Abstract
Decaprenylphosphoryl-β-D-ribose oxidase (DprE1) plays important roles in the biosynthesis of mycobacterium cell wall. DprE1 inhibitors have shown great potentials in the development of new regimens for tuberculosis (TB) treatment. In this study, an integrated molecular modeling strategy, which combined computational bioactivity fingerprints and structure-based virtual screening, was employed to identify potential DprE1 inhibitors. Two lead compounds (B2 and H3) that could inhibit DprE1 and thus kill Mycobacterium smegmatis in vitro were identified. Moreover, compound H3 showed potent inhibitory activity against Mycobacterium tuberculosis in vitro (MICMtb = 1.25 μM) and low cytotoxicity against mouse embryo fibroblast NIH-3T3 cells. Our research provided an effective strategy to discover novel anti-TB lead compounds.
Similar content being viewed by others
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), is the leading death cause from a single infectious agent worldwide. TB at its early drug sensitive stage is capable of being effectively treated, but normally requires a combination of multiple drugs and needs a duration of 6–20 months. Inappropriate treatment possibly leads to multidrug-resistant TB (MDR-TB), which is resistant to the two most potent anti-TB drugs (isoniazid and rifampin), or even extensively drug-resistant TB (XDR-TB), which is a subgroup of MDR-TB with additional resistance to any fluoroquinolone or at least one of the second-line injectable drugs. In 2019, 7.1 million people with TB were newly diagnosed, and 206,030 people with MDR-TB or rifampin-resistant TB (RR-TB) were detected [1]. Therefore, it is quite urgent to develop novel regimens for MDR/XDR/RR-TB [2,3,4].
Decaprenylphosphoryl-β-D-ribose oxidase (DprE1) is an enzyme involved in the synthesis of arabinogalactan, an essential constituent of the Mtb cell wall [5]. DprE1 inhibitors can block arabinan synthesis, thus provoking cell lysis and bacterial death [6]. Several phenotypic screenings to discover potent DprE1 inhibitors have been reported, and both of covalent and non-covalent inhibitors have been identified [7,8,9,10,11,12]. Currently, four DprE1 inhibitors have been pushed into clinical trials, including BTZ043, Macozinone (formerly PBTZ169), TBA-7371 and OPC-167832 (Fig. S1). BTZ043 and Macozinone are benzothiazinones and they can inhibit DprE1 by forming covalent bonds with the active-site Cys387 residue [13]. They are both active against MDR-TB [14]. Macozinone could act synergistically with the new anti-TB drug bedaquiline and the repurposed anti-leprosy drug clofazimine [15, 16], therefore raising a great potential to elaborate a macozinone-containing regimen for the treatment of MDR-TB [17]. TBA-7371 and OPC-167832 are non-covalent inhibitors with potent antimycobacterial activities [18, 19]. TBA-7371 did not show cross-resistance to BTZ043 [20]. OPC-167832 exhibited significant combination effects in 2-drug combinations with delamanid, bedaquiline, or levofloxacin. Apparently, DprE1 inhibitors may become a promising component of new regimens for TB treatment.
High throughput screening, molecular docking, functional genomics and protein-ligand cocrystallography have facilitated the development of DprE1 inhibitors [21]. However, as a powerful computational approach for the identification of lead compounds with novel structural scaffolds, virtual screening has rarely been utilized in the discovery of DprE1 inhibitors [22,23,24]. In 2019, Gao et al. screened a database containing ~6,200,000 molecules toward DprE1 using the ICM docking algorithm [25]. A total of 63 compounds were selected for antibacterial activity test, and one compound (compound 50) with a minimal inhibitory concentration against Mtb (MICMtb) of 9.75 μM was identified.
Scaffold hopping is a widely used technique for the exploration of the chemical space of known active compounds [26]. Various computational methods have been developed for scaffold hopping, such as pharmacophore searching, shape searching, fingerprint- or structure-based similarity searching [27]. Recently, our group has proposed the computational bioactivity fingerprint (CBFP), a novel descriptor to characterize the biological space of a molecule by combining the predictions from multiple quantitative structure-activity relationship (QSAR) models for 832 proteins [28]. The CBFP-based similarity searching tends to find compounds with similar biological profiles rather than compounds with similar structures. The CBFP representation demonstrates outstanding scaffold hopping capability for searching novel inhibitors toward poly [ADP-ribose] polymerase 1 [28].
In this study, an integrated molecular modeling strategy by combining the CBFP-based scaffold hopping and structure-based virtual screening (SBVS) was employed to identify potential DprE1 inhibitors. By screening the ChemDiv chemical library, a total of 93 potential compounds were identified and submitted to bioassays. Two compounds (B2 and H3) were identified to inhibit Mycobacterium smegmatis (M. smegmatis) with MIC50 values less than 1 μM. Further MIC shift assay, thermal shift assay and structure-activity relationship (SAR) analysis illustrated that both compounds are DprE1 inhibitors. The antibacterial activities of compounds B2 and H3 against Mtb strain H37Ra were further evaluated. Compound H3 turned out to be a significantly effective bactericidal agent against Mtb in vitro (MICMtb = 1.25 μM). Furthermore, it is noteworthy that no obvious toxicity was observed for compound H3 in cytotoxicity testing.
Materials and methods
CBFP-based similarity searching
In our study, the Chemdiv library containing 833,569 commercially available compounds was virtually screened by the CBFP-based similarity searching method [28]. The CBFP for each molecule in ChemDiv was generated by the following steps. At first, three sets of molecule descriptors (i.e., CATS, MACCS, and MOE2D) were calculated for each molecule. Then, the molecule was predicted by the established 832 × 9 QSAR models for 832 proteins. For each molecule, there were nine possible values for each protein and the average value was computed to represent the potential activity against the protein target. Finally, the predictions of the molecule for all the proteins were assembled as an 832-bit vector and each bit is in a scale of 0–1. This feature vector was translated into a standard binary fingerprint by using a cutoff of 0.5, where 1 means that this molecule has high possibility to bind to this protein and 0 has very low possibility. The 832-bit CBFP descriptor can be used to represent the bioactivity space of this small molecule.
A total of 3 non-covalent and 6 covalent DprE1 inhibitors (Fig. 1a) were chosen as the query molecules in the CBFP-based similarity searching. The CBFP descriptors for the query molecules were calculated. Then, the similarity between each query molecule and each screened molecule was evaluated by the Tanimoto similarity coefficient based on the CBFP representation. For each query molecule, the top 5000 hits were saved, and then a total of 45,000 molecules (named as the DprE1_CBFP dataset) were saved for the nine query molecules.
Structure-based virtual screening
The crystal structure of DprE1 in complex with the non-covalent inhibitor Ty38c (PDB code: 4P8K [9]) was selected as the template for SBVS. The protein structure was prepared by using the Protein Preparation Wizard [29] in Schrödinger 2018 [30]. The grid box of the protein for docking centered on the co-crystallized ligand (Ty38c) in the binding pocket was generated. The small molecules in the DprE1_CBFP dataset processed by LigPrep were docked into the prepared structure by using the Glide module [31], and the binding energies were scored and ranked by the Glide SP scoring mode. The top-ranked 5000 compounds were filtered by the Lipinski’s rule-of-five and Oprea’s rules, and then the remaining molecules were clustered based on the 2D similarities (Tanimoto coefficient) of the MACCS fingerprints. The binding poses of the clustered compounds were carefully checked and filtered. Finally, 93 compounds (Table S1) were purchased for the subsequent bioassays.
Molecular dynamics (MD) simulations
The MD simulations were performed with Amber18 [32]. The structure of DprE1 bound with B2 predicted by molecular docking was used as the initial conformation for the MD simulations. The ff14SB [33] force field was used for the proteins, and the AMBER general force field [34] was used for the ligands with the AM1-BCC charges. The complex was solvated into a TIP3P water cubic box (10 Å), and Na+ ions were added to neutralize the net charge of the system. The Particle Mesh Ewald (PME) algorithm [35] was employed for the long-range electrostatic interaction, and the cut-off for the real-space interactions was set to 12.0 Å.
Subsequently, each system was subjected to the four-step energy minimizations: (1) the system was optimized with a force restraint of 50 kcal·mol−1·Å−2 on the protein and ligand atoms; (2) the system was optimized with a force restraint of 10 kcal·mol–1·Å−2 on the protein and ligand atoms; (3) the protein atoms were restrained by a 10 kcal·mol−1·Å−2 force constant and the other atoms were minimized; (4) the whole system was minimized without any restraint. Each minimization stage consists of 1000 steps of steepest descent and 2000 cycles of conjugate gradient optimizations. The sander.MPI engine in the AMBER18 package was used to run the energy minimizations.
Using weak-coupling thermostats, the minimized complexes were then heated to 300 K under the NVT ensemble during 30 ps. Then, the systems were equilibrated without any restraint for 110 ps under the NPT (P = 1 atm, T = 300 K) ensemble. Finally, the unrestrained production simulations were performed in the NPT (P = 1 atm, T = 300 K) ensemble. The length of the production simulation was 500 ns with a time step of 2 fs, and the conformations were saved per 10 ps. The pmemd.cuda module was employed for the MD production simulations. The root-mean-square deviation (RMSD) and distance analyses were carried out by the cpptraj module in AmberTools18.
MM/GBSA binding free energy calculations
The last 50 ns MD simulation trajectory with 100 snapshots was submitted to the MM/GBSA binding free energy calculation (∆Gbind) and free energy decomposition as the following equations:
where ΔEMM, ΔGsol, –TΔS represent the changes of the gas phase molecular mechanism (MM) energy, the solvation free energy, and the conformational entropy upon ligand binding, respectively. ΔEMM is the sum of the internal energy (ΔEint), the van der Waals energy ΔEvdw, and the electrostatic energy (ΔEele). ΔGsol is the sum of the polar (ΔGGB) and non-polar contributions (ΔGSA). ΔGsol was estimated by the generalized Born model (GBOBC1) developed by Onufriev and co-workers [36] and ΔGSA was measured by the solvent accessible surface area (SASA). –TΔS is usually neglected due to its high computational cost and low prediction accuracy [37]. The exterior (solvent) dielectric constant was set to 80 and the interior (solute) dielectric constant was set to 1.
Covalent docking
Compound H3 containing NO2 was subjected to covalent docking using CovDock [38] of Schrödinger 2018. Before covalent docking, NO2 was changed to NO, and the compound was prepared by the ligPrep module in Schrödinger 2018. The crystal structure of DprE1 covalently bound with BTZ043 (PDB code: 4F4Q [13]) was prepared using the Protein Preparation Wizard in Schrödinger 2018. The ligand (BTZ043) in the crystal structure of DprE1 was retained and used for grid generation. “Nucleophilic Addition to a Double Bond” was selected as the reaction type to match the reactive groups on both the ligands (N=O) and protein (Cys394). The residues that have any atoms within 5 Å distance of any atom in the ligand were included in the minimization. The Prime MM-GBSA energy was calculated and used to filter the poses.
H3d processed by LigPrep was docked into the prepared BTZ043-DprE1 (PDB code: 4F4Q) structure by using the Glide module. It is worth noting that in order to remove the effect of Cys394 on non-covalent docking, Cys394 was mutated to Ala394 in the H3d docking.
The compounds for bioassays
The positive control compounds PBTZ169 (CAS 1377239-83-2) and TBA-7371 (CAS 1494675-86-3) were purchased from MedChemExpress (purity ≥99% by HPLC). The negative control compound rifampin was also purchased from MedChemExpress (purity ≥99% by HPLC). Each compound was dissolved in 100% dimethyl sulfoxide (DMSO) as a 20 mM stock solution. The final DMSO concentration in each reaction was less than 1%. All the 93 tested compounds were purchased from Topscience (Shanghai, China).
Strain and growth conditions
Considering the severe infectiousness of Mtb, M. smegmatis mc2155, the model bacteria of Mtb widely used in TB-related studies, was utilized in this study. M. smegmatis were grown in Luria-Bertani (LB) medium containing 0.05% Tween 80 (Sigma-Aldrich). Culture media were incubated for 48 h at 37 °C at 120 r/min in a shaker incubator, and the growth was monitored by measuring the absorbance at OD600 nm using a spectrophotometer (Bioteck Eon, Winooski, VT).
In vitro antibacterial activity test using M. smegmatis
All the virtual screening hits were initially screened against the M. smegmatis strain at a single concentration of 10 μM in triplicate in a 96-well plate. The active compounds which exhibited more than 80% inhibition at 10 μM were further tested for minimum inhibitory concentration (MIC) using the broth microdilution assay. The 96-well plates were incubated at 37 °C for 48 h and the growth was monitored by measuring the absorbance at OD600 nm using a spectrophotometer (Bioteck Eon, Winooski, VT). The MIC and MIC50 are defined as the concentrations at which >99 and 50% bacterial growth can be inhibited in contrast to the drug-free control plates, respectively. Furthermore, the bactericidal effects (minimum bactericidal concentration, MBC) of the active compounds were assessed. A total of 0.1 mL of M. smegmatis suspension from each well tested at the concentrations ≥MIC were plated onto LB agar medium, and the resulting bacterial counts were counted after 7 days of incubation at 37 °C. The MBC is defined as the minimal concentration at which at least 99% of the viable counts are effectively reduced compared with the drug-free control plates. INH was used as the positive control.
In vitro antibacterial activity test using Mtb
The antitubercular activities of the compounds were evaluated by using previously reported procedures against the autoluminescent Mtb H37Ra strain [39, 40]. Bacteria growth was conveniently monitored by means of the bioluminescence intensity. RLU-based minimum inhibitory concentration (MICMtb) is defined as the lowest concentration that can inhibit >90% RLUs compared with the negative control.
Overexpression and purification of DprE1
DprE1 (Rv3790) was coexpressed with chaperones from Escherichia coli (GroES) and Mtb (CPN60.2) in E. coli BL21 (DE3). BL21 cells were grown at 37 °C in LB broth (Sigma) supplemented with chloromycetin (100 μg/mL) and kanamycin (50 μg/mL). When the OD600 nm value of the culture reached 0.6, the expressions of DprE1 and chaperones were induced by 119 μg/mL isopropyl-β-D-thiogalactopyranoside and 250 μg/mL arabinose, followed by incubation overnight at 16 °C. The cells were then harvested and resuspended in 40 mL of 60 mM NaH2PO4 (pH 8), 300 mM NaCl, and 10 mM imidazole (buffer A), supplemented with EDTA-free protease inhibitor mixture (Roche). The suspension was sonicated and centrifuged (27,000 × g, 30 min, 4 °C), and the supernatant passed through a preequilibrated (buffer A) 1-mL Ni-NTA Resin (Thermo Scientific™; USA). The protein was eluted with a 50–300 mM gradient of imidazole. The purity of the protein was checked by SDS-page gel. The protein was stored at −80 °C until used.
Construction of DprE1 overexpressing M. smegmatis strain
Gene Rv3790 encoding DprE1 was inserted into plasmid pMV261. Then, pMV261-Rv3790 was transformed into the wild-type M. smegmatis. The genotype of the resulting strain was verified by PCR. The MICs of the test compounds toward the recombinant strains were determined as described above. Resazurin was added to all the wells and the color conversions were recorded. A blue color in the well was interpreted as no growth, and a pink color was interpreted as growth.
Fluorescence-based thermal shift assay
A fluorescence-based thermal shift assay was used to probe the effects of compounds on the thermal stability of DprE1. The fluorescence-based thermal shift assay was assessed by the Protein Thermal Shift™ Dye Kit (Applied Biosystems, Cat #4461146). The DprE1 protein was purified as previously described. The purified DprE1 protein was mixed with serial dilutions of the tested compounds, RNase-free water, and Protein Thermal Shift™ mix in a 96-well plate. Experiments were carried out in the StepOnePlus™ Real-Time PCR System (Applied Biosystems™, California, USA).
Cytotoxic activity assay
Mouse embryo fibroblast NIH-3T3 cells were cultivated in RPMI-1640 medium supplemented with 10% fetal bovine serum. The cells were seeded at a density of 3000 cells/well into 96-well plates and placed in an incubator with 5% CO2 at 37 °C. After 24 h, the cells were treated with serial dilutions of the tested compounds for 3 days. Afterward, 10 μL of 5 mg/mL MTT solution was added into each well and incubated for an additional 4 h. Then, 100 μL of triplex 10% SDS-5% isobutyl alcohol-0.012 mol/L HCl (w/v/v) solution was added to dissolve the formazan crystals. The absorbance at 570 nm was measured with the reference wavelength at 650 nm using a spectrophotometer (Bioteck Eon, Winooski, VT).
Data and statistical analysis
Statistical analyses were performed with the GraphPad Prism software v 6.0 (San Diego, CA, USA). All data were presented as means ± SD. A P-value less than 0.05 was statistically significant.
Results and discussion
Discovery of novel hit compounds B2 and H3
The workflow of the SBVS protocol used in this study is presented in Fig. 1a. Nine DprE1 inhibitors were chosen as the query molecules in the CBFP-based similarity search. The similarities between the query molecule and the Chemdiv molecules were calculated. A total of 45,000 molecules with the highest similarities (DprE1_CBFP) were retained. Then the molecules in the DprE1_CBFP data set were docked into the prepared protein 4P8K using Glide with the SP scoring mode. Through structural clustering, druglike analysis and binding mode inspection, a total of 93 potential DprE1 inhibitors were screened out and submitted to bioassays (Table S1).
At first, the antibacterial activities of the 93 compounds against M. smegmatis were tested at 10 μM (Fig. 1b), and most compounds exhibited antibacterial activities. Among them, eight compounds (B2, D8, D9, E5, F4, F15, H3, and I13) showed more than 50% inhibition, suggesting that their MIC50 values were lower than 10 μM. In addition, these potent compounds showed diverse structural scaffolds (Fig. 1b) though they shared some fragments with the query molecules, which confirmed the admirable performance of CBFP in searching for novel scaffolds. However, we also observed that the 21 potential hits (I1-I21) identified by the similarity search based on the covalent DprE1 inhibitor macozinone (I) as the query had higher CBFP similarities than those based on the other query molecules, but only one compound (I13) exhibited weak antibacterial bioactivity. It seems that although CBFP has superior performance in scaffold hopping, the hit rate toward the molecules identified by the CBFP-based similarity search depends on the query molecule. Therefore, in virtual screening, the CBFP-based similarity search can be used to prioritize compounds, and then docking-based screening can be used to enrich bioactive candidates.
Among the active molecules, two promising compounds (B2 and H3 in Fig. 1b) can completely inhibit the growth of M. smegmatis at 10 μM. The antibacterial activities of compounds B2 and H3 were further evaluated. As shown in Fig. 2a, the MIC50 values of B2 and H3 were 0.38 and 0.33 μM, respectively, and those of TBA-7371 and rifampin were 2.31 and 1.09 μM, respectively. Then, the bactericidal effects (minimum bactericidal concentration, MBC) of B2 and H3 were assessed. As shown in Fig. S2, the two compounds showed good bactericidal activities on replicating M. smegmatis (MBCB2 = 5 μM and MBCH3 = 1.25 μM).
a The antibacterial activity of compound B2 and H3 against M. smegmatis. The TBA-7371 and rifampin were used as positive controls. b Overexpression of Mt-DprE1 in M. smegmatis confers 4-fold, 16-fold, 8-fold and 8-fold increase in resistance to PBTZ169, TBA-7371, H3 and B2, but no resistance to rifampin. The compound concentrations expressed as fold-× of the MIC of each compound. Pink indicates viable cells, and blue indicates nonviable cells. WT strain refers to wild-type M. smegmatis that did not overexpress Mt-DprE1. c The thermal shift melt curves for DprE1 with rifampi. d The thermal shift melt curves for DprE1 with compound B2. e The thermal shift melt curves for DprE1 with PBTZ169. f The thermal shift melt curves for DprE1 with compound H3.
Validation of the mode of action via DprE1 overexpression and thermal shift assay
The overexpression of DprE1 in bacteria is supposed to counteract the attack of DprE1 inhibitors. Shifts of MIC values caused by targeting protein overexpression can provide a practical measure of target engagement at the cellular level. Many studies have used the MIC modulation observed in the DprE1-overexpressing strain to confirm DprE1 as the target [7, 41]. We therefore established a M. smegmatis strain which overexpresses Mtb DprE1 (Mt-DprE1). As we expected, both B2 and H3 showed 8-fold MIC shifts on the DprE1 strain (Fig. 2b). These results were in agreement with the observations for other known DprE1 inhibitors such as PBTZ169, TBA-7371 and BTZ043 [7]. The MIC shifts caused by the DprE1 overexpression provided a basic evidence for the target engagement of compounds B2 and H3.
The fluorescence-based thermal shift assay is well accepted for studying the binding of a specific ligand to a certain protein [42]. The fluorescent dye was used to monitor protein thermal unfolding, from which a melting temperature (Tm) can be derived. When a ligand binds to a protein, the Tm will shift, producing a thermal shift (ΔTm). It has been reported that ΔTm correlates well with the binding constants measured by other methods [43, 44]. Then, the thermal shift assay was carried out to further confirm the interactions between DprE1 and the two DprE1 inhibitors. As shown in Fig. 2c–e and Table S2, compound B2 displayed a melting curve different from that of the rifampin control, stabilizing DprE1 by 1.0 °C at 300 μM, similar to PBTZ169, whereas the ΔTm by rifampin is quite small. Compound H3 showed significant stabilization effect at 300 μM, stabilizing DprE1 by 1.5 °C (Fig. 2f). Compared with B2 and PBTZ169, compound H3 induced the stabilization of DprE1 more significantly, indicating that the complex of H3-Dpre1 is more compact. Further analysis showed that B2, H3 and PBTZ169 could dose-dependently stabilize DprE1, confirming that their inhibitions on M. smegmatis were achieved by targeting DprE1.
Cell toxicities of compounds B2 and H3
The inherent toxicities of B2 and H3 on mouse embryo fibroblast NIH-3T3 cells were tested to verify their safeties. The cells were treated with different concentrations of compounds and the anti-proliferative effects were evaluated. As shown in Fig. 3a, the IC50 of B2 for 3T3 cells was 31.55 μM, which was more than 80 times higher compared with its MIC50 against M. smegmatis. At 25 μM, the effect of compound B2 on cell proliferation was less than that of PBTZ169. The IC50 value of H3 for 3T3 cells was higher than 50 μM, which was more than 150 times higher than its MIC50 against M. smegmatis. Unlike PBTZ169, B2 showed no cytotoxicity against NIH-3T3 cells even at a high dose of 50 μM. Thus, one can conclude that B2 and H3 are nontoxic at their effective doses against M. smegmatis, highlighting the possibility of B2 and H3 as the candidates of DprE1 inhibitors.
a Cytotoxicity tests of compound B2 and H3. The cytotoxicities of compound B2 and H3 were measured on mouse embryo fibroblast NIH-3T3 cells. b The 10 top-ranked residues in DprE1 responsible for the binding of B2 predicted by MM/GBSA. c The structural analysis of the key residues to the binding of B2. d The thermal shift melt curves for DprE1 with compound B2a. e The energy differences between B2 and B2a. f Alignment of the representative structures of B2 (orange) and B2a (cyan) bound to DprE1. The residues that form stronger interaction with B2 than with B2a are highlighted.
Structure-activity relationship (SAR) for sulfonamide series
Compound B2 possessed an N-(4-hydroxy-3-mercaptonaphthalen-1-yl) sulfonamide scaffold, which has never been reported in any DprE1 inhibitor. To guide the structural optimization of B2, MD simulations were performed to investigate the dynamic behavior between DprE1 and B2. Firstly, the RMSDs of the heavy atoms of DprE1 and B2 as a function of the simulation time were computed to monitor the stability of the DprE1-B2 complex during the MD simulations. As shown in Fig. S3a, the RMSDs of the heavy atoms of DprE1 fluctuated between 2.5 and 3.0 Å, while B2 tended to converge after ~100 ns with the RMSD fluctuations <0.5 Å, suggesting that B2 could stably bind to DprE1.
The 100 snapshots extracted from the 450 to 500 ns MD trajectories were used for the structural and energetic analyses. The per-residue MM/GBSA decomposition showed that 10 residues made major contributions to the binding of B2 to DprE1, including Lys418, Arg325, Lys134, Val365, Trp230, Leu363, Pro316, Met319, Cys387 and Gln334 (Fig. 3b). The structural analysis indicated that the carboxyl group (R1) formed H-bonds with Lys418 and Arg325 (Fig. 3c). The naphthalene ring of B2 was stably bound in a hydrophobic pocket formed by Val365, Trp230, Leu363, Cys387 and Pro316. The sulfonamide of B2 formed a H-bond with Lys134. The benzene ring (R2) exhibited favorable van der Waals interaction with Met319. According to the above structural analysis, the naphthol group may be vital for antibacterial activity. The R1 and R2 moieties have been modified to study the SAR of naphthol compounds (Table 1). Accordingly, 7 analogues of B2 were submitted to bioassays to verify our hypotheses.
The MIC50 values of the 7 analogues against M. smegmatis were shown in Table 1, and a preliminary SAR was analyzed. The compounds with the naphthol group (B2a–B2d) maintained the inhibitory effects with MIC50 from 1.62 to 6.98 μM, while the analogues with distinct scaffolds (B2e–B2g) did not show obvious inhibitory effect against M. smegmatis (MIC50 > 50 μM), suggesting that the middle naphthol group exerts an important influence on the inhibitory activities of compounds. The compound without the carboxyl group (B2b, MIC50 = 1.62 μM) still showed moderate inhibitory activity, which provides a direction for the modification of the R1 group.
As shown in Fig. 3d, when compound B2a was added to DprE1, the melting curve shifted significantly, indicating that the inhibition of B2a to M. smegmatis was achieved by targeting DprE1. Compared with B2, the inhibitory activity of compound B2a (MIC50 = 2.5 μM) decreased by 6 times, indicating that the ortho-substitution on the benzene ring was not conducive to the activity. The MD simulations were performed to investigate the dynamic behavior of DprE1-B2a (Fig. S3b). The binding free energy of B2a predicted by MM/GBSA was −28.27 kcal/mol, which was higher than that of B2 (−66.32 kcal/mol). To further characterize the energetic differences between B2 and B2a, the energetic differences between the binding spectra of B2 and B2a (ΔΔG = ΔGB2 − ΔGB2a) were calculated. It can be observed that the residues of Pro316, Met319, Arg325, and Lys418 formed stronger interactions with B2 than B2a (Fig. 3e). The structural analysis showed that the binding structures of B2 (orange) and B2a (cyan) in the active site were different (Fig. 3f). Compared with compound B2a, the carboxyl group of compound B2 was closer to Arg325 and Lys418 (Fig. S3c, d), and therefore the residues of Arg325 and Lys418 can form stronger H-bond interactions with B2 than with B2a. According to the above energetic and structural analyses, the ortho substitution on the benzene ring caused the carboxyl group to stretch away from Arg325 and Lys418, which was not conducive to improving the binding affinity.
Generally, based on the MIC shift assay, thermal shift assay, MD simulations and SAR analysis, the sulfonamide scaffold was proved to be a novel structure of DprE1 inhibitors. Then the best one in our sulfonamides (B2) was submitted for the Mtb H37Ra test, but it did not show good inhibitory effects (MICMtb > 10 μM). The possible reason is that some Mtb proteins lack conserved orthologs in M. smegmatis [45]. Due to the high infectiousness of Mtb, model species (i.e., M. smegmatis and M. bovis BCG) are often used for preliminary screening, but this may limit the potential for identifying new inhibitors with efficacy against Mtb [46]. In addition, among the approaches employed for antibacterial discovery, target-based programs have achieved limited success due to the lack of the correlation between the target binding activities and the MIC values [47]. But the results still demonstrated that the CBFP-based similarity search was competent to identify novel DprE1 inhibitors.
Compound H3 is a promising candidate against tuberculosis
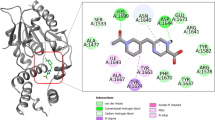
Compound H3 was discovered through the scaffold hopping based on DNB1. Both H3 and DNB1 have a 3,5-dinitrobenzamide fragment, but the middle linker of compound H3 is a dihydrazide group, not an ester chain amide. It was suggested that the nitro group of DNB1 can interact with Cys387 of DprE1 [8]. Therefore, compound H3 may also be a covalent inhibitor of DprE1. To further analyze its predicted binding mode, compound H3 was subjected to covalent docking with Schrödinger. Fig. 4a showed the predicted binding mode of compound H3 in the substrate binding site of DprE1. Compound H3 reacted with Cys387 and formed an adduct with DprE1. This was in agreement with the mechanism of other covalent inhibitors of DprE1 [48]. The dihydrazide group formed H-bonds with Trp24, Tyr67, Asp396 and Gln341. The benzene ring without the nitro group exhibited lipophilic interactions with Phe339.
To demonstrate the potential of the scaffold for hit-to-lead optimization, five analogues of compound H3 in the Chemdiv database were submitted to bioassay. As shown in Table 2, two analogues (H3a and H3d) showed more than 50% inhibition against M. smegmatis at 50 μM. The best analogue H3d showed a moderate antibacterial activity with MIC50 of 3.72 μM. Compared with H3 (MIC50 = 0.33 μM), the inhibitory activity of compound H3d decreased by 11 times. Molecular docking was performed to investigate the binding mode of H3d (Fig. 4b). It was observed that the N atom of compound H3d formed an H-bond with Gln341 but lost the H-bonds interactions with Trp24, Tyr67 and Asp396. The residue Arg26 formed the H-bond interaction with the carboxyl group of compound H3d. Compared with compound H3, compound H3d did not contain nitro, and the dihydrazide group was replaced by hydrazide in this case. The preliminary SAR analysis further confirmed the importance of the dihydrazide and nitro groups. In addition, compound H3c–e possessed the same scaffold, but compound H3d showed a higher inhibitory effect against M. smegmatis, indicating that addition of H-bond donors to the benzene ring may increase activity.
Compound H3 presented a comparable activity to that of isoniazid (MICMtb = 0.16 μM), with an MICMtb of 1.25 μM (Fig. 4c, d). But the MICMtb of H3 was higher than that of the other DprE1 covalent inhibitors, such as PBTZ169 [15]. Therefore, further chemical optimization was required.
Conclusion
CBFP is a highly discriminate structural descriptor that incorporates the predictive bioactivities of multiple QSAR models to characterize the bioactivity space of compounds [28]. To test the scaffold hopping ability of the CBFP representation, we combined the CBFP-based scaffold hopping and SBVS to discover novel DprE1 inhibitors, and a total of 93 potential inhibitors were submitted to bioassay. Among these compounds, B2 and H3 were identified as novel DprE1 inhibitors with 8-fold shifts on MIC values when Mt-DprE1 was overexpressed in M. smegmatis. The thermal shift assays further proved their direct binding to DprE1. Our study demonstrated a successful application of CBFP in searching novel bioactive compounds for drug discovery.
To explore the SAR of B2 and H3, seven analogues of B2 and five analogues of H3 were purchased and submitted to bioassays. Limited by the compound source, no analogue is more active than the leads. Nevertheless, it further confirmed that B2 and H3 are DprE1 inhibitors, and provided helpful information for further structural optimization. The two novel DprE1 inhibitors B2 and H3 were then submitted for the Mtb H37Ra test. Although B2 did not exhibit good inhibitory activity, it manifested a remarkable new scaffold as DprE1 inhibitors. H3 showed commendable activity against Mtb H37Ra in vitro comparable with the first-line anti-TB drugs and no obvious toxicity, providing a prospective lead compound against TB.
References
WHO, Global Tuberculosis Report 2020. Geneva: World Health Organization. 2020, Licence: Cc by-Nc-Sa 3.0 Igo.
Balganesh TS, Alzari PM, Cole ST. Rising standards for tuberculosis drug development. Trends Pharmacol Sci. 2008;29:576–81.
Pietersen E, Ignatius E, Streicher EM, Mastrapa B, Padanilam X, Pooran A, et al. Long-term outcomes of patients with extensively drug-resistant tuberculosis in South Africa: a cohort study. Lancet. 2014;383:1230–9.
Zhang MW, Zhou L, Zhang Y, Chen B, Peng Y, Wang F, et al. Treatment outcomes of patients with multidrug and extensively drug-resistant tuberculosis in Zhejiang, China. Eur J Med Res. 2021;26:31.
Manina G, Pasca MR, Buroni S, De Rossi E, Riccardi G. Decaprenylphosphoryl-beta-D-ribose 2 ‘-epimerase from Mycobacterium Tuberculosis is a magic drug target. Curr Med Chem. 2010;17:3099–108.
Makarov V, Manina G, Mikusova K, Mollmann U, Ryabova O, Saint-Joanis B, et al. Benzothiazinones kill Mycobacterium Tuberculosis by blocking arabinan synthesis. Science. 2009;324:801–4.
Batt SM, Cacho Izquierdo M, Castro Pichel J, Stubbs CJ, Vela-Glez Del Peral L, Perez-Herran E, et al. Whole cell target engagement identifies novel inhibitors of Mycobacterium Tuberculosis decaprenylphosphoryl-beta-D-ribose oxidase. ACS Infect Dis. 2015;1:615–26.
Christophe T, Jackson M, Jeon HK, Fenistein D, Contreras-Dominguez M, Kim J, et al. High content screening identifies decaprenyl-phosphoribose 2’ epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathog. 2009;5:e1000645.
Neres J, Hartkoorn RC, Chiarelli LR, Gadupudi R, Pasca MR, Mori G, et al. 2-Carboxyquinoxalines kill Mycobacterium Tuberculosis through noncovalent inhibition of Dpre1. ACS Chem Biol. 2015;10:705–14.
Panda M, Ramachandran S, Ramachandran V, Shirude PS, Humnabadkar V, Nagalapur K, et al. Discovery of pyrazolopyridones as a novel class of noncovalent Dpre1 inhibitor with potent anti-mycobacterial activity. J Med Chem. 2014;57:4761–71.
Rogacki MK, Pitta E, Balabon O, Huss S, Lopez-Roman EM, Argyrou A, et al. Identification and profiling of hydantoins-a novel class of potent antimycobacterial Dpre1 inhibitors. J Med Chem. 2018;61:11221–49.
Wang F, Sambandan D, Halder R, Wang J, Batt SM, Weinrick B, et al. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. Proc Natl Acad Sci USA. 2013;110:E2510–7.
Neres J, Pojer F, Molteni E, Chiarelli LR, Dhar N, Boy-Rottger S, et al. Structural basis for benzothiazinone-mediated killing of Mycobacterium Tuberculosis. Sci Transl Med. 2012;4:150ra21.
Pasca MR, Degiacomi G, Ribeiro ALDL, Zara F, De Mori P, Heym B, et al. Clinical isolates of Mycobacterium tuberculosis in four european hospitals are uniformly susceptible to benzothiazinones. Antimicrob Agents Chemother. 2010;54:1616–8.
Makarov V, Lechartier B, Zhang M, Neres J, van der Sar AM, Raadsen SA, et al. Towards a new combination therapy for tuberculosis with next generation benzothiazinones. EMBO Mol Med. 2014;6:372–83.
Lechartier B, Cole ST. Mode of action of clofazimine and combination therapy with benzothiazinones against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2015;59:4457–63.
Lupien A, Vocat A, Foo CS, Blattes E, Gillon JY, Makarov V, et al. Optimized background regimen for treatment of active tuberculosis with the next-generation benzothiazinone macozinone (Pbtz169). Antimicrob Agents Chemother. 2018;62:e00840–18.
Shirude PS, Shandil RK, Manjunatha MR, Sadler C, Panda M, Panduga V, et al. Lead optimization of 1,4-azaindoles as antimycobacterial agents. J Med Chem. 2014;57:5728–37.
Hariguchi N, Chen X, Hayashi Y, Kawano Y, Fujiwara M, Matsuba M, et al. Opc-167832, a novel carbostyril derivative with potent antituberculosis activity as a Dpre1 inhibitor. Antimicrob Agents Chemother. 2020;64:e02020–19.
Shirude PS, Shandil R, Sadler C, Naik M, Hosagrahara V, Hameed S, et al. Azaindoles: noncovalent Dpre1 inhibitors from scaffold morphing efforts, kill Mycobacterium tuberculosis and are efficacious in vivo. J Med Chem. 2013;56:9701–8.
Chikhale RV, Barmade MA, Murumkar PR, Yadav MR. Overview of the development of Dpre1 inhibitors for combating the menace of tuberculosis. J Med Chem. 2018;61:8563–93.
Wang Z, Sun H, Shen C, Hu X, Gao J, Li D, et al. Combined strategies in structure-based virtual screening. Phys Chem Chem Phys. 2020;22:3149–59.
Hu XP, Chai X, Wang XW, Duan MJ, Pang JP, Fu WT, et al. Advances in the computational development of androgen receptor antagonists. Drug Discov Today. 2020;25:1453–61.
Timo GO, Reis R, Melo AF, Costa TVL, Magalhaes PO, Homem-de-Mello M. Predictive power of in silico approach to evaluate chemicals against M. Tuberculosis: a systematic review. Pharmaceuticals (Basel). 2019;12:135.
Gao Y, Xie J, Tang R, Yang K, Zhang Y, Chen L, et al. Identification of a pyrimidinetrione derivative as the potent Dpre1 inhibitor by structure-based virtual ligand screening. Bioorg Chem. 2019;85:168–78.
Schneider G, Neidhart W, Giller T, Schmid G. “Scaffold-Hopping” by topological pharmacophore search: a contribution to virtual screening. Angew Chem-Int Ed. 1999;38:2894–6.
Hu Y, Stumpfe D, Bajorath J. Recent advances in scaffold hopping. J Med Chem. 2017;60:1238–46.
Xiong GL, Zhao Y, Liu L, Ma ZY, Lu AP, Cheng Y, et al. Computational bioactivity fingerprint similarities to navigate the discovery of novel scaffolds. J Med Chem. 2021;64:7544–54.
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–34.
Schrödinger Release 2018-1: Maestro; Schrödinger, Llc: New York, Ny, USA, 2018.
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49.
Case DC, Cheateham T, Darden T, Duke R, Giese T, Gohlke H, et al. AMBER 18. San Francisco: University of California; 2018.
Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. Ff14sb: improving the accuracy of protein side chain and backbone parameters from Ff99sb. J Chem Theory Comput. 2015;11:3696–713.
Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–74.
Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–92.
Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins Struct Funct Genet. 2004;55:383–94.
Hou T, Wang J, Li Y, Wang W. Assessing the performance of the Mm/Pbsa and Mm/Gbsa methods: I. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51:69–82.
Zhu K, Bonelli KW, Greenwood JR, Day T, Abel R, Farid RS, et al. Docking covalent inhibitors: a parameter free approach to pose prediction and scoring. J Chem Inf Model. 2014;54:1932–40.
Zhang T, Li SY, Nuermberger EL. Autoluminescent Mycobacterium tuberculosis for rapid, real-time, non-invasive assessment of drug and vaccine efficacy. PLoS One. 2012;7:e29774.
Zhang NN, Liu ZY, Liang J, Tang YX, Qian L, Gao YM, et al. Design, synthesis, and biological evaluation of M-amidophenol derivatives as a new class of antitubercular agents. MedChemComm. 2018;9:1293–304.
Landge S, Ramachandran V, Kumar A, Neres J, Murugan K, Sadler C, et al. Nitroarenes as antitubercular agents: stereoelectronic modulation to mitigate mutagenicity. ChemMedChem. 2016;11:331–9.
Martinez Molina D, Nordlund P. The cellular thermal shift assay: a novel biophysical assay for in situ drug target engagement and mechanistic biomarker studies. Annu Rev Pharmacol Toxicol. 2016;56:141–61.
Fedorov O, Marsden B, Pogacic V, Rellos P, Muller S, Bullock AN, et al. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci USA. 2007;104:20523–8.
Larsson EA, Jansson A, Ng FM, Then SW, Panicker R, Liu B, et al. Fragment-based ligand design of novel potent inhibitors of tankyrases. J Med Chem. 2013;56:4497–508.
Prasanna AN, Mehra S. Comparative phylogenomics of pathogenic and non-pathogenic mycobacterium. PLoS One. 2013;8:e71248.
Stanley SA, Grant SS, Kawate T, Iwase N, Shimizu M, Wivagg C, et al. Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS Chem Biol. 2012;7:1377–84.
Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40.
Tiwari R, Moraski GC, Krchnak V, Miller PA, Colon-Martinez M, Herrero E, et al. Thiolates chemically induce redox activation of Btz043 and related potent nitroaromatic anti-tuberculosis agents. J Am Chem Soc. 2013;135:3539–49.
Acknowledgements
This work was financially supported by Natural Science Foundation of China of Zhejiang Province (LZ19H300001), National Natural Science Foundation of China (21907084, 81302679, 81973372, 21920102003), Fundamental Research Funds for the Central Universities (2020QNA7003), and “Double Top-Class” University Project (181201*194232101).
Author information
Authors and Affiliations
Contributions
TJH, DL and DSC initiated and supervised the research. XPH and LY designed the experiments. XPH and LY conducted virtual screening, compound validations and biological assays. XC, YXL, MSA, CS, DJJ, ZW, ZYL, LX, KLW, TYZ, and YLY helped performing part of biological assays. LL helped performing the CBFP-based similarity search. XPH, LY, DL and TJH wrote the manuscript, and the other authors contributed specific parts of the manuscript. TJH, DL and DSC assume responsibility for the manuscript in its entirety. All authors have critically revised the manuscript and approved its final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Hu, Xp., Yang, L., Chai, X. et al. Discovery of novel DprE1 inhibitors via computational bioactivity fingerprints and structure-based virtual screening. Acta Pharmacol Sin 43, 1605–1615 (2022). https://doi.org/10.1038/s41401-021-00779-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-021-00779-1