Abstract
Aspirin as a chemopreventive agent is able to restrict the tumor growth. Phosphoglycerate mutase 1 (PGAM1) is a key enzyme of glycolysis, playing an important role in the development of cancer. However, the underlying mechanism by which aspirin inhibits the proliferation of cancer cells is poorly understood. This study aims to identify the effects of aspirin on modulating PGAM1 enzymatic activities in liver cancer. Here, we found that aspirin attenuated the PGAM1 succinylation to suppress the PGAM1 enzymatic activities and glycolysis in hepatoma cells. Mechanically, aspirin remarkably reduced the global succinylation levels of hepatoma cells, including the PGAM1 succinylation, which led to the block of conversion from 3-phosphoglycerate (3-PG) to 2-phosphoglycerate (2-PG) in cells. Interestingly, RNA-seq analysis identified that aspirin could significantly decrease the levels of histone acetyltransferase 1 (HAT1), a writer of PGAM1 succinylation, in liver cancer. As a target of aspirin, NF-κB p65 could effectively up-regulate the expression of HAT1 in the system, resulting in the increase of PGAM1 enzymatic activities. Moreover, we observed that the PGAM1-K99R mutant failed to rescue the aspirin-induced inhibition of PGAM1 activities, glycolysis, and proliferation of hepatoma cells relative to PGAM1-WT. Functionally, aspirin down-regulated HAT1 and decreased the PGAM1 succinylation levels in the tumor tissues from mice treated with aspirin in vivo. Thus, we conclude that aspirin modulates PGAM1K99 succinylation to restrict the PGAM1 activities and glycolysis through NF-κB p65/HAT1/PGAM1 signaling in liver cancer. Our finding provides new insights into the mechanism by which aspirin inhibits glycolysis in hepatocellular carcinoma.
Similar content being viewed by others
Introduction
Aspirin as a nonsteroidal anti-inflammatory drug (NSAID) is most commonly used for the secondary prevention of cardiovascular diseases (CVD) [1, 2]. Nowadays, more and more evidence indicates the potential roles of aspirin against cancer [3, 4]. Aspirin has a protective effect against the development of colorectal cancer (CRC) by several inter-related mechanisms, such as inhibition of Wnt-β-catenin signaling, inhibition of prostaglandin-endoperoxide synthase 2 (PTGS2), inactivation of protein phosphatase 2A (PP2A) [5,6,7]. Glycolysis is primarily characterized biology aspect in cancer cells [8, 9]. Our group reported that aspirin attenuated the glycolysis and proliferation of hepatoma cells by 2-hydroxyisobutyrylation of ENO1 [10]. However, the underlying mechanism by which aspirin decreases the glycolysis of hepatoma cells need to be further clarified.
Phosphoglycerate mutase 1 (PGAM1) is critical in cancer metabolism by catalyzing the conversion from 3-phosphoglycerate (3-PG) to 2-phosphoglycerate (2-PG) during aerobic glycolysis [11, 12]. However, the effect of aspirin on PAGM1 in liver cancer cells is elusive. Posttranslational modifications (PTMs) play important roles in the regulation of protein activity and function, which are involved in the development of cancer [13,14,15]. Aspirin is able to acetylate biomolecules, which plays potential roles in the development of cancers [16, 17]. As a target of aspirin, nuclear factor-κB (NF-κB, especially one of the subunits, p65) serves as a major factor controlling the proliferation of cancer cells [18]. Lysine succinylation occurs frequently among the newly identified posttranslational modifications of human histones and is critical for tumor development [19]. Our group reported that the succinylation of PGAM1 catalyzed by HAT1 played critical roles in promoting tumor progression [20]. However, the effects of aspirin on the HAT1 and PGAM1K99 succinylation in cancer cells need to be further studied.
In this study, we are interested in the roles of aspirin in modulation of PGAM1 succinylation in liver cancer. Interestingly, we observed that 4 mM aspirin decreased the enzymatic activities of PGAM1 and the succinylation levels of PGAM1 (termed PGAM1 Succ), in which NF-κB and HAT1 were involved in the event. Aspirin down-regulated the expression of HAT1 and decreased the levels of PGAM1 succinylation, contributing to the suppression of growth of liver cancer. Our finding provides new insights into the mechanism by which aspirin inhibits the glycolysis and growth of liver cancer.
Materials and methods
Cell culture and treatment
Human hepatoma cell lines HepG2 and Huh7 were cultured with Dulbecco′s modified Eagle′s medium (DMEM, Gibco, USA) containing 10% fetal bovine serum (FBS, Gibco, USA), 100 U/mL penicillin and 100 mg/mL streptomycin. All cell lines were cultured in 5% CO2 and 37 °C conditions. For aspirin or PDTC treatment, hepatoma cells lines were treated with 2 mM and/or 4 mM aspirin [21] (Sigma-Aldrich, USA) or 50 μM and/or 100 μM PDTC (MCE, USA) for 48 h [22]. The supernatant or cells were collected for further study.
Measurement of glucose consumption and lactate acid production
The protocols of analyses of glucose consumption and lactate acid were conducted as previously described [20, 23]. The consumption of glucose in the supernatant of the hepatoma cells was measured by using GOPOD format kit (Mlbio, China). The levels of lactate acid in the supernatant of the hepatoma cells were carried out by using LA ELISA kit (Mlbio, China).
Measurement of the levels of 3-PG, 2-PG and PGAM1 activities
The protocols of analyses of 3-PG and 2-PG were conducted as previously described [20, 23]. The packed cell pellets were homogenized in 1.5 mL of hypotonic lysis buffer (20 mM HEPES (pH 7.0), 5 mM KCl, 1 mM MgCl2, 5 mM DTT, and protease inhibitor cocktail), determining cellular concentration of 2-PG and 3-PG. The homogenates were centrifuged in a cold room at 4 °C for 10 min at maximum speed, and the supernatants were applied to Amicon Ultra tubes with 10 kDa cutoff filter (Millipore, USA). The levels of 3-PG and 2-PG in hepatoma cells were measured by 3-PG Kit (Mlbio, China) and 2-PG Kit (Mlbio, China) according to the instructions of producers, respectively. The activities of PGAM1 were measured by PGAM1 Kit (Mlbio, China) according to the instructions of producers.
Western blot analysis
Total protein lysates were extracted from cells with RIPA buffer. The boiled proteins with loading buffer were subjected to SDS-PAGE and then transferred to a nitrocellulose membrane. The 8% non-fat milk was used to block the membrane. Two hours later, the membrane was incubated with first antibody for 1 h at room temperature. After incubation with the secondary antibody for 1 h at 37 °C, the membrane was visualized by ECL Western Blotting Detection Kit from GE Healthcare (Waukesha, USA). The primary antibodies included mouse anti-β-actin (Sigma, USA), rabbit anti-PGAM1 (Abcam, UK), rabbit anti-p65 (Proteintech, USA), rabbit anti-p-p65 (Proteintech, USA), rabbit anti-HAT1 antibody (Abcam, UK), rabbit anti-Pan-succinylation (PTM Biolabs, China), rabbit anti-BCL2 (Proteintech, USA), rabbit anti-XIAP (Proteintech, USA). Protein bands were visualized and quantified using an Odyssey Infrared Imaging System (LiCor Biosciences).
RNA extraction and RT-qPCR
Total RNA from hepatoma cells was isolated according to the Trizol extraction method and reverse-transcription was performed using the HiFair III 1st Strand cDNA Synthesis SuperMix from YEASEN (Shanghai, China). RT-qPCRs were carried out using Hieff qPCR SYBR Green Master Mix from YEASEN. Fold changes of gene expression were calculated as 2-ΔΔCt. GAPDH was used as an internal control for normalization. HAT1 primer: forward primer, 5′-GAAATGGCGGGATTTGGTGC-3′; reverse primer, 5′-TGTAGCCTACGGTCGCAAAG-3′.
Immunoprecipitation (IP) assays
The IP protocol was described in detail in previously published articles [24]. After the treatment with 4 mM aspirin or 100 μM PDTC, the cells lysates were harvested and then subjected to overnight immunoprecipitation at 4 °C using 2–5 μg of PGAM1 antibodies (Abcam, UK). Immune complexes were incubated with protein A/G agarose beads (Bimake, USA) at 4 °C. The precipitates were washed six times with ice-cold lysis buffer and then resuspended in the PBS. The IP products were resolved by SDS-PAGE followed by Western blot analysis.
Plasmid construct, mutagenesis and siRNA
The coding sequence (CDS) regions of PGAM1 (765 bp) was cloned into pCMV-3Tag-1A; forward primer, 5′-ATGGCCGCCTACAAACTGG-3′; reverse primer, 5′-TCACTTCTT GGCCTTGCC-3′. PGAM1 mutant (K99R) was cloned into pCMV-3Tag-1A; forward primer, 5′-AGCAGTTTCTGCTCTATTGAGACCGGTTAGACCC-3′, reverse primer, 5′-GGGTCTA ACCGGTCTCAATAGAGCAGAAACTGCT-3′. The two plasmids were named as PGAM1-WT and PGAM1-K99R, respectively. CDS regions of HAT1 (1260 bp) was cloned into pCMV-3Tag-1A [20]; forward primer, 5′-ATGGCGGGATTTGGTGCTATG-3′; reverse primer, 5′-TTACTCTTGAGCAAGTCGTTC-3′. CDS regions of p65 (1656 bp) was cloned into pCMV-3Tag-1A; forward primer, 5′-ATGGACGAACTGTTCC-3′; reverse primer, 5′-TT AGGAGCTGATCTGACTCAG-3′. The siRNA sequence targeting p65 mRNA, forward sequence GATGAGATCTTCCTACTGT, was synthesized by Sangon Biotech (Shanghai, China).
CCK-8 assays
Hepatoma cells were seeded into 96-well plates (2000 cells/well) for 12 h before transfection. CCK-8 (YEASEN, China) assays were used to assess the cell proliferation every 24 h from the first day until 72 h after transfection.
EdU incorporation assays
Hepatoma cells were seeded into 24-well plates (8000 cells/well) for 12 h before transfection. The 5-ethynyl-2′-deoxyuridine incorporation assays were carried out using the Cell-LightTM EdU imaging detecting kit according to the manufacturer′s instructions (RiboBio, China) after 48 h of transfection.
Colony formation assays
The 1000 viable hepatoma cells were plated in six-well plates after 24 h transfection, and cultured in complete medium for 2 weeks. Colonies were fixed with methanol and stained with 0.1% crystal violet (Beyotime, China).
RNA- seq analysis
Total RNA was extracted from HepG2 cells treated with 4 mM aspirin or DMSO, and subjected to RNA-seq analysis performed by Shanghai Majorbio Bio-pharm Technology Co. Ltd. The data were analyzed on the free online platform of Majorbio Cloud Platform (www.majorbio.com).
Chromatin immunoprecipitation (ChIP)
For the enrichment of NF-κB p65 on the promoter region of HAT1 gene, ChIP assays were performed in HepG2 and Huh7 cells transfected with p65-pCMV-3Tag-1A according to the manufacturer’s protocol (Cell Signaling Technology, USA). The primary antibodies included rabbit anti-p65 (Proteintech, USA). The qPCR primer: forward primer, 5′-GGGTTCAAGCGATTCTCCTG-3′; reverse primer, 5′-GCCTGAGCTCAGGAGTTCGAG-3′.
In vivo tumorigenicity assays
HepG2 cells were harvested and suspended at 1 × 107 cells/mL in 0.2 ml of PBS in separate sets of mice, and then injected into the subaxillary region of 6-week-old male BALB/c athymic nude mice, six mice per group. A week later, the mice in aspirin groups were daily treated with intragastric gavage with aspirin (suspended in physiological saline, 75 mg/kg). The DMSO group mice were daily treated with intragastric gavage with DMSO (suspended in physiological saline). After 0 days from the treatment, the tumor growth was measured every 5 days. Blind measurements were carried out to avoid unconscious biases. After 20 days, mice were euthanized and tumors were sectioned, which were used to examine the expression of HAT1 and the PGAM1 Succ levels. Tumor volume was monitored by measuring the length (L) and width (W) with calipers and calculated with the formula (L × W2) × 0.5. The Institute Research Ethics Committee at Tianjin Medical University Cancer Institute and Hospital approved the study protocol.
Statistical analysis
Each experiment was repeated at least three times unless otherwise indicated. All analysis and graphs were generated with GraphPad Prism 8. Statistical significance was assessed by comparing mean values (±SD) using a Student‘s t-test for independent groups.
Results
Aspirin suppresses the enzymatic activities of PGAM1 in hepatoma cells
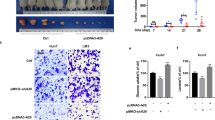
Our group reported that aspirin attenuated the glycolysis of hepatoma cells [10]. In this study, we try to identify the novel mechanisms by which aspirin modulates glycolysis in liver cancer cells. Firstly, we validated that 4 mM aspirin attenuated the consumption of glucose and the production of lactate acid in HepG2 and Huh7 cells (Fig. 1a, b) [21]. Considering that PGAM1 as key enzyme catalyzes the conversion of 3-PG to 2-PG during aerobic glycolysis [11], we supposed that PGAM1 might be involved in the glycolysis mediated by aspirin. Interestingly, we observed that aspirin significantly decreased the enzymatic activities of PGAM1 in hepatoma cells, but not the expression of PGAM1 (Fig. 1c, d). Furthermore, aspirin remarkably increased the levels of 3-PG and reduced the levels of 2-PG in HepG2 and Huh7 cells (Fig. 1e, f), suggesting that aspirin can attenuate the catalyzing activities of PGAM1 in cells. Thus, we conclude that aspirin suppresses the enzymatic activities of PGAM1 to attenuate glycolysis in hepatoma cells.
a The levels of glucose consumption were measured by using GOPOD format assays in HepG2 and Huh7 cells treated with 4 mM aspirin for 48 h. b The levels of lactate acid production were examined by LA ELISA assays in HepG2 and Heh7 cells treated with 4 mM aspirin for 48 h. c The activities of PGAM1 in HepG2 and Huh7 cells treated with 4 mM aspirin for 48 h were detected by ELISA. d The protein levels of PGAM1 were examined by Western blot analysis in HepG2 and Huh7 cells treated with 4 mM aspirin for 48 h. e, f The levels of 3-PG and 2-PG in HepG2 and Huh7 cells treated with 4 mM aspirin for 48 h were measured by ELISA. The mean ± SD of at least three experiments is shown. Statistically significant differences are indicated as follows: Student‘′s t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
Aspirin decreases the levels of PGAM1 Succ in hepatoma cells
Next, we try to identify the mechanism by which aspirin decreases the enzymatic activities of PGAM1 in hepatoma cells. Based on that PGAM1 Succ played important roles in promoting tumor progression [20], we hypothesized that aspirin might affect the levels of PGAM1 Succ in the event. Surprisingly, we observed that aspirin could reduce the global levels of succinylation by Western blot analysis using pan anti-succinylation antibody (Fig. 2a, b), suggesting that aspirin is able to attenuate the succinylation levels of proteins in cells. Furthermore, IP assays and Western blot analysis demonstrated that the levels of PGAM1 Succ were significantly decreased in HepG2 and Huh7 cells treated by aspirin (Fig. 2c, d). Thus, we conclude that aspirin inhibits the succinylation levels of PGAM1 in liver cancer cells.
a, b Total cell lysate from HepG2 and Huh7 cells treated with 4 mM aspirin was used to analyze the succinylation levels by using Western blot analysis with the indicated pan anti-succinylation antibody. Total protein levels were measured by Coomassie blue staining. c, d IP assays were performed by using PGAM1 antibody in HepG2 and Huh7 cells treated with 4 mM aspirin for 48 h. The levels of PGAM1 Succ were detected by Western blot analysis.
HAT1 is required for the aspirin-mediated decrease of PGAM1 Succ levels and PGAM1 enzymatic activities
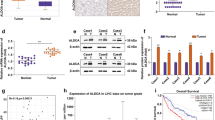
To identify the underlying mechanism by which aspirin inhibits hepatoma cells, we performed the RNA-seq analysis using HepG2 cells treated with 4 mM aspirin. The KEGG pathway analysis revealed that aspirin modulated a variety of biology aspects, including cancer-related human diseases, cell growth and death processes, signal transduction, lipid metabolism and amino acid metabolism (Fig. 3a). Next, we try to identify the potential succinyltransferase mediated by aspirin in the regulation of PGAM1 Succ. Surprisingly, we found that aspirin down-regulated the expression of HAT1, a succinyltransferase (Fig. 3b). Considering that HAT1 served as a succinyltransferase catalyzing the PGAM1 Succ [20], we supposed that aspirin might inhibit the levels of PGAM1 Succ through modulating HAT1 in cells. We further validated the aspirin-mediated inhibition of HAT1 mRNA with the data of RNA-seq in the HepG2/Huh7 cells treated with aspirin (Fig. 3c and Fig. S1a). Moreover, we revealed that aspirin down-regulated the protein levels of HAT1 in a dose-dependent manner in HepG2 and Huh7 cells (Fig. 3d). Next, the overexpression levels of HAT1 were examined in hepatoma cells (Fig. S1b, c). We found that the overexpression of HAT1 could significantly rescue the succinylation levels of PGAM1 which was decreased by aspirin in HepG2 and Huh7 cells (Fig. 3e), suggesting that aspirin inhibits the levels of PGAM1 Succ through down-regulating HAT1 in cells. Furthermore, we demonstrated that the overexpression of HAT1 could remarkably rescue the aspirin-attenuated PGAM1 enzymatic activities in HepG2 and Huh7 cells (Fig. 3f). Surprisingly, HAT1 could decrease the levels of 3-PG enhanced by aspirin in cells (Fig. 3g). Oppositely, the aspirin-blocked 2-PG levels were significantly increased by the overexpression of HAT1 (Fig. 3h) in the system, suggesting that HAT1 is involved in the conversion of 3-PG to 2-PG blocked by aspirin. Thus, we conclude that HAT1 is involved in the aspirin-mediated decrease of PGAM1 Succ levels and PGAM1 enzymatic activities (Fig. S1d).
a KEGG pathway analysis for RNA-seq in HepG2 cells treated with 4 mM aspirin. b The volcano analysis of RNA-seq of 4 mM aspirin-treated HepG2 cells. c The expression of HAT1 in RNA-seq of 4 mM aspirin-treated HepG2 cells. d The expression levels of HAT1 were examined by Western blot analysis in HepG2 and Huh7 cells treated with DMSO, 2 mM, and 4 mM aspirin for 48 h. e IP assays were performed by using PGAM1 antibody in HepG2 and Huh7 cells treated with DMSO, 4 mM aspirin, both 4 mM aspirin and 2 μg HAT1-pCMV-3Tag-1A for 48 h, respectively. The levels of PGAM1 succinylation and HAT1 were detected by Western blot analysis. f The activities of PGAM1 in HepG2 and Huh7 cells treated with DMSO, 4 mM aspirin, co-treatment of 4 mM aspirin, and 2 μg HAT1-pCMV-3Tag-1A for 48 h were measured by ELISA, respectively. g, h The levels of 3-PG and 2-PG in HepG2 and Huh7 cells treated with DMSO, 4 mM aspirin, both 4 mM aspirin and 2 μg HAT1-pCMV-3Tag-1A for 48 h were examined by ELISA, respectively. The mean ± SD of at least three experiments is shown. Statistically significant differences are indicated as follows: Student‘s t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
NF-κB activates HAT1 to modulate the levels of PGAM1 Succ and PGAM1 enzymatic activities
Based on that aspirin attenuates the activities of NF-κB to mediate the progression of cancers [25, 26], we speculated that the NF-κB may contribute to the aspirin-mediated modulation of HAT1 in cells. Interestingly, we observed that the treatment with PDTC, an inhibitor of NF-κB, could down-regulate both the mRNA and protein levels of HAT1 in a dose-dependent manner in HepG2 and Huh7 cells. Meanwhile, the NF-κB-targeted genes, such as BCL2 and XIAP, were inhibited by PDTC in a dose-dependent manner in both HepG2 and Huh7 cells (Fig. 4a and Fig. S2a, b). Moreover, konckdown of p65 by siRNA significantly down-regulated the expression of HAT1 in cells (Fig. 4b and Fig. S2c). Oppositely, we showed that the overexpression of p65, a key subunit of NF-κB, significantly up-regulated HAT1 in a dose-dependent manner in cells (Fig. 4c and Fig. S2d). Next, we predicted the NF-κB p65 binding elements (relative score over 80%) in the promoter regions of HAT1 gene (Fig. 4d) by JASPAR database, and confirmed that NF-κB p65 could significantly enrich on the promoter of HAT1 by ChIP-qPCR assays (Fig. 4e), suggesting that NF-κB p65 may regulate the expression of HAT1 by enriching on the promoter of HAT1 gene. It has been reported that the phosphorylation of p65 is a marker of NF-κB activation [27]. We confirmed that in our system (Fig. S2e). Surprisingly, our data showed that p65 could remarkably rescue the expression levels of HAT1 decreased by aspirin in cells (Fig. 4f and Fig. S2f), suggesting that aspirin down-regulates HAT1 by targeting NF-κB p65. Furthermore, our data revealed that HAT1 could remarkably rescue the levels of PGAM1 Succ decreased by PDTC in HepG2 and Huh7 cells (Fig. 4g and Fig. S2g). The overexpression of HAT1 significantly rescue the enzymatic activities of PGAM1 and the conversion of 3-PG to 2-PG decreased by PDTC in hepatoma cells (Fig. 4h–j), suggesting that NF-κB increases the PGAM1 Succ and PGAM1 enzymatic activities by modulating HAT1. Thus, we conclude that aspirin modulate the succinylation of PGAM1 to restrict the activities of PGAM1 by NF-κB p65/HAT1/PGAM1 signaling in a model (Fig. 4k).
a The protein levels of HAT1, BCL2 and XIAP were examined by Western blot analysis in HepG2 cells treated with DMSO, 50 μM and 100 μM PDTC for 48 h, respectively. b The expression levels of HAT1 and p65 were measured by Western blot analysis in HepG2 cells transfected with 50 nM and 100 nM sip65 for 48 h. c The expression levels of HAT1 and p65 were measured by Western blot analysis in HepG2 cells transfected with 1 μg and 2 μg p65-pCMV-3Tag-1A for 48 h. d The NF-κB p65 sequence motif logos and the binding elements in the promoter regions of HAT1 gene were analyzed by JASPAR database with a relative score higher than 80%. e The enrichment of NF-κB p65 on the HAT1 region of −813bp~−669bp was examined by ChIP-qPCR assays in HepG2 and Huh7 cells transfected with p65-pCMV-3Tag-1A. f The expression levels of HAT1, PGAM1, and p65 were measured by Western blot analysis in HepG2 cells treated with 4 mM aspirin or both 4 mM aspirin and 2 μg p65-pCMV-3Tag-1A for 48 h. g IP assays were performed by using PGAM1 antibody in HepG2 cells treated with DMSO, 100 μM PDTC, both 100 μM PDTC and 2 μg HAT1-pCMV-3Tag-1A for 48 h, respectively. The levels of PGAM1 Succ and HAT1 were detected by Western blot analysis. h The activities of PGAM1 in HepG2 and Huh7 cells treated with DMSO, 100 μM PDTC, both 100 μM PDTC and 2 μg HAT1-pCMV-3Tag-1A for 48 h were measured by ELISA, respectively. i, j The levels of 3-PG and 2-PG in HepG2 and Huh7 cells treated with DMSO, 100 μM PDTC, both 100 μM PDTC and 2 μg HAT1-pCMV-3Tag-1A for 48 h were examined by ELISA, respectively. k A model of aspirin-mediated NF-κB confers HAT1 to modulate the levels of PGAM1 Succ and PGAM1 enzymatic activities. The mean ± SD of at least three experiments is shown. Statistically significant differences are indicated as follows: Student‘s t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
PGAM1K99 de-Succ contributes to the aspirin-mediated depression of glycolysis in hepatoma cells
Considering that the PGAM1K99 Succ site is involved in the stimulation of glycolytic flux in cancer cells [20], we speculated that aspirin might inhibit glycolysis by modulating the levels of PGAM1K99 Succ in hepatoma cells. Thus, we generated a mutant of PGAM1 in which K99 site was mutated to R99 which was deficient in succinylation, termed PGAM1-K99R mutant. We validated the overexpression efficiency of PGAM1-WT and PGAM1-K99R in Fig. S3a, b. Surprisingly, we observed that PGAM1-K99R mutant failed to block the accumulation of 3-PG and failed to rescue the levels of 2-PG in HepG2 and Huh7 cells treated with aspirin, relative to the PGAM1-WT (Fig. 5a–c), suggesting that aspirin decreases the catalytic activities of PGAM1 by inhibiting the levels of PGAM1K99 Succ, termed PGAM1K99 de-Succ. Moreover, we showed that PGAM1-WT could significantly rescue the levels of consumption of glucose and production of lactate acid blocked by aspirin, but the PGAM1-K99R mutant failed to work in cells (Fig. 5d, e and Fig. S3c, d), suggesting that aspirin results in the defective glycolysis by restricting the levels of PGAM1K99 Succ. Accordingly, PGAM1-WT, rather than the PGAM1-K99R mutant, could rescue the levels of consumption of glucose and production of lactate acid mediated by PDTC in cells (Fig. 5f, g and Fig. S3e, f). Thus, we conclude that PGAM1K99 de-Succ contributes to the aspirin-mediated depression of glycolysis in hepatoma cells (Fig. 5h).
a–c The concentration of 3-PG and 2-PG were measured by ELISA in HepG2 and Huh7 cells with DMSO, 4 mM aspirin, both 4 mM aspirin and 2 μg PGAM1-WT, both 4 mM aspirin and 2 μg PGAM1-K99R mutant for 48 h, respectively. d The levels of glucose consumption were measured by using GOPOD format assays in HepG2 cells treated with DMSO, 4 mM aspirin, both 4 mM aspirin and 2 μg PGAM1-WT, both 4 mM aspirin and 2 μg PGAM1-K99R mutant for 48 h, respectively. e The levels of lactate acid production were examined by LA ELISA assays in HepG2 cells with DMSO, 4 mM aspirin, both 4 mM aspirin and 2 μg PGAM1-WT, both 4 mM aspirin and 2 μg PGAM1-K99R mutant for 48 h, respectively. f The levels of glucose consumption were measured by using GOPOD format assays in HepG2 cells treated with DMSO, 100 μM PDTC, both 100 μM PDTC and 2 μg PGAM1-WT, both 100 μM PDTC and 2 μg PGAM1-K99R mutant for 48 h, respectively. g The levels of lactate acid production were examined by LA ELISA assays in HepG2 cells with DMSO, 100 μM PDTC, both 100 μM PDTC and 2 μg PGAM1-WT, both 100 μM PDTC and 2 μg PGAM1-K99R mutant for 48 h, respectively. h A model of aspirin modulates succinylation of PGAM1K99 to restrict the activities of PGAM1 and glycolysis. The mean ± SD of at least three experiments is shown. Statistically significant differences are indicated as follows: Student‘s t-test. **P < 0.01; ***P < 0.001.
Aspirin down-regulates HAT1 and induces PGAM1K99 de-Succ to suppress the growth of hepatoma cells
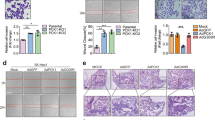
It has been reported that the succinylation plays an important role in the progression of cancers [20, 28, 29]. To explore the significance of PGAM1K99 de-Succ in the proliferation of hepatoma cells mediated by aspirin, we examined the effect of PGAM1-K99R mutant on the proliferation of hepatoma cells treated by aspirin. Notably, we observed that the PGAM1-K99R mutant failed to rescue the aspirin-induced inhibition of hepatoma cell proliferation, relative to PGAM1-WT (Fig. 6a and Fig. S4a). Furthermore, same result was shown by using EdU incorporation and colony formation assays in HepG2 and Huh7 cells (Fig. 6b, c and Fig. S4b, c), suggesting that aspirin suppresses the proliferation of hepatoma cells through modulating PGAM1K99 de-Succ (Fig. 6d). Next, we performed tumorigenesis in nude mice to explore the function of aspirin on the proliferation of hepatoma cells in vivo. Our data revealed that the treatment with 75 mg/kg aspirin could significantly decrease the tumor volume of mice (Fig. 6e). Moreover, aspirin significantly blocked the conversion of 3-PG to 2-PG in the tumor tissues from mice (Fig. 6f). Western blot analysis demonstrated that aspirin down-regulated the expression of HAT1 (Fig. 6g) in the tumors. Accordingly, the activity of NF-κB, the expression of BCL2 and XIAP were decreased in aspirin-treated tumors from mice (Fig. 6g). Moreover, IP assays showed that the levels of PGAM1 succinylation were significantly decreased in the aspirin group (Fig. 6h), suggesting that aspirin inhibits the expression of HAT1 and levels of PGAM1 Succ in vivo. In conclusion, aspirin down-regulates HAT1 and induces PGAM1K99 de-Succ to suppress the proliferation of hepatoma cells.
a The cell proliferation was measured by CCK8 assays in HepG2 cells treated with DMSO, 4 mM aspirin, both 4 mM aspirin and 2 μg PGAM1-WT, both 4 mM aspirin and 2 μg PGAM1-K99R mutant for 48 h, respectively. b EdU positive rates were determined in HepG2 and Huh7 cells treated with DMSO, 4 mM aspirin, both 4 mM aspirin and 2 μg PGAM1-WT, both 4 mM aspirin and 2 μg PGAM1-K99R mutant for 48 h, respectively. c Colony formation assays and colony formation numbers were examined in HepG2 and Huh7 cells treated with DMSO, 4 mM aspirin, both 4 mM aspirin and 2 μg PGAM1-WT, both 4 mM aspirin and 2 μg PGAM1-K99R mutant for 48 h, respectively. d A model of PGAM1K99 de-Succ contributes to the aspirin-mediated inhibition of proliferation of hepatoma cells. e The representative tumor volume of aspirin or DMSO treated xenograft mice was measured. f The levels of 3-PG and 2-PG were measured by ELISA in tumor tissues from aspirin or DMSO-treated xenograft mice. g The levels of HAT1, p-p65, p65, BCL2 and XIAP were explored by Western blot analysis in tumor tissues from aspirin or DMSO-treated xenograft mice. h IP assays and Western blot analysis were performed to examine the succinylation levels of PGAM1 in tumor tissues from aspirin or DMSO-treated xenograft mice. The mean ± SD of at least three experiments is shown. Statistically significant differences are indicated as follows: Student‘s t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
It has been reported that aspirin markedly attenuates the proliferation of cancer cells [30, 31], but the underlying mechanism is poorly understood. In this study, we are interested in the effect of aspirin on PGAM1 Succ in hepatoma cells.
Given that PGAM1 plays important roles in glycolysis by catalyzing the conversion of 3-PG to 2-PG during aerobic glycolysis [11, 32, 33], we explored the effect of aspirin on PGAM1 activities in hepatoma cells. Interestingly, our data revealed that aspirin suppressed the enzymatic activities of PGAM1 to attenuate glycolysis in hepatoma cells. Considering that the PGAM1 Succ plays important roles in promoting glycolysis [20], we supposed that the PGAM1 Succ might be involved in the depression of PGAM1 enzymatic activities mediated by aspirin. As expected, we observed that aspirin reduced the global levels of succinylation of proteins by using pan anti-succinylation antibody, including the PGAM1 Succ in hepatoma cells. It suggests that aspirin inhibits the levels of PGAM1 Succ in liver cancer cells.
Next, we performed RNA-seq to explore the mechanism by which aspirin regulated PGAM1. It has been reported that sirtuin 5 (SIRT5) and sirtuin 7 (SIRT7) regulate protein desuccinylation, and ketoglutarate dehydrogenase complex (KGDHC), carnitine palmitoyltransferase 1A (CPT1A) and HAT1 modulate the protein succinylation [34,35,36,37]. Interestingly, our RNA-seq analysis revealed that aspirin down-regulated the expression of HAT1, but not other succinylation-related proteins. Then, we showed that aspirin decreased the protein levels of HAT1 in a dose-dependent manner in hepatoma cells. It suggests that aspirin down-regulates HAT1 in liver cancer. Considering that HAT1 can induce the succinylation of PGAM1 in liver cancer [20], we supposed that aspirin might inhibit the levels of PGAM1 Succ through modulating HAT1. Our data validated that in our system. It suggests that aspirin decreases the levels of PGAM1 Succ and PGAM1 enzymatic activities by down-regulating HAT1 in liver cancer cells. In this study, we identified that aspirin could affect the global levels of succinylation of proteins, implying that the other potential aspirin-mediated succinylation of proteins might contribute to the inhibition of PGAM1 enzymatic activities in liver cancer cells, which needs to be further studied.
It has been reported that NF-κB is a key target of aspirin in many biology progressions of cancers [25, 26]. Thereby, we speculated that NF-κB might be involved in the HAT1-mediated deficiency of PGAM1 Succ levels and PGAM1 enzymatic activities. At present, the effect of NF-κB on HAT1 has not been reported. In this study, ChIP-qPCR assays showed that NF-κB p65 could significantly enrich on the promoter of HAT1. It suggests that NF-κB p65 may modulate the transcription of HAT1. Moreover, we found that the treatment with PDTC, an inhibitor of NF-κB, could decrease the expression levels of HAT1. Moreover, we showed that the overexpression of NF-κB p65 could significantly up-regulate HAT1 in cells. Meanwhile, the overexpression of NF-κB p65 could remarkably rescue the expression levels of HAT1 decreased by aspirin in cells. It suggests that aspirin down-regulates HAT1 by targeting NF-κB. Potentially, the other NF-κB-modulated targets might be involved in the event, which needs to be further investigated. Next, we found that aspirin restricts the levels of PGAM1 Succ and enzymatic activities through down-regulating NF-κB-mediated HAT1 in liver cancer cells, implying that a NF-κB/HAT1/PGAM1 signaling might contribute to the suppression of glycolysis mediated by aspirin in cells. It has been reported that HAT1 is a multi-modification enzyme with acetylation, succinylation and isobutyrylation [20, 38, 39]. Potentially, the other HAT1-modified targets might be involved in the event, which needs to be further studied.
It has been reported that the PGAM1K99 Succ is involved in the stimulation of glycolytic flux in cancer cells [20]. Therefore, we supposed that the PGAM1K99 Succ might be involved in the event that aspirin inhibits glycolysis in hepatoma cells. Surprisingly, we observed that the PGAM1-K99R mutant failed to block the accumulation of 3-PG (or decrease of 2-PG) in cells treated with aspirin, relative to the overexpression of PGAM1-WT. It suggests that aspirin attenuates the catalytic activities of PGAM1 by inhibiting the levels of PGAM1K99 Succ. Functionally, we observed that PGAM1-WT significantly increased the consumption of glucose (production of lactate acid) attenuated by aspirin, but the PGAM1-K99R mutant failed to work in hepatoma cells. It suggests that PGAM1K99 de-Succ confers the aspirin-mediated defect of glycolysis in the liver cancer. Next, we try to explore the significance of PGAM1K99 Succ in the proliferation of hepatoma cells mediated by aspirin. Our data showed that the overexpression of PGAM1-WT, but not PGAM1-K99R mutant, could remarkably enhance the proliferation of hepatoma cells inhibited by aspirin. It suggests that the PGAM1K99 de-Succ contributes to the inhibition of hepatoma cells proliferation mediated by aspirin. It has been reported that aspirin could acetylate many proteins, which plays potential roles in cancer progression [16, 40]. However, whether aspirin can acetylate PGAM1K99 and its potential function in glycolysis and proliferation of hepatoma cells needs to be further studied. Next, we evaluated the effect of aspirin on HAT1 and PGAM1 Succ in animal. Our data revealed that the treatment with 75 mg/kg aspirin significantly suppressed the tumor volume of mice, supporting that aspirin suppresses the growth of liver cancer. Consistently, we observed that aspirin down-regulated the expression of HAT1 and decreased the levels of PGAM1 succinylation in vivo, contributing to the inhibition of growth of liver cancer. It suggests that aspirin restricts the growth of liver cancer through modulating HAT1 and PGAM1 succinylation. In conclusion, aspirin modulates PGAM1K99 succinylation to restrict the glycolysis through NF-κB p65/HAT1/PGAM1 signaling in liver cancer. It has been reported that NF-κB inhibits the apoptosis of cancer cells through regulating the expression of several anti-apoptotic members, such as BCL2 and XIAP [41]. In this study, we observed that BCL2 and XIAP were down-regulated in the PDTC-treated hepatoma cells and the tumor tissues from mice treated by aspirin. It suggests that apoptosis is also involved in the aspirin-mediated inhibition of liver cancer cells by targeting NF-κB.
Taken together, we summarize a model that aspirin attenuates PGAM1K99 succinylation (Fig. 7). In this model, aspirin down-regulates HAT1 by targeting NF-κB to induce PGAM1K99 de-succinylation, leading to the decrease of PGAM1 enzymatic activities. Functionally, the aspirin-mediated PGAM1K99 de-Succ suppresses the glycolysis and growth of hepatoma cells. Our finding provides new insights into the mechanism by which aspirin inhibits glycolysis in hepatocellular carcinoma.
References
Ricciotti E, Wangensteen KJ, FitzGerald GA. Aspirin in hepatocellular carcinoma. Cancer Res. 2021;81:3751–61.
Capodanno D, Angiolillo DJ. Aspirin for primary cardiovascular risk prevention and beyond in diabetes mellitus. Circulation. 2016;134:1579–94.
Hua H, Zhang H, Kong Q, Wang J, Jiang Y. Complex roles of the old drug aspirin in cancer chemoprevention and therapy. Med Res Rev. 2019;39:114–45.
Joharatnam-Hogan N, Cafferty F, Hubner R, Swinson D, Sothi S, Gupta K, et al. Aspirin as an adjuvant treatment for cancer: feasibility results from the Add-Aspirin randomised trial. Lancet Gastroenterol Hepatol. 2019;4:854–62.
Khan FU, Owusu-Tieku NYG, Dai X, Liu K, Wu Y, Tsai HI, et al. Wnt/β-catenin pathway-regulated fibromodulin expression is crucial for breast cancer metastasis and inhibited by aspirin. Front Pharmacol. 2019;10:1308–24.
McQuillan A, Eikelboom JW. Cyclooxygenase inhibitors and the antiplatelet effects of aspirin. N Engl J Med. 2002;346:1589–90.
Ou YC, Li JR, Wang JD, Chen WY, Kuan YH, Yang CP, et al. Aspirin restores ABT-737-mediated apoptosis in human renal carcinoma cells. Biochem Biophys Res Commun. 2018;502:187–93.
Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:152–62.
Akins NS, Nielson TC, Le HV. Inhibition of glycolysis and glutaminolysis: an emerging drug discovery approach to combat cancer. Curr Top Med Chem. 2018;18:494–504.
Yuan Y, Yuan HF, Geng Y, Zhao LN, Yun HL, Wang YF, et al. Aspirin modulates 2-hydroxyisobutyrylation of ENO1K281 to attenuate the glycolysis and proliferation of hepatoma cells. Biochem Biophys Res Commun. 2021;560:172–8.
Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600.
Sharif F, Rasul A, Ashraf A, Hussain G, Younis T, Sarfraz I, et al. Phosphoglycerate mutase 1 in cancer: A promising target for diagnosis and therapy. IUBMB Life. 2019;71:1418–27.
Xiao M, Li X, Su Y, Liu Z, Han Y, Wang S, et al. Kinetochore protein MAD1 participates in the DNA damage response through ataxia-telangiectasia mutated kinase-mediated phosphorylation and enhanced interaction with KU80. Cancer Biol Med. 2020;17:640–51.
Yan F, Qian M, He Q, Zhu H, Yang B. The posttranslational modifications of Hippo-YAP pathway in cancer. Biochim Biophys Acta Gen Subj. 2020;1864:129397–404.
Wang WH, Yuan T, Qian MJ, Yan FJ, Yang L, He QJ, et al. Post-translational modification of KRAS: potential targets for cancer therapy. Acta Pharmacol Sin. 2021;42:1201–11.
Chen Z, Li W, Qiu F, Huang Q, Jiang Z, Ye J, et al. Aspirin cooperates with p300 to activate the acetylation of H3K9 and promote FasL-mediated apoptosis of cancer stem-like cells in colorectal cancer. Theranostics. 2018;8:4447–61.
Ou YQ, Zhu W, Li Y, Qiu PX, Huang YJ, Xie J, et al. Aspirin inhibits proliferation of gemcitabine-resistant human pancreatic cancer cells and augments gemcitabine-induced cytotoxicity. Acta Pharmacol Sin. 2010;31:73–80.
Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6.
Tong Y, Guo D, Yan D, Ma C, Shao F, Wang Y, et al. KAT2A succinyltransferase activity-mediated 14-3-3ζ upregulation promotes β-catenin stabilization-dependent glycolysis and proliferation of pancreatic carcinoma cells. Cancer Lett. 2020;469:1–10.
Yang G, Yuan Y, Yuan H, Wang J, Yun H, Geng Y, et al. Histone acetyltransferase 1 is a succinyltransferase for histones and non-histones and promotes tumorigenesis. EMBO Rep. 2021;22:e50967.
Dai J, Huang YJ, He X, Zhao M, Wang X, Liu ZS, et al. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell. 2019;176:1447–60.e14.
Saha S, Mukherjee S, Khan P, Kajal K, Mazumdar M, Manna A, et al. Aspirin suppresses the acquisition of chemoresistance in breast cancer by disrupting an NFκB-IL6 signaling axis responsible for the generation of cancer stem cells. Cancer Res. 2016;76:2000–12.
Meng Q, Peng X, Zhao S, Xu T, Wang S, Liu Q, et al. Hypoxic storage of erythrocytes slows down storage lesions and prolongs shelf-life. J Cell Physiol. 2019;234:22833–44.
Lee HW, Kyung T, Yoo J, Kim T, Chung C, Ryu JY, et al. Real-time single-molecule co-immunoprecipitation analyses reveal cancer-specific Ras signalling dynamics. Nat Commun. 2013;4:1505–13.
Liao D, Zhong L, Duan T, Zhang RH, Wang X, Wang G, et al. Aspirin suppresses the growth and metastasis of osteosarcoma through the NF-κB pathway. Clin Cancer Res. 2015;21:5349–59.
Jiang W, Yan Y, Chen M, Luo G, Hao J, Pan J, et al. Aspirin enhances the sensitivity of colon cancer cells to cisplatin by abrogating the binding of NF-κB to the COX-2 promoter. Aging. 2020;12:611–27.
Zhang W, Zhangyuan G, Wang F, Jin K, Shen H, Zhang L, et al. The zinc finger protein Miz1 suppresses liver tumorigenesis by restricting hepatocyte-driven macrophage activation and inflammation. Immunity. 2021;54:1168–85.e8.
Li X, Zhang C, Zhao T, Su Z, Li M, Hu J, et al. Lysine-222 succinylation reduces lysosomal degradation of lactate dehydrogenase a and is increased in gastric cancer. J Exp Clin Cancer Res. 2020;39:172–88.
Chen XF, Tian MX, Sun RQ, Zhang ML, Zhou LS, Jin L, et al. SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep. 2018;19:e45124.
Peng F, Wang JH, Fan WJ, Meng YT, Li MM, Li TT, et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene. 2018;37:1062–74.
Li S, Dai W, Mo W, Li J, Feng J, Wu L, et al. By inhibiting PFKFB3, aspirin overcomes sorafenib resistance in hepatocellular carcinoma. Int J Cancer. 2017;141:2571–84.
Huang K, Liang Q, Zhou Y, Jiang LL, Gu WM, Luo MY, et al. A novel allosteric inhibitor of phosphoglycerate mutase 1 suppresses growth and metastasis of non-small-cell lung cancer. Cell Metab. 2019;30:1107–19.e8
Liu X, Tan X, Liu P, Wu Y, Qian S, Zhang X. Phosphoglycerate mutase 1 (PGAM1) promotes pancreatic ductal adenocarcinoma (PDAC) metastasis by acting as a novel downstream target of the PI3K/Akt/mTOR pathway. Oncol Res. 2018;26:1123–31.
Yang Y, Gibson GE. Succinylation links metabolism to protein functions. Neurochem Res. 2019;44:2346–59.
Liu J, Shangguan Y, Tang D, Dai Y. Histone succinylation and its function on the nucleosome. J Cell Mol Med. 2021;25:7101–9.
Kwon OK, Bang IH, Choi SY, Jeon JM, Na AY, Gao Y, et al. SIRT5 is the desuccinylase of LDHA as novel cancer metastatic stimulator in aggressive prostate cancer. Genomics Proteom Bioinforma. 2022;S1672-0229:00018–3.
Yu HB, Cheng ST, Ren F, Chen Y, Shi XF, Wong VKW, et al. SIRT7 restricts HBV transcription and replication through catalyzing desuccinylation of histone H3 associated with cccDNA minichromosome. Clin Sci. 2021;135:1505–22.
Gruber JJ, Geller B, Lipchik AM, Chen J, Salahudeen AA, Ram AN, et al. HAT1 coordinates histone production and acetylation via H4 promoter binding. Mol Cell. 2019;75:711–24.e5.
Zhu Z, Han Z, Halabelian L, Yang X, Ding J, Zhang N, et al. Identification of lysine isobutyrylation as a new histone modification mark. Nucleic Acids Res. 2021;49:177–89.
Alfonso L, Ai G, Spitale RC, Bhat GJ. Molecular targets of aspirin and cancer prevention. Br J Cancer. 2014;111:61–7.
Beug ST, Cheung HH, LaCasse EC, Korneluk RG. Modulation of immune signalling by inhibitors of apoptosis. Trends Immunol. 2012;33:535–45.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82103066).
Author information
Authors and Affiliations
Contributions
YFW, LNZ, YG, HFY, CYH, and HHZ performed the experiments; XDZ and GY conceived and designed the project. YFW, XDZ, and YG wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, Yf., Zhao, Ln., Geng, Y. et al. Aspirin modulates succinylation of PGAM1K99 to restrict the glycolysis through NF-κB/HAT1/PGAM1 signaling in liver cancer. Acta Pharmacol Sin 44, 211–220 (2023). https://doi.org/10.1038/s41401-022-00945-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-022-00945-z