6 interaction and develop anti-tumor anti-CMTM6 nanobody. Acta Pharmacol Sin 44, 1095–1104 (2023). https://doi.org/10.1038/s41401-022-01020-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-022-01020-3