Abstract
The ubiquitin-proteasome system (UPS) is essential for maintaining cell homeostasis by orchestrating the protein degradation, but is impaired in various diseases, including cancers. Several proteasome inhibitors, such as bortezomib, are currently used in cancer treatment, but associated toxicity limits their widespread application. Recently metal complex-based drugs have attracted great attention in tumor therapy; however, their application is hindered by low water-solubility and poor absorbency. Herein, we synthesized a new type of gold (I) complex named Na-AuPT, and further characterized its anticancer activity. Na-AuPT is highly water-soluble (6 mg/mL), and it was able to potently inhibit growth of a panel of 11 cancer cell lines (A549, SMMC7721, H460, HepG2, BEL7402, LNCap, PC3, MGC-803, SGC-7901, U266, and K562). In A549 and SMMC7721 cells, Na-AuPT (in a range of 2.5–20 μM) inhibited the UPS function in a dose-dependent fashion by targeting and inhibiting both 20 S proteasomal proteolytic peptidases and 19 S proteasomal deubiquitinases. Furthermore, Na-AuPT induced caspase-dependent apoptosis in A549 and SMMC7721 cells, which was prevented by the metal chelator EDTA. Administration of Na-AuPT (40 mg · kg−1 · d−1, ip) in nude mice bearing A549 or SMMC7721 xenografts significantly inhibited the tumor growth in vivo, accompanied by increased levels of total ubiquitinated proteins, cleaved caspase 3 and Bax protein in tumor tissue. Moreover, Na-AuPT induced cell death of primary mononuclear cells from 5 patients with acute myeloid leukemia ex vivo with an average IC50 value of 2.46 μM. We conclude that Na-AuPT is a novel metal-based proteasome inhibitor that may hold great potential for cancer therapy.
Similar content being viewed by others
Introduction
The ubiquitin-proteasome system (UPS) is responsible for the degradation of intracellular misfolded or mis-assembled proteins in eukaryotic cells. The proteasome is composed of a 20 S proteolytic core and two 19 S regulatory caps, which binds to both ends of the 20 S core to form a 26 S complex [1, 2]. The constitutive 20 S proteasome core has three distinct catalytic activities, chymotrypsin-like (CT-L), trypsin-like (T-L), and caspase-like (C-L), derived from β5, β2, and β1 subunits, respectively. A target protein to be degraded is first polyubiquitinated by several enzymes, called E1, E2, and E3. After that, the 19 S cap distinguishes and removes the ubiquitinated chain from the target protein, and sends it to the 20 S core for degradation. Inhibition of the proteasome activity in cancer cells results in growth arrest and cell death or apoptosis [3, 4]. Compared with normal cells, cancer cells usually have a higher level of proteasome activity. These characteristics cause cancer cells to be more sensitive to apoptosis induced by proteasome inhibitors than normal cells [5].
The development of proteasome inhibitors has been widely explored as a cancer therapeutic strategy. As an example, bortezomib (Velcade) is the first proteasome inhibitor approved by the United States FDA for the treatment of multiple myeloma and mantle cell lymphoma [6, 7]. Although the clinical success of bortezomib-based therapy is encouraging, a large amount of patients suffers severe toxicities associated with its clinical use. In addition, dose-limiting toxicities, including painful peripheral neuropathy and thrombocytopenia, have been reported in patients treated with bortezomib [8]. The second generation 20 S proteasome inhibitor carfilzomib (PR-171) specifically targets and inhibits the CT-L activity of 20 S proteasome and is effective for treating some patients with multiple myeloma who did not respond to bortezomib previously [9]. The consecutive day dosing regimen with carfilzomib is well tolerated and exhibits antitumor activity in hematologic malignancies, including patients who have previously received bortezomib treatment [10]. However, the emergence of primary or secondary carfilzomib resistance remains as a clinical challenge [11]. Therefore, the development of novel proteasome inhibitors to overcome resistance to conventional 20 S proteasome inhibitors would provide patients with alternatives, including more flexible dosages and better responses. The proteasome-associated deubiquitinases (DUBs) as a target for the development of next-generation UPS inhibitors has recently attracted great interest. It is believed that use of proteasomal DUB inhibitors may overcome resistance to 20 S proteasome inhibitors [12]. Better understanding of properties and functions of different UPS inhibitors may provide further clinical possibilities for precision treatment of cancers.
We and others have previously reported that some metal-based compounds have strong activity in inhibiting the UPS machinery and inducing cytotoxicity in cancer cells [13,14,15,16,17,18,19]. However, most of these agents have poor water-solubility, which reduces their dissolution rate and bioavailability. In this study, we designed a new metal-based proteasome inhibitor named Na-AuPT, a water-soluble gold(I) complex. We then showed that Na-AuPT can suppress tumor growth in vitro and in vivo by blocking activities of both 20 S proteasome proteolytic peptidases and 19 S proteasome-associated DUBs. Our preclinical studies have provided evidence for Na-AuPT as a novel proteasome inhibitor and potential anticancer agent.
Materials and methods
Materials
Na-AuPT was synthesized in our laboratory and stored as a 10 mM stock solution in distilled and deionized water (ddH2O) at −20 °C. N-ethylmaleimide (NEM, 04259) was purchased from Sigma-Aldrich, St. Louis, MO, USA. Pan-caspase inhibitor z-VAD-FMK (S7023), cisplatin (S1166), and bortezomib (S1013) were purchased from Selleckchem, Houston, TX, USA. Suc-Leu-Leu-Val-Tyr-AMC (S-280), Z-Leu-Leu-Glu-AMC (S-230), Boc-Leu-Arg-Arg-AMC (S-300), b-AP15 (4566), 20 S proteasome (E-360), 26 S proteasome (E-365), and HA-Ubiquitin-Vinyl Sulfone (HA-Ub-VS, U-212) were obtained from Boston Biochem, Cambridge, MA, USA. Antibodies used in this study were purchased from the following sources: anti-ubiquitin (Santa Cruz Biotechnology, sc-8017), anti-GFP (Santa Cruz Biotechnology, sc-9996), anti-K11-linkage specific polyubiquitin (MERCK, MABS107-1), anti-caspase 3 (Cell Signaling Technology, 9665), anti-caspase 8 (Cell Signaling Technology, 9746), anti-caspase 9 (Cell Signaling Technology, 9508), anti-cleaved caspase-3 (Cell Signaling Technology, 9661), anti-cleaved caspase-8 (Cell Signaling Technology, 9496), anti-PARP (Cell Signaling Technology, 9532), anti-K48-linkage specific polyubiquitin (Cell Signaling Technology, 8081), anti-K63-linkage specific polyubiquitin (Cell Signaling Technology, 5621), anti-Bax (Cell Signaling Technology, 5023), anti-phospho-ATM (Ser1981) (Cell Signaling Technology, 5883), anti-ATM (Cell Signaling Technology, 2873), anti-γH2AX (Cell Signaling Technology, 9718), anti-H2AX (Cell Signaling Technology, 7631) and anti-GAPDH (Bioworld Technology, BS60630).
Structure analysis of Na-AuPT
Crystal structure of complex Na-AuPT (crystal size: 0.3 × 0.2 × 0.15 mm) was determined by single-crystal X-ray analysis. The diffraction data were collected on a Rigaku Mercury CCD using a graphite monochromator Mo Kα radiation (λ = 0.071073 nm) at 293(2) K. A total of 17,258 reflections were collected with 3316 unique one (Rint = 0.060 0) for complex Na-AuPT. Lorentz factor, polarization, air absorption, and absorption due to variations in the path length through the detector faceplate were used to correct the data sets. Absorption correction based on multi-scan method was also employed. The structure of Na-AuPT was solved via direct methods (SHELXTL) and refined by full-matrix least-square techniques with atomic coordinates and anisotropic temperature factors for all non-hydrogen atoms. The hydrogen atoms attached to C and O atoms were added according to theoretical models, and their positions and thermal parameters were fixed during the structure refinement. For complex Na-AuPT, the final R1 = 0.0592, and wR2 = 0.1134, S = 1.012, (▵ρ) max = 0.919 and (▵ρ) min = −1.242 e ·Å−3 for 2 844 observed reflections (I > 2σ (I)) with 253 parameters.
Cell culture
Human lung cancer A549 and human liver cancer SMMC7721 cell lines were obtained from American Type Culture Collection, Manassas, VA, USA. A549 and SMMC7721 cell lines were cultured in RPMI-1640. Both of these media were supplemented with 10% fetal bovine serum (FBS), 100 units/mL of penicillin, and 100 μg/mL of streptomycin. Cell cultures were maintained at 37 °C and 5% CO2. All cells were mycoplasma-free and authenticated using short tandem repeat DNA profiling analysis.
Cell viability assay
The CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS assay, Promega, G3582) was performed to test cell viability as previously reported [16]. Briefly, A549 and SMMC7721 cells were plated onto 96-well plates at total of 8000 cells/well, and then treated with either vehicle or indicated complexes for 24 and 48 h before performing the MTS assay. MTS assay reagent (20 μL) was added to each well of the 96 well plate 3 h before the termination of the experiments. The absorbance at 490 nm was detected using a plate reader (Varioskan Flash 3001, Thermo, Waltham, MA, USA). The concentrations of Na-AuPT that induce 50% inhibition of cell growth (IC50) values were derived.
Cell death assay
Apoptotic rates were assessed by flow cytometry using Annexin V-fluoroisothiocyanate (FITC)/propidium iodide (PI) double staining kit (Keygen, KGA108). After treatment with Na-AuPT at various concentrations for 24 h, cells were collected and washed with binding buffer, then incubated with Annexin V-FITC followed by the addition of PI prior to analysis. In addition, cells were submitted to Annexin V-FITC /PI staining in situ, then imaged with an inverted fluorescence microscope equipped with a digital camera (Axio Obsever Z1, Zeiss, Germany).
Western blot analysis
Whole cells were lysed in RIPA buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) and supplemented with freshly added 10 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× Protease Inhibitor Cocktail (Roche, 4693116001). Western blot assay was performed as we described previously [15]. In brief, an equal amount of the total protein extracted from cultured cells was fractionated by 12% SDS–PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. After blocking with 5% non-fat milk for 1 h, the designated proteins were detected using specific primary antibodies (1:1000) and appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5000). Blots were reacted with the Enhanced chemiluminescence detection reagents (Santa Cruz Biotechnology, sc-2048) and revealed by X ray films (Kodak, Japan).
Measurement of reactive oxygen species generation
Cancer cells were treated with Na-AuPT or positive control in the absence or presence of antioxidant vitamin C for 1 h. The cells were harvested and incubated with the free serum medium with addition of DCFH-DA (10 μM) for 20 min at 37 °C in the dark. In the presence of reactive oxygen species (ROS), DCFH penetrates the cells and is in turn oxidized to DCF. DCF fluorescence was detected by flow cytometry using FACScanto (BD Biosciences, Franklin Lakes, NJ, USA). The data were processed and analyzed through FlowJo v7.6 (Ashland, OR, USA).
Sample collection and cell culture
Written informed consent was obtained from all participants, and the study was approved by the Ethical Committee of the Second Affiliated Hospital of Guangzhou Medical University. Five patients with acute myeloid leukemia (AML) and five healthy volunteers were recruited in this preclinical study. Blood samples of patients with AML were obtained from the Department of Hematology (the Second Affiliated Hospital of Guangzhou Medical University). Peripheral blood samples of normal control individuals were obtained from Guangzhou Blood Center. Ficoll-Paque (GE, 17-1440-02) was used to isolate mononuclear cells from either peripheral blood samples by density gradient. The mononuclear cells were cultured in RPMI-1640 culture medium with 15% FBS as described previously [16].
Proteasomal peptidase activity assay
Briefly, purified human 20 S proteasome was incubated with 2.5, 5, 10, or 20 μM Na-AuPT or 100 nM bortezomib for 60 min at 37 °C before the addition of fluorogenic substrates. The 20 S proteasome activity was measured by the release of hydrolyzed AMC groups using a fluorescence microplate reader (Varioskan Flash 3001, Thermo, USA) as previously described [15]. The fluorogenic substrate used for CT-L (β5), C-L (β1), and T-L (β2) activity is Suc-Leu-Leu-Val-Tyr-AMC, Z-Leu-Leu-Glu-AMC, Boc-Leu-Arg-Arg-AMC, respectively.
Proteasomal deubiquitinase (DUB) activity assay
Proteasomal DUB activity was performed as previously reported. After pretreatment with Na-AuPT (2.5, 5, 10, or 20 μM) or NEM (2 mM), 26 S proteasome (25 nM) was dissolved in ice-cold DUB buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 250 mM sucrose, and 1 mM PMSF) for 15 min. Then, ubiquitin 7-amido-4-methylcoumarin (Ub-AMC, U-550) was added and the mixtures (at a reaction volume of 100 μl) were incubated at 25 °C. A fluorescence microplate reader (Varioskan Flash 3001, Thermo, USA) was used to record the release of AMC from substrate cleavage.
DUB active-site-directed labeling assays
Purified 26 S proteasomes (25 nM) were dissolved in DUB buffer (25 mM Tris-HCl pH 7.4, 5 mM MgCl2, 20 mM NaCl, 200 μM ATP) and then treated with Na-AuPT or b-AP15 for 10 min before they were incubated with HA-Ub-VS for 1 h at 37 °C, followed by boiling in the reducing sample buffer and fractionated via SDS–PAGE. After transferred to PVDF membranes, HA-Ub-VS-labeled DUBs were immunodetected using anti-HA antibodies.
Nude mouse xenograft model
Male nude Balb/c mice aged 5 weeks were obtained from Guangdong Animal Center. The use and care of experimental animals were approved by the Institutional Animal Care and Use Committee of Guangzhou Medical University. The mice were bred in a barrier facility with a 12 h light dark cycle, with food and water available ad libitum. Approximately 1 × 107 of A549 or SMMC7721 cells in a final volume of 100 μL were inoculated subcutaneously on the flanks of each male mouse. After 3 days of inoculation, mice were treated with either vehicle (ddH2O2, intraperitoneal injection) or Na-AuPT (40 mg · kg−1 · d−1, intraperitoneal injection) for a total of 13 days in A549 and 15 days in SMMC7721 xenografts. Tumor volumes were measured and calculated every 2 days with the use of calipers as we reported previously [15]. After Na-AuPT treatment, the mice were sacrificed, and tumor xenografts were removed, weighed, stored, and fixed for biochemical or histological analyses, respectively.
Immunohistochemical staining
Xenografts were formalin-fixed, embedded in paraffin and sectioned with standard techniques as previously indicated [15]. Immunostaining of tumor xenograft sections (4 μm) was performed using MaxVisionTM reagent (Maixin Biol, Fuzhou, China) according to the manufacturer’s instructions. After incubation with the primary antibodies, the slides were treated with 0.05% diaminobenzidine and 0.03% H2O2 in 50 mM Tris-HCl (pH 7.6) and counterstained with hematoxylin.
Statistical analysis
All the results were expressed as Mean ± SEM, with at least three individual measurements in all experiments. GraphPad Prism 5.0 software (GraphPad Software, USA) was used for statistical analysis. Two tailed unpaired Student’s t-test was used to evaluate the statistical significance of the difference between two groups in all experiments. The P < 0.05 was considered statistically significant.
Results
Na-AuPT induces apoptosis in cancer cells
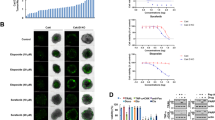
Metal complexes, including gold(I) complexes, show potential anticancer effects, but their clinical application could be hindered by poor water-solubility [20, 21]. To overcome this shortcoming, we synthesized a new water-soluble gold(I) complex, named Na-AuPT. The crystal and structure of Na-AuPT are shown in Table 1 and Fig. 1a. Indeed, Na-AuPT had higher water-solubility than several reported metal-containing agents, including auranofin [22], aumdubin [23], CdPT [24], AgDT [25], and PdPT [16] (Table 2). To determine anticancer activity of Na-AuPT, we first evaluated dose and time effects of Na-AuPT on cell growth in A549 (human lung cancer) and SMMC7721 (human liver cancer) cells. Cell viability assay using the MTS method showed that Na-AuPT significantly inhibited growth of A549 and SMMC7721 cells (Fig. 1b). The potency of Na-AuPT was also determined in several other cancer cell lines, including lung cancer (H460), liver cancer (HepG2 and BEL7402), prostate cancer (LNCap and PC3), gastric cancer (MGC-803 and SGC-7901), multiple myeloma (U266) and leukemia (K562). The results showed that Na-AuPT had a wide spectrum of anticancer activity (Supplementary Fig. S1). Next, to determine whether growth inhibition induced by Na-AuPT is due to increased cell death, we used Annexin V-FITC/PI double staining to assay levels of apoptosis and necrosis in A549 and SMMC7721 cells following treatment with Na-AuPT (2.5–20 μM) for 24 h. Flow cytometry or fluorescence microscope image analysis showed that Na-AuPT caused a dose-dependent increase in Annexin V/PI-positive cells (Fig. 1c, d), indicating that Na-AuPT induced apoptosis. Subsequent Western blot analysis of caspase cleavage (a key biochemical event of apoptosis) further confirmed that Na-AuPT is a strong inducer of apoptosis, as shown by increased levels of cleavage of caspase-3, -8, and -9 (Fig. 1e). Consistently, Na-AuPT induced an increase in cleavage of poly (ADP-ribose) polymerase (PARP), a classic substrate protein of caspase activation (Fig. 1e). Importantly, the Na-AuPT-induced cell death was prevented by addition of a pan-caspase inhibitor z-VAD-FMK (Fig. 1f), supporting the conclusion that caspase-dependent apoptosis is responsible for the anticancer activity of Na-AuPT in vitro.
a The structure of Na-AuPT. Single crystals of Na-AuPT were scanned by single-crystal X-ray diffraction. The structure of Na-AuPT was solved using direct methods (SHELXTL) and refined using the full-matrix least-square technique with atomic coordinates and anisotropic temperature factors for all non-hydrogen atoms. b A549 and SMMC7721 cells were treated with Na-AuPT at the indicated doses for 24 or 48 h. Cell viability of A549 and SMMC7721 was determined by MTS assay. c, d A549 and SMMC7721 cells were treated with Na-AuPT at the indicated doses for 24 h. Cells were stained with Annexin V-FITC/PI and analyzed by flow cytometry (c) or microscopy (d) detection. e A549 and SMMC7721 cells were treated with Na-AuPT at the indicated doses for 24 h. The indicated proteins were detected using Western blot analysis. f A549 and SMMC7721 cells were exposed to Na-AuPT (10 μM) alone or combined with caspase inhibitor z-VAD-FMK (50 μM) for 12 h. Cells were stained with Annexin V-FITC/PI, followed by microscopy detection and statistical analysis of cell death. Mean ± SEM (n = 5). *P < 0.05.
Na-AuPT represses UPS function
Several metal complexes (e.g., cisplatin) exert their cytotoxic effect by inducing DNA damage [26]. To investigate whether Na-AuPT could induce cell death through induction of DNA damage response pathway, levels of DNA damage-related proteins were measured in the treated A549 and SMMC7721 cells. We found that cisplatin, but not Na-AuPT, induced expression of p-ATM and γH2AX in A549 and SMMC7721 cells (Supplementary Fig. S2a). This indicates that Na-AuPT’s anticancer activity seems not to involve induction of DNA damage. Another possible mechanism of metal complex-induced apoptosis is induction of ROS [27]. We next investigated whether Na-AuPT could induce ROS in cancer cells. We found that Na-AuPT has minimal effects to induce the increase of ROS in A549 cells (Supplementary Fig. S2b). Consistently, Na-AuPT-induced cytotoxicity was not inhibited by an antioxidant vitamin C (Supplementary Fig. S2c), which is an effective ROS scavenger in A549 cells (Supplementary Fig. S2b). Thus, induction of ROS may not be a main cause of Na-AuPT-mediated apoptosis in cancer cells.
It has been reported that anticancer activity of metal complexes is associated with inhibition of proteasome function [13,14,15,16,17,18,19]. We then investigated whether Na-AuPT can affect UPS function. We first estimated abundance of ubiquitinated proteins in A549 and SMMC7721 cells treated with Na-AuPT or bortezomib (as a positive control). Lysine 48 (K48)-linked polyubiquitin chain is a signal for the proteasomal degradation, while lysine 63 (K63)- and lysine 11 (K11)-linked polyubiquitin chains are involved in nonproteolytic events and endoplasmic reticulum-associated degradation, respectively [28]. Similar to bortezomib, Na-AuPT triggered a conspicuous upregulation in both total and K48-linked ubiquitinated proteins in a dose- and time-dependent manner (Fig. 2a, b). However, Na-AuPT had no apparent effect on K63-linked and K11-linked ubiquitinated proteins (Supplementary Fig. S3a, b). In addition, Na-AuPT induced accumulation of GFPu, a surrogate UPS substrate that is commonly used as an indicator of degradation function of proteasome [29], in HEK293 cells (Fig. 2c, d). These findings support the conclusion that Na-AuPT is an inhibitor of the UPS pathway.
a A549 and SMMC7721 cells were exposed to the indicated doses of Na-AuPT or bortezomib (100 nM) for 6 h. The total ubiquitinated proteins (Ub-prs) and K48-linked ubiquitinated proteins (K48-) were assessed by Western blot. b A549 and SMMC7721 cells were administered 10 μM Na-AuPT for various time points. Ub-prs., K48- and PARP were estimated by Western blot. c, d HEK293-GFPu cells stably expressing GFPu, a UPS surrogate substrate, were treated with 2.5 and 5 μM Na-AuPT or 100 nM bortezomib. The Ub-prs and GFPu levels were assessed by Western blot (c). d Microscopy analysis was performed to detect GFPu. e A549 and SMMC7721 cells were exposed to Na-AuPT (10 μM) alone or combined with caspase inhibitor z-VAD-FMK (50 μM) for 12 h. Total ubiquitinated proteins (Ub-prs) and PARP were detected via Western blot.
We noted that in the Na-AuPT-treated cells, the appearance of ubiquitination accumulation was earlier than PARP cleavage (Fig. 2b), indicating that Na-AuPT-induced ubiquitin-specific protease (USP) inhibition might be an upstream signal of apoptosis induction. Consistent with this hypothesis, the Na-AuPT-mediated cleavage of PARP, but not the accumulation of ubiquitinated proteins, was inhibited by Z-VAD-FMK (Fig. 2e). Thus, Na-AuPT is potent to inhibit ubiquitination-mediated protein degradation that leads to apoptosis induction in cancer cells.
Na-AuPT inhibits activities of proteolytic peptidases and 19 S proteasomal DUBs
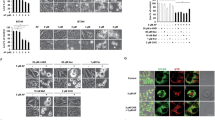
To study whether Na-AuPT inhibits the proteasome activity, we first examined whether Na-AuPT has a direct effect on the proteolysis peptidase activity of 20 S proteasome. As well known, bortezomib inhibits proteasome function by suppressing the CT-L (β5) and C-L (β1) but not the T-like (β2) activity [30]. When Na-AuPT was incubated with purified 20 S proteasome, we observed that Na-AuPT caused a dose-dependent decrease in β5 and β1, instead of β2, peptidase activities of purified 20 S proteasome, similar to bortezomib (Fig. 3a). The activities of Na-AuPT and bortezomib were further compared using proteasomes derived from A549 cell extracts. Na-AuPT inhibited all β1, β2, and β5 activities of the proteasome in A549 cell extracts in a concentration-dependent manner, which is different from the inhibitory effect of bortezomib that inhibited only β1 and β5 activities (Fig. 3b). These biochemical analyses indicate that Na-AuPT may have a different mechanism to inhibit the 20 S proteasome from bortezomib.
a, b Effect of Na-AuPT on 20 S proteasome activity. Purified 20 S proteasomes (a) or A549 cell extracts (b) were exposed to various doses of Na-AuPT or 100 nM bortezomib. The caspase-like (β1), trypsin-like (β2) and chymotrypsin-like (β5) peptidase activities were assessed by special fluorogenic substrates, while bortezomib was positive control. Mean ± SEM (n = 5). *P < 0.05. c Effect of Na-AuPT on proteasome deubiquitinases activity. Purified 26 S proteasome was exposed to various doses of Na-AuPT or 2 mM N-ethylmaleimide (NEM) for 15 min prior to incubation with Ub-AMC. The deubiquitinases activity was shown via fold change of fluorescence relative to initiated point fluorescence reading. d Active-site-directed labeling of proteasome deubiquitinases. Purified 26 S proteasomes were incubated with Na-AuPT (20 or 200 μM) or 50 μM b-AP15 (a positive control) for 10 min, labeled with HA-Ub-VS for 60 min 37 °C, and then the deubiquitinases activity was analyzed via HA antibody with Western blot.
Next, effect of Na-AuPT on 19 S proteasome-associated DUB activities was determined by measuring the cleavage of Ub-AMC, which is a fluorogenic substrate for DUBs, including ubiquitin carboxy-terminal hydrolases (UCHs) and USPs. Na-AuPT completely abrogated the cleavage of Ub-AMC by purified 26 S proteasome at the doses of 2.5, 5, 10, and 20 μM, similar to a general DUB inhibitor NEM (Fig. 3c). These findings suggest that Na-AuPT is able to inhibit the DUB activity of the proteasome.
We then tested whether Na-AuPT can bind to the proteasomal DUBs UCHL5 and USP14 by using a probe of DUB activities HA-Ub-VS, which covalently binds to the active sites of UCHs and USPs [31]. Similar to the reported proteasomal DUB inhibitor b-AP15, high doses of Na-AuPT (20 and 200 μM) were able to competitively disturb binding of HA-Ub-VS to UCHL5 or USP14 of a purified 26 S proteasome (Fig. 3d). Altogether, these findings demonstrate that Na-AuPT inhibits both 20 S proteasome and 19 S proteasome-associated DUBs, UCHL5 and USP14.
EDTA blocks Na-AuPT-induced apoptosis and proteasome inhibition
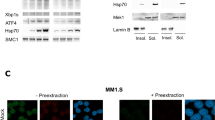
To verify whether Au+ is indispensable to Na-AuPT-induced effects, ethylenediaminetetraacetic acid (EDTA), a widely used metal chelating agent was used to chelate Au+ of Na-AuPT, followed by investigating changes in levels of Na-AuPT-induced cell death and proteasome inhibition. A co-treatment of EDTA with Na-AuPT inhibited cell death induced by Na-AuPT alone in A549 and SMMC7721 cells (Fig. 4a). Consistent with the hypothesis that Na-AuPT-induced apoptosis requires Au+, EDTA also blocked Na-AuPT-induced PARP cleavage in A549 cells (Fig. 4b). Furthermore, EDTA prevented two other Na-AuPT-induced events, accumulation of polyubiquitinated proteins in A549 cells (Fig. 4b) and increase of GFPu in HEK293 cells (Fig. 4c, d). Collectively, these data support the conclusion that Au+ plays a significant role in mediating Na-AuPT activity in terms of apoptosis induction and proteasome inhibition.
a A549 and SMMC7721 cells were treated with Na-AuPT (10 μM) in the absence or presence of EDTA (100 μM). Apoptotic cells were stained with Annexin V-FITC/PI, followed by flow cytometry. b A549 cells were treated with Na-AuPT in the absence or presence of EDTA. Western blot was employed to analyze of Ub-prs and PARP expression. c, d GFPu-HEK293 cells, a clonal HEK293 cell line stably transfected with GFPu, were exposed to Na-AuPT in absence or presence of EDTA. Microscopy analysis was performed to detect GFPu (c). The Ub-prs and GFPu levels were assessed by Western blot (d).
The anticancer activity of Na-AuPT in vivo
To evaluate the anticancer effect of Na-AuPT in vivo, immunodeficient Balb/c nude mice were employed to create xenograft models by subcutaneous injection of A549 or SMMC7721 cells. Three days after inoculation, these mice were administered either vehicle or Na-AuPT (40 mg · kg−1d−1) via intraperitoneal injection for up to 13 and 15 days in A549 and SMMC7721 xenografts, respectively. Compared with the vehicle group, the tumor volume and tumor size in the Na-AuPT treatment group were reduced (Fig. 5a, b). Moreover, Na-AuPT treatment resulted in reduction in tumor weights of both models, 54.3% in SMMC7721 xenografts and 35.2% in A549 tumors (Fig. 5c). There were no significant differences in body weights between the control group and the Na-AuPT-treated group (Fig. 5d), indicating that Na-AuPT is not toxic under the used in vivo conditions. Immunohistochemistry staining revealed that levels of total ubiquitinated proteins, cleaved caspase 3 and Bax (a pro-apoptosis protein that can be degraded by the UPS [32]) were all significantly increased in the Na-AuPT-treated group (Fig. 5e). These animal studies demonstrate that the anticancer activity of Na-AuPT in vivo was accompanied by the accumulation of pro-apoptotic Bax protein and induction of apoptosis in cancer cells.
BALB/c nude mice bearing A549 and SMMC7721 tumors were administered vehicle or Na-AuPT (40 mg · kg−1d−1, intraperitoneal injection) for 13 (A549) and 15 (SMMC7721) days, respectively. Tumor growth curves were recorded every 2 days. Tumor size (a), tumor images (b) and tumor weight (c), and body weight (d) were shown. Mean ± SEM (n = 6). *P < 0.05; ns no significance. e Immunohistochemistry staining of total ubiquitinated proteins (Ub-prs), Bax, and cleaved caspase 3 in tumor tissues. Images collected at a magnification of ×200 are shown.
Ex vivo effect of Na-AuPT on primary cancer cells from leukemia patients
Given that proteasome inhibitors (e.g., bortezomib) have been proven effective in treating some leukemia patients [33], we next sought to determine the ex vivo antitumor effect of Na-AuPT and cytotoxic effect on primary mononuclear cells isolated from five patients with AML and five healthy volunteers. We found that primary mononuclear cells from AML patients were more sensitive to Na-AuPT-induced cell death, as evidenced by an average IC50 value of 2.46 μM, which was much lower than normal controls (average IC50: 38.97 μM) (Fig. 6a). Consistently, Annexin V/PI staining followed by flow cytometry and fluorescent microscope analysis revealed that Na-AuPT caused more robust apoptosis in primary mononuclear cells isolated from AML patients than normal controls (Fig. 6b, c). Like bortezomib, Na-AuPT also increased the levels of total and K48-linked ubiquitinated proteins in the primary mononuclear cells driven from AML patients (Fig. 6d). Collectively, these ex vivo results suggest that Na-AuPT is much more cytotoxic to primary cancer cells than normal cells.
a Cancer cells from five AML patients and peripheral blood mononuclear cells from five healthy volunteers were exposed to Na-AuPT at the indicated concentrations for 24 h. Cell viability was determined by MTS assay. The IC50 values in each group are shown in the scatter plot. Mean ± SEM (n = 5). *P < 0.05, normal controls versus AML patients. b, c Peripheral mononuclear cells from AML patients were treated with Na-AuPT at the indicated concentrations for 24 h. Cell death was detected by Annexin V/PI staining, followed by flow cytometry (b) or immunofluorescence (c). d Peripheral mononuclear cells from AML patients were treated with Na-AuPT at the indicated concentrations or 100 nM bortezomib for 6 h. Western blots of the indicated proteins are shown.
Discussion
The development of new proteasome inhibitors represents a multifaceted challenge but provides exciting opportunities for disease treatment [34]. In this study, we designed and synthesized Na-AuPT as a new water-soluble proteasome inhibitor, which shows extensive anticancer activity in a variety of liver, lung, and AML tumor models. Our in vivo and ex vivo studies also showed that Na-AuPT has low toxicity to normal tissues or cells. Mechanically, Na-AuPT-mediated proteasome inhibition triggers the activation of multiple caspases involving extrinsic and intrinsic apoptotic signaling pathways. The fact that the anticancer activity of Na-AuPT can be prevented by using the metal chelating agent EDTA highlights that Na-AuPT is a metal-based proteasome inhibitor. Further understanding on structural basis of Na-AuPT activity and kinetics of its metabolism in vivo may lead to development of new metal-based proteasome inhibitor drugs.
Proteasome inhibitors have various mechanisms of action on the UPS, which may cause various toxic effects, thereby affecting their clinical application. Compared with bortezomib, carfilzomib, a next-generation proteasome inhibitor for the treatment of multiple myeloma has lower toxicity. Bortezomib reversibly inhibits the β5 subunit but has peripheral neuropathy side effects [35]. Carfilzomib exerts its effect by irreversibly inhibiting the β5 subunit, which gave less neurotoxicity; however, patients can also develop drug resistance in carfilzomib clinical applications [36].
Toward the goal of developing new proteasome inhibitors, in the current study, we compared, and demonstrated similarities and differences between Na-AuPT and bortezomib on proteasome inhibition. It is generally believed that K48-linked ubiquitination marks protein degradation by the proteasome [37]. We found that Na-AuPT can induce the accumulation of both total and K48-linked ubiquitinated proteins in cancer cells, further supporting that Na-AuPT is a proteasome inhibitor. Na-AuPT inhibits both β1 and β5 (but not β2) activities of purified 20 S proteasomes, while it inhibits all three proteasome activities of A549 cell lysis (Fig. 3). This difference could be due to the 19 S proteasome or other proteases in cell lysis that may affect the binding between Na-AuPT and β2 subunits or that might use the same peptide substrate which is inhibitable by Na-AuPT. Other non-protease endogenous proteins may also competitively affect the binding between Na-AuPT and β2 subunits. In addition to inhibiting 20 S proteasome, we also demonstrate that Na-AuPT can directly target and inhibit 19 S proteasome-associated UCHL5 and USP14. Thus, the dual inhibition on both 19 S proteasomal DUBs and 20 S proteasome by Na-AuPT may offer great promise to overcome resistance to conventional 20 S proteasome inhibitors (e.g., bortezomib), and this hypothesis, however, should be verified in the future studies.
There are three DUBs associated with the 19 S proteasome, USP14, UCHL5, and RPN11 [38]. UCHL5 and USP14 are two cysteine protease DUBs, while RPN11 is a JAB1/MPN/Mov34 (JAMM) domain-containing metalloprotease. By detecting the cleavage of Ub-AMC, we revealed that Na-AuPT has strong effects in limiting the activities of UCHL5 and USP14 of purified 26 S proteasomes and in cancer cells (Fig. 3). Interestingly, inhibition of either UCHL5 or USP14 promotes proteasomal proteolytic function, while inhibition of both of these two DUBs block proteasomal proteolytic function [39]. Therefore, similar to the effect of b-AP15 [40], the UPS function damage caused by Na-AuPT is likely to be the result of inhibiting both UCHL5 and USP14. Due to lack of commercially available probes for monitoring activity of RPN11, our current study cannot rule out effect of Na-AuPT on RPN11. Nonetheless, identification of direct protein substrates in the UPS pathway affected by Na-AuPT is important for further optimization of anticancer strategies.
In summary, we have identified that gold (I) complex Na-AuPT is a new type of anticancer agent, which triggers apoptotic cell death by inhibiting multiple components of the protein degradation machinery. Our research not only provides new insights for understanding the mechanism of action of metal-based drugs, but also highlights the importance of high absorption and low toxicity of Na-AuPT in tumor therapy, which supports the necessity of its further clinical tests and applications.
References
Budenholzer L, Cheng CL, Li Y, Hochstrasser M. Proteasome structure and assembly. J Mol Biol. 2017;429:3500–24.
Gerards WL, de Jong WW, Boelens W, Bloemendal H. Structure and assembly of the 20S proteasome. Cell Mol Life Sci. 1998;54:253–62.
Guo N, Peng Z. MG132, a proteasome inhibitor, induces apoptosis in tumor cells. Asia Pac J Clin Oncol. 2013;9:6–11.
Orlowski RZ. The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death Differ. 1999;6:303–13.
Mukherjee S, Sparks R, Metcalf R, Brooks W, Daniel K, Guida WC. Cupriphilic compounds to aid in proteasome inhibition. Bioorg Med Chem Lett. 2016;26:3826–9.
Mujtaba T, Kanwar J, Wan SB, Chan TH, Dou QP. Sensitizing human multiple myeloma cells to the proteasome inhibitor bortezomib by novel curcumin analogs. Int J Mol Med. 2012;29:102–6.
Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr Cancer Drug Targets. 2011;11:239–53.
Zhang L, Mager DE. Population-based meta-analysis of bortezomib exposure-response relationships in multiple myeloma patients. J Pharmacokinet Pharmacodyn. 2020;47:77–90.
Parlati F, Lee SJ, Aujay M, Suzuki E, Levitsky K, Lorens JB, et al. Carfilzomib can induce tumor cell death through selective inhibition of the chymotrypsin-like activity of the proteasome. Blood. 2009;114:3439–47.
Pautasso C, Bringhen S, Cerrato C, Magarotto V, Palumbo A. The mechanism of action, pharmacokinetics, and clinical efficacy of carfilzomib for the treatment of multiple myeloma. Expert Opin Drug Metab Toxicol. 2013;9:1371–9.
Lee MJ, Bhattarai D, Yoo J, Miller Z, Park JE, Lee S, et al. Development of novel epoxyketone-based proteasome inhibitors as a strategy to overcome cancer resistance to carfilzomib and bortezomib. J Med Chem. 2019;62:4444–55.
Selvaraju K, Mazurkiewicz M, Wang X, Gullbo J, Linder S, D’Arcy P. Inhibition of proteasome deubiquitinase activity: a strategy to overcome resistance to conventional proteasome inhibitors? Drug Resist Updat. 2015;21-22:20–9.
Zhao C, Chen X, Zang D, Lan X, Liao S, Yang C, et al. Platinum-containing compound platinum pyrithione is stronger and safer than cisplatin in cancer therapy. Biochem Pharmacol. 2016;116:22–38.
Liu N, Liu C, Li X, Liao S, Song W, Yang C, et al. A novel proteasome inhibitor suppresses tumor growth via targeting both 19S proteasome deubiquitinases and 20S proteolytic peptidases. Sci Rep. 2014;4:5240.
Li X, Huang Q, Long H, Zhang P, Su H, Liu J. A new gold(I) complex-Au(PPh3)PT is a deubiquitinase inhibitor and inhibits tumor growth. EBioMedicine. 2019;39:159–72.
Yang L, Chen X, Yang Q, Chen J, Huang Q, Yao L, et al. Broad spectrum deubiquitinase inhibition induces both apoptosis and ferroptosis in cancer cells. Front Oncol. 2020;10:949.
Verani CN. Metal complexes as inhibitors of the 26S proteasome in tumor cells. J Inorg Biochem. 2012;106:59–67.
Lan X, Zhao C, Chen X, Zhang P, Zang D, Wu J, et al. Nickel pyrithione induces apoptosis in chronic myeloid leukemia cells resistant to imatinib via both Bcr/Abl-dependent and Bcr/Abl-independent mechanisms. J Hematol Oncol. 2016;9:129.
Zhao C, Chen X, Yang C, Zang D, Lan X, Liao S, et al. Repurposing an antidandruff agent to treating cancer: zinc pyrithione inhibits tumor growth via targeting proteasome-associated deubiquitinases. Oncotarget. 2017;8:13942–56.
Notaro A, Gasser G, Castonguay A. Note of caution for the aqueous behaviour of metal-based drug candidates. ChemMedChem. 2020;15:345–8.
Lee YH, Tuyet PT. Synthesis and biological evaluation of quercetin-zinc (II) complex for anti-cancer and anti-metastasis of human bladder cancer cells. Vitr Cell Dev Biol Anim. 2019;55:395–404.
Chen X, Shi X, Zhao C, Li X, Lan X, Liu S, et al. Anti-rheumatic agent auranofin induced apoptosis in chronic myeloid leukemia cells resistant to imatinib through both Bcr/Abl-dependent and -independent mechanisms. Oncotarget. 2014;5:9118–32.
Yan D, Li X, Yang Q, Huang Q, Yao L, Zhang P, et al. Regulation of Bax-dependent apoptosis by mitochondrial deubiquitinase USP30. Cell Death Discov. 2021;7:211.
Chen X, Wu J, Yang Q, Zhang X, Zhang P, Liao S, et al. Cadmium pyrithione suppresses tumor growth in vitro and in vivo through inhibition of proteasomal deubiquitinase. Biometals. 2018;31:29–43.
Chen X, Yang Q, Chen J, Zhang P, Huang Q, Zhang X, et al. Inhibition of proteasomal deubiquitinase by silver complex induces apoptosis in non-small cell lung cancer cells. Cell Physiol Biochem. 2018;49:780–97.
Cohen SM, Lippard SJ. Cisplatin: from DNA damage to cancer chemotherapy. Prog Nucleic Acid Res Mol Biol. 2001;67:93–130.
Jungwirth U, Kowol CR, Keppler BK, Hartinger CG, Berger W, Heffeter P. Anticancer activity of metal complexes: involvement of redox processes. Antioxid Redox Signal. 2011;15:1085–127.
Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45.
Hope AD, de Silva R, Fischer DF, Hol EM, van Leeuwen FW, Lees AJ. Alzheimer’s associated variant ubiquitin causes inhibition of the 26S proteasome and chaperone expression. J Neurochem. 2003;86:394–404.
Kisselev AF, Callard A, Goldberg AL. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J Biol Chem. 2006;281:8582–90.
Rivard C, Bazzaro M. Measurement of deubiquitinating enzyme activity via a suicidal HA-Ub-VS probe. Methods Mol Biol. 2015;1249:193–200.
Li B, Dou QP. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc Natl Acad Sci USA. 2000;97:3850–5.
Robak P, Robak T. Bortezomib for the treatment of hematologic malignancies: 15 years later. Drugs R D 2019;19:73–92.
Manasanch EE, Orlowski RZ. Proteasome inhibitors in cancer therapy. Nat Rev Clin Oncol. 2017;14:417–33.
Lu S, Yang J, Song X, Gong S, Zhou H, Guo L, et al. Point mutation of the proteasome beta5 subunit gene is an important mechanism of bortezomib resistance in bortezomib-selected variants of Jurkat T cell lymphoblastic lymphoma/leukemia line. J Pharmacol Exp Ther. 2008;326:423–31.
Moore TA, Brodersen P, Young EWK. Multiple myeloma cell drug responses differ in thermoplastic vs PDMS microfluidic devices. Anal Chem. 2017;89:11391–8.
Tsuchiya H, Ohtake F, Arai N, Kaiho A, Yasuda S, Tanaka K, et al. In vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 axis contributes to K48-linked chain specificity of the proteasome. Mol Cell. 2017;66:488–502 e7.
D’Arcy P, Linder S. Proteasome deubiquitinases as novel targets for cancer therapy. Int J Biochem Cell Biol. 2012;44:1729–38.
Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–82.
D’Arcy P, Brnjic S, Olofsson MH, Fryknas M, Lindsten K, De Cesare M, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1636–40.
Acknowledgements
The study was supported by the National Natural Science Foundation of China (81802405), the National Funds for Developing Local Colleges and Universities (B16056001), Natural Science Foundation Research Team of Guangdong Province (2018B030312001), the Science and Technology Program of Guangzhou (201604020001), Innovative Academic Team of Guangzhou Education System (1201610014), and the Research Team of Department of Education of Guangdong Province (2017KCXTD027).
Author information
Authors and Affiliations
Contributions
JBL and XC designed the study; DX, LY, and PQZ conducted the experiments and analyzed the data. DY, QX, QTH, XFL, and YLH participated in the experiments. DCX, LY, XC, and JL wrote the manuscript. DLT and QPD assisted in data interpretation and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Xu, Dc., Yang, L., Zhang, Pq. et al. Pharmacological characterization of a novel metal-based proteasome inhibitor Na-AuPT for cancer treatment. Acta Pharmacol Sin 43, 2128–2138 (2022). https://doi.org/10.1038/s41401-021-00816-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-021-00816-z