Abstract
The acquired drug resistance by Mycobacterium tuberculosis (M. tuberculosis) to antibiotics urges the need for developing novel anti-M. tuberculosis drugs that possess novel mechanism of action. Since traditional drug discovery is a labor-intensive and costly process, computer aided drug design is highly appreciated tool as it speeds up and lower the cost of drug development process. Herein, Asinex antibacterial compounds were virtually screened against thioesterase domain of Polyketide synthase 13, a unique enzyme that forms α-alkyl β-ketoesters as a direct precursor of mycolic acids which are essential components of the lipid-rich cell wall of M. tuberculosis. The study identified three drug-like compounds as the most promising leads; BBB_26582140, BBD_30878599 and BBC_29956160 with binding energy value of − 11.25 kcal/mol, − 9.87 kcal/mol and − 9.33 kcal/mol, respectively. The control molecule binding energy score is -9.25 kcal/mol. Also, the docked complexes were dynamically stable with maximum root mean square deviation (RMSD) value of 3 Å. Similarly, the MM-GB\PBSA method revealed highly stable complexes with mean energy values < − 75 kcal/mol for all three systems. The net binding energy scores are validated by WaterSwap and entropy energy analysis. Furthermore, The in silico druglike and pharmacokinetic investigation revealed that the compounds could be suitable candidates for additional experimentations. In summary, the study findings are significant, and the compounds may be used in experimental validation pipeline to develop potential drugs against drug-resistant tuberculosis.
Similar content being viewed by others
Introduction
Tuberculosis (TB) is a deadly infectious disease caused by Mycobacterium tuberculosis (M. tuberculosis) and listed as the 13th leading cause of mortality worldwide and the 2nd infectious leading killer after COVID-19. According to the WHO, it was estimated that 10 million people were infected with TB in 2020, of which 1.5 million people died. Global reports indicate that the incidence rate is rising in all countries and among all ages and further aggravated by latent infections. Furthermore, it is estimated that 5–10% of world population will develop active TB during their lifetime1. The standard treatment for drug sensitive TB requires 6–9 months regimen of antibiotics combination. Essentially, four first-line antibiotics used in the treatment of TB (isoniazid–INH, rifampicin–RMP, ethambutol–EMB, pyrazinamide–PZA) and some other drugs which can be administered to patients infected with resistant strains M. tuberculosis Although TB is curable and preventable, multidrug-resistant TB (MDR-TB) and extensively drug-resistant strains of M. tuberculosis (XDR-TB) remain a global burden rendering the standard treatment ineffective2. It is estimated that about 0.5 million of MDR-TB cases reported annually worldwide and nearly 14,000 XDR-TB cases were originated from 81 countries3,4. Additionally, the management of TB was further aggravated as MDR-TB developed resistant to two of the first-line drugs; RMP and INH, while XDR-TB is resistant to the four essential antitubercular drugs and at least one of the injectable second-line drugs such as amikacin, capreomycin, or kanamycin5,6. The current guideline of treating MDR-TB and XDR-TB involve pretomanid, and bedaquiline (approved by the US FDA in 2019 and 2012, respectively) along with linezolid as the first line tuberculosis therapy1,7. In consequence to the high number of TB resistant cases, the demand to introduce promising antitubercular drugs are urgently needed particularly those that are designed to target tuberculosis biomolecules essential for various biological processes.
The cell wall of M. tuberculosis is regarded a rich source of molecular targets that can be exploited to design and develop novel antitubercular drugs8,9. It is a unique structure that contains mycomembrane and peptidoglycan layer. The former comprised long chains of fatty acids termed as mycolic acids (MAs). These molecules are specific lipid components that are essential for the survival and virulence of M. tuberculosis and bind to arabinogalactan via ester bonds10,11. The inhibition of MAs biosynthesis is the main mechanism for the effect of the forefront and effective anti-tubercular drug INH12. Hence, much interest have been devoted to deciphering the biosynthesis of MAs to recruit attractive targets for development of unconventional antitubercular drugs to tackle the drug-resistant TB.
Polyketide synthase 13 (Pks13) plays a crucial role in the biosynthesis of MAs in which it mediates the condensation of two fatty acids to form mycolic β-ketoester, a direct precursor for MAs. It comprises five distinct catalytic domains, of which thioesterase domain (TE) plays a dual function where it first acts as a hydrolase to break the thioester bond and form an ester bond between mycolic β-ketoesters and the hydroxyl group of Ser1533 of the TE domain. In the second function, TE acts as an acyltransferase to form trehalose monomycolate (TMM) which is then transported and attached to arabinogalactan13,14. The thioesterase domain of Pks13 (Pks13-TE) has been recognized as a druggable target for developing anti-tubercular drugs15,16. As a result, several potential compounds have been proposed to interact with the Pks13-TE. These compounds include thiophenes17, benzofurans18, and β-lactones19.
Conventional drug development is a daunting, time-consuming, and costly task that suffers from high failure rate20,21,22,23. In recent times, the computer aided drug design (CADD) techniques have gained considerable attention and proved indispensable in drug discovery process24. CADD methods are in silico approach that can accelerate the drug discovery and shorten the time needed for leads identification to drug marketing. Additionally, these computational methods are critical in estimating biological activities of chemical compounds against any given biological target25,26. CADD can also be helpful in determining the binding affinity of compounds when it is docked to any target of interest as well as predicting the physicochemical properties of compounds27,28,29,30,31. The CAAD methods have successfully used in the past for discovery of drugs such as nelfinavir, imatinib, and zanamivir32. Considering the vast applications of CADD techniques in drug discovery, we herein targeted Pks13-TE domain using a multi-pronged in silico approach. The findings gathered in this study might be useful in successful identification of leads that can be further optimized for biological activity as novel anti-tubercular therapeutics.
Materials and methods
The study was based on in silico techniques that can be split into following phases;
Asinex library preparation
The Asinex antibacterial library was retrieved from Asinex web available at https://www.asinex.com/. The library contains 6208 compounds of natural product-like scaffolds providing diversity and accessibility for experimental studies. The library was imported to LigandScout software33 where the library was filtered based on Lipinski rule of five34. The filtered library was then subjected to PyRx 0.835 to energy minimized the compounds using MM2 force field36 followed by conversion to .pdbqt format to make the compounds ready for docking studies.
Pks13-TE enzyme preparation
For potential leads identification against Pks13-TE, a protein data bank (PDB) ID of “7M7V” was chosen based on the fact that the structure is most recent with good resolution of 2.29 Å37. The enzyme structure was then subjected to energy minimization process in UCSF Chimera 1.1638. The energy minimization process was done in two steps: first by steepest descent for 1000 steps followed by conjugate gradient algorithm for 1500 rounds. Missing hydrogen atoms were added to the enzyme and charge assignment was done through gasteiger method. The RMSD of minimized and non-minimized Pks13-TE was 0.24 Å. After energy minimization, the enzyme was saved into .pdb format and converted afterward into .pdbqt format for utilization in virtual docking studies.
Structure based virtual screening process
To identify potential leads against Pks13-TE, structure based virtual screening process was performed. This was done using coordinates information of Tyr1663 which is considered a focus point of the thioesterase domain. The virtual screening process was achieved using PyRx 0.8 by employing AutoDock Vina 4.035,39. The grid box was centered covering coordinates of 4.32 Å (x-axis), 35.85 Å (y-axis) and 6.77 Å (z-axis) with dimensions of 25 Å on XYZ planes. Each molecule in the library was docked with the enzyme 100 times and the best posed molecules were ranked based on binding energy score which was measured in kcal/mol. Molecules with lowest binding energy score in reference to the control molecule, Benzofuran 137, were reported. The control molecules extracted from the crystal structure and redocked to the enzyme to validate the docking procedure. Another round of docking procedure revalidation was achieved by using GOLD docking software considering the same set of parameters described above. The best docked complexes were used in Discovery Studio Visualizer 2021 to examine the docked intermolecular conformation and observed chemical interactions network40.
Molecular dynamic simulation (MDS)
MDS was done to get information about physical movements of docked complexes. The MDS was performed using AMBER20 package41. The complexes were processed through Antechamber program42. For compounds, Amber general force field (GAFF) was employed to generate parameter files while for Pks13-TE, FF14SB was considered43,44. The systems were solvated into TIP3P water model and then counter ions were added to neutralize the net charge. The padding distance set between the complex and water box edge was 12 Å45,46,47. Energy minimization was then carried out for complexes using steepest descent and conjugate gradient algorithms for total of 3000 steps. After that, the complexes were heated to 310 K for 500 ps. The systems were then equilibrated and subjected to production run of 300 ns. This time scale was found sufficient to get well converged systems. The long term interactions were treated via particle mesh Ewald method48. The hydrogen bonds were constrained via SHAKE algorithm while for temperature control, Langevin algorithm was used49,50. The simulation trajectories processed through CPPTRAJ module to investigate structure stability of the systems51. Plots were made using XMGRACE52. Further, MM-GB\PSA binding free energy was employed to get confidence on the intermolecular binding affinity53,54. For this purpose, 1500 frames were opted from the entire simulation trajectories at equal time interval to investigate intermolecular binding energies of complexes along the simulation time length. The extracted frames were subjected to MMPBSA.py script to estimate net binding free energy55. The net binding free energy was estimated through the following equation;
Validation on the MM-GB\PSA was done through more sophisticated WaterSwap method that uses reaction coordinates to swap water cluster equal to size of the ligand molecule and swap it with the ligand56. The WaterSwap was defaulted for 1000 iterations. Three algorithms were used in WaterSwap: thermodynamic integration, Bennett’s, and free energy perturbation.
Entropy energy analysis
The binding entropy energy of docked compound was also estimated to get real binding affinity score of complexes. For this, AMBER normal mode analysis was employed54. The method is computationally expensive; therefore, only several snapshots (10 frames) from the entire simulation trajectories were selected. In this method, several energies were determined such as translational, vibrational and rotational57,58.
SwissADME and pkCSM analysis
The computational druglikeness and pharmacokinetics were predicted to ensure selection of only promising molecules59,60. The absorption, distribution, metabolism, and excretion (ADME) and toxicity of compounds were estimated using SwissADME and pkCSM28,61.
Results and discussion
Docking studies
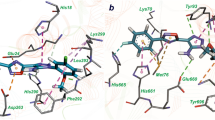
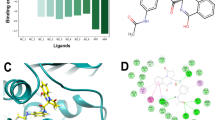
Structure based virtual screening was conducted to prioritized chemical compound that bind best to the Pks13-TE. The combined approach of virtual screening and MDS have been used in the past to successfully screen inhibitor molecules against biological targets45,62,63,64. The Lipinski rule of five revealed total of 26,024 molecules as druglike compounds. These molecules were used in virtual screening process against Pks13-TE. Three compounds have stable binding affinity and identified as promising leads. These compounds are BBB_26582140, BBD_30878599 and BBC_29956160 with binding energy value of − 11.25 kcal/mol, − 9.87 kcal/mol and − 9.33 kcal/mol, respectively. The control molecule binding energy score was − 9.25 kcal/mol. The binding affinity of the compounds/control at the active pocket of polyketide synthase Pks13 was revisited by GOLD docking software. The GOLD fitness score of BBB_26582140 was 78.62, BBD_30878599 has GOLD score of 75.83 while the GOLD docked affinity of BBC_29956160 was 72.55. The control molecule secures a GOLD fitness score of 73.00. The BBB_26582140 is 2-((1-acetylpiperidin-3-yl)methyl)-5-carboxypyrazine-1,4-diium. The compounds achieved deep floor binding at Pks13 functional domain and accomplished rich interacting network. The oxygen atom of 1-(piperidin-1-yl)ethanone formed closed distance hydrogen bond of 2.01 Å with His1699. Another hydrogen bond was revealed with Asp1644 at distance of 1.88 Å via 2-carboxypyrazine-1,4-diium ring. The binding mode and interactions of compound BBB_26582140 is given in Fig. 1.
The BBD_30878599 is N-(2-hydroxyethyl)-5-(piperidin-2-yl)-2,5-dihydro-1H-pyrazole-3-carboxamide. The compound interactions are balanced by both van der Waals and hydrogen bonds. The N-(2-hydroxyethyl)formamide is mainly engaged in hydrogen bonds with Asp1644 and Ala1667. The compounds like the first compound docked deep inside the pocket and gained access to the pocket bottom (Fig. 2).
The BBC_29956160 is 3-(aminomethyl)-N-methyl-2,5-dihydro-1,2,4-oxadiazole-5-carboxamide. This compound accommodates across the pocket length and forms hydrogen bond with Asp1644 at distance of 1.87 Å. Also, the compound produces His1699 and Asn1640 at bond distance of 2.63 Å and 1.91 Å, respectively (Fig. 3).
The control compound interactions with the enzyme is mainly dominated by hydrophobic contact and only one hydrogen bond was witnessed with Asp1644 (Fig. 4).
Dynamic studies of docked complexes
Insights about physical movements of docked compound with receptor were accomplished using molecular dynamic simulation assay. This was essential as docking studies only provide one snapshot view while biomolecules behave in time dependent dynamics milieu. The first analysis performed in this regard was root mean square deviation (RMSD), followed by root mean square fluctuation (RMSF). Both these analyses were conducted considering carbon alpha atoms of the complexes. Compared to control, all the complexes disclosed stable dynamic conformation of polyketide synthase Pks13. The RMSD of all complexes reported stable behavior in the presence of docked compounds. The most stable system is of BBD_30878599 with mean RMSD of 1.07 Å. The control system shown higher RMSD with regular variations throughout the simulation length. The mean RMSD noted for control complex was greater than 3 Å. Similarly, the RMSF complements the RMSD analysis. The N-terminal of the receptor bimolecular showed higher RMSF than the C-terminal. The RMSD and RMSF plots are given in Fig. 5. Previously, it was reported that Coumestan derivatives were able to bind well with Pks13 and induce colony-forming unit (CFU) reduction of 1.0/ml of the bacterial culture65. Further, it was demonstrated that Benzofuran derivatives inhibit the Pks13 enzyme both in vitro and in vivo37. The radius of gyration (RoG) analysis was further performed to examine the compact behavior of docked complexes66,67. The receptors compact behavior is important to maintain strong intermolecular docked conformation and interactions. The RoG of studied systems is given in Fig. 6. All the three systems revealed RoG value ~ 37–38 Å, revealing highly stable RoG. Similarly, solvent accessible surface area (SASA) analysis was conducted for the systems68,69. The SASA for BBB_26582140, BBD_30878599, BBC_29956160 and control is given S-Fig. 1, S-Fig. 2, S-Fig. 3 and S-Fig. 4, respectively. The analysis demonstrated that the intermolecular interactions is highly intact and the water molecules accessibility is low.
MDS free energy analysis
The MM-GBSA and MM-PBSA binding free energy are undoubtedly promising methods in predicting binding affinity of compounds docked to a particular biomolecule. These methods are appreciated in term of computational speed demands and reliability in predicting compounds interaction strength compared to experimental data. In comparison to control, it has been revealed that molecule BBD_30878599 is the most promising binder to Pks13-TE with net energy value of − 79.75 kcal/mol in MM-GBSA and − 79.31 kcal/mol in MM-PBSA. Similarly, the BBB_26582140 and BBC_29956160 have also demonstrated better binding to Pks13-TE in reference to the control. The gas phase energy was found dominating, whereas van der Waals played a major role in compounds binding with Pks13-TE. This was followed by electrostatic energy. The negative contribution was seen polar solvation energy. The data reported by both MM-GBSA and MM-PBSA techniques noticed the compounds showed considerable compounds binding to the receptor biological molecule. Details about each parameter energy contribution to the net MM-GBSA and MM-PBSA are presented in Table 1. Furthermore, the binding energy score of hotspot residues involved in binding with the compounds is given in Table 2. These hotspot residues have vital contribution in overall intermolecular conformational and interaction stability66,70.
WaterSwap binding energy
Additional confidence was accomplished on the MM-GBSA and MM-PBSA results using WaterSwap method. This method is regarded more accurate due to its ability to consider the role of water molecules particularly those that bridge the ligand with the protein residues. The WaterSwap based energy calculation was done using three algorithms; TI, FEP and Bennett’s. The difference of value 1 kcal/mol among the algorithms demonstrated the systems well converged. As can be noticed, the systems are significantly stable in term of intermolecular binding interactions and binding conformations. The WaterSwap based binding free energies are given in Fig. 7.
Entropy energy calculation
Binding entropy estimation was performed to determine freedom energy of docked molecules with Pks13-TE. It was demonstrated that net entropy energy is negative in all systems, indicating that the systems significantly hold non-favorable energy that contribution to the instability of the systems. However, this energy is much less than the net total of MM-GBSA and MM-PBSA which indicates that systems are stable enough to ensure intermolecular docked conformation. The systems entropy energy is provided in Fig. 8.
Pharmacokinetic analysis
In silico pharmacokinetic analysis is vital to shed light on what the body will do with the selected drugs. The study can guide how the drugs will be subjected to absorption, bioavailability, distribution, metabolism, and excretion once administered into the host body. In pharmacokinetic, the most significant factor to evaluate is the oral bioavailability and adsorption of the drugs in gastrointestinal tract. Except BBD_30878599, the selected compounds were predicted to show high gastrointestinal absorption including the control molecule. This indicates that high proportions of drugs can be available for therapeutic affect at the target sites. Determination of compounds permeability to the central nervous system (CNS) is prerequisite in drug discovery. All the compounds as well as the control were found non-permeable to the CNS. The P-glycoprotein 1 (Pgp-1) is a multi-drug resistance protein that pumps out foreign substances from the cells. It was found that the BBD_30878599 and control are non-substrate to Pgp-1 and thus they are unlikely to be excluded from the cells. Similarly, all the compounds are non-inhibitors of CYP1A2, CYP2C19, CYP2C9, CYP2D6 and CYP3A4. These cytochrome proteins are involved in metabolism of drugs and xenobiotic. The control is inhibitor for majority of these cytochromes. The compounds skin permeation value ranges from − 7.61 to − 8.17 cm/s compared to control (− 7.19 cm/s). The pharmacokinetic analysis of compounds is presented in Table 3. From water solubility perspective, the compounds were classified as good water soluble. The control molecule was predicted as moderate water soluble.
Druglikeness/medicinal chemistry analysis
Druglike compounds have higher chances of reaching market and can be branded into a successful drug. There are certain rules that are available to check whether a compound is drug like or not. The most important is Lipinski rule of five. According to this rule, all the compound fulfills Lipinski rule of five and are classified as drug like. This rule defines a drug molecule to have molecular weight < 500 Dalton, hydrogen bond donors < 5, hydrogen bond acceptors < 10, and topological polar surface area < 140 Å2 and LogP value < 5. Likewise, except control molecule, Ghose predicted all the screened molecules to be non-drug as they have disagreed on the LogP and molecular weight. Similarly, the compounds agree on Veber, Egan and Muegge rules. Also, the compounds have good bioavailability score. The compounds except control was found to have zero alert for pan-assay interference (PAINS) compounds and likely to interact with one specific biomolecule. The BBB_26582140 was the only compound that has lead-like structure and thus can be structurally optimized to enhance the biological potency. The synthetic accessibility predicted the compounds can be easily synthesized for experimental analysis. All the druglikeness/medicinal chemistry analysis are presented in Table 4. The compounds are also classified as non-mutagenic, non-carcinogenic and non-AMES toxic. Further, the compounds showed less hepatotoxicity.
Conclusions
In this study, three compounds; BBB_26582140, BBD_30878599 and BBC_29956160 were identified as best binders of Pks13-TE. The compounds were noticed to docked deep inside the binding pocket of the enzyme and formed rich pattern of both hydrophobic and hydrophilic contacts. The compounds binding conformations with the enzyme were found highly stable. This was validated by the different binding free energy methods used in the study. Also, the compounds were found to have promising druglike and pharmacokinetic properties. In a nutshell, the compounds identified herein were promising in term of affinity for the Pks13-TE and thus can be subjected to experimental evaluations. Further, the compounds might provide starting leads to speed up drug discovery and development against thioesterase domain of Polyketide synthase 13 of M. tuberculosis.
Data availability
All data generated or analysed during this study are included in this published article.
References
Organization, W. H. & others. WHO Consolidated Guidelines on Drug-Resistant Tuberculosis Treatment. (World Health Organization, 2019).
Stoffels, K. et al. From multidrug-to extensively drug-resistant tuberculosis: Upward trends as seen from a 15-year nationwide study. PLoS ONE 8, e63128 (2013).
Chakaya, J. et al. The WHO global tuberculosis 2021 report—Not so good news and turning the tide back to End TB. Int. J. Infect. Dis. 124, S26–S29 (2022).
Minias, A. et al. Early drug development and evaluation of putative antitubercular compounds in the—Omics era. Front. Microbiol. 11, 618168 (2021).
Seung, K. J., Keshavjee, S. & Rich, M. L. Multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis. Cold Spring Harb. Perspect. Med. 5, a017863 (2015).
Dagne, B. et al. The Epidemiology of first and second-line drug-resistance Mycobacterium tuberculosis complex common species: Evidence from selected TB treatment initiating centers in Ethiopia. PLoS ONE 16, e0245687 (2021).
FDA, U. S. FDA approves new drug for treatment-resistant forms of tuberculosis that affects the lungs. US FDA Silver Spring, MD, USA (2019).
Favrot, L. & Ronning, D. R. Targeting the mycobacterial envelope for tuberculosis drug development. Expert Rev. Anti. Infect. Ther. 10, 1023–1036 (2012).
Chen, H. et al. The mycobacterial membrane: A novel target space for anti-tubercular drugs. Front. Microbiol. 9, 1627 (2018).
Barry, C. E., Crick, D. C. & McNeil, M. R. Targeting the formation of the cell wall core of M. tuberculosis. Infect. Disord Targets Former. Curr. Drug Targets-Infect. Disord. 7, 182–202 (2007).
Marrakchi, H., Lanéelle, M.-A. & Daffé, M. Mycolic acids: Structures, biosynthesis, and beyond. Chem. Biol. 21, 67–85 (2014).
Takayama, K., Wang, L. & David, H. L. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2, 29–35 (1972).
Zhao, G. et al. The structure-based virtual screening of non-benzofuran inhibitors against M. tuberculosis Pks13-TE for anti-tuberculosis phenotypic discovery. New J. Chem. 45, 1286–1300 (2021).
Gavalda, S. et al. The polyketide synthase Pks13 catalyzes a novel mechanism of lipid transfer in mycobacteria. Chem. Biol. 21, 1660–1669 (2014).
Ioerger, T. R. et al. Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS ONE 8, e75245 (2013).
Jeffrey North, E., Jackson, M. & E Lee, R. New approaches to target the mycolic acid biosynthesis pathway for the development of tuberculosis therapeutics. Curr. Pharm. Des. 20, 4357–4378 (2014).
Wilson, R. et al. Antituberculosis thiophenes define a requirement for Pks13 in mycolic acid biosynthesis. Nat. Chem. Biol. 9, 499–506 (2013).
Aggarwal, A. et al. Development of a novel lead that targets M. tuberculosis polyketide synthase 13. Cell 170, 249–259 (2017).
Lehmann, J. et al. An antibacterial β-lactone kills Mycobacterium tuberculosis by disrupting mycolic acid biosynthesis. Angew. Chemie Int. Ed. 57, 348–353 (2018).
Shaker, B., Ahmad, S., Lee, J., Jung, C. & Na, D. In silico methods and tools for drug discovery. Comput. Biol. Med. 137, 104851 (2021).
Wahedi, H. M., Ahmad, S. & Abbasi, S. W. Stilbene-based natural compounds as promising drug candidates against COVID-19. J. Biomol. Struct. Dyn. https://doi.org/10.1080/07391102.2020.1762743 (2020).
Hassan-Baig, M. et al. Computer aided drug design: Success and limitations. Curr. Pharm. Des. 22, 572–581 (2016).
Ahmad, S., Waheed, Y., Ismail, S., Abbasi, S. W. & Najmi, M. H. A computational study to disclose potential drugs and vaccine ensemble for COVID-19 conundrum. J. Mol. Liq. 324, 114734 (2021).
Yu, W. & MacKerell, A. D. Computer-aided drug design methods. In Antibiotics 85–106 (Springer, 2017).
Sydow, D., Morger, A., Driller, M. & Volkamer, A. TeachOpenCADD: A teaching platform for computer-aided drug design using open source packages and data. J. Cheminform. 11, 29 (2019).
Ahmad, S., Raza, S., Uddin, R. & Azam, S. S. Binding mode analysis, dynamic simulation and binding free energy calculations of the MurF ligase from Acinetobacter baumannii. J. Mol. Graph. Model. 77, 72–85 (2017).
Meng, X.-Y., Zhang, H.-X., Mezei, M. & Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided. Drug Des. 7, 146–157 (2011).
Daina, A., Michielin, O. & Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 1–13 (2017).
Ahmad, F. et al. Discovery of potential antiviral compounds against hendra virus by targeting its receptor-binding protein (G) using computational approaches. Molecules 27, 554 (2022).
Javed, N., Ahmad, S., Raza, S. & Azam, S. S. Subtractive proteomics supported with rational drug design approach revealed ZINC23121280 as a potent lead inhibitory molecule for multi-drug resistant Francisella tularensis: Drug designing for multidrug-resistant Francisella tularensis. Proc. Pakistan Acad. Sci. B. Life Environ. Sci. 58, 1–42 (2021).
Ahmad, S., Raza, S., Abro, A., Liedl, K. R. & Azam, S. S. Toward novel inhibitors against KdsB: A highly specific and selective broad-spectrum bacterial enzyme. J. Biomol. Struct. Dyn. 37, 1326–1345 (2019).
Talele, T. T., Khedkar, S. A. & Rigby, A. C. Successful applications of computer aided drug discovery: Moving drugs from concept to the clinic. Curr. Top. Med. Chem. 10, 127–141 (2010).
Wolber, G. & Langer, T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 45, 160–169 (2005).
Lipinski, C. A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 1, 337–341 (2004).
Dallakyan, S. & Olson, A. J. Small-molecule library screening by docking with PyRx. In Chemical Biology 243–250 (Springer, 2015).
Halgren, T. A. Merck molecular force field. J. Comput. Chem. 17, 490–519 (1996).
Wilson, C. et al. Optimization of TAM16, a benzofuran that inhibits the thioesterase activity of Pks13; evaluation toward a preclinical candidate for a novel antituberculosis clinical target. J. Med. Chem. 65, 409–423 (2021).
Pettersen, E. F. et al. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Biovia, D. S. Discovery studio visualizer. San Diego, CA, USA (2017).
Case, D. A. et al. Amber 2020. (2020).
Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Antechamber: An accessory software package for molecular mechanical calculations. J. Am. Chem. Soc. 222, U403 (2001).
Sprenger, K. G., Jaeger, V. W. & Pfaendtner, J. The general AMBER force field (GAFF) can accurately predict thermodynamic and transport properties of many ionic liquids. J. Phys. Chem. B 119, 5882–5895 (2015).
Maier, J. A. et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Singh, V., Dhankhar, P., Dalal, V., Tomar, S. & Kumar, P. In-silico functional and structural annotation of hypothetical protein from Klebsiella pneumonia: A potential drug target. J. Mol. Graph. Model. 116, 108262 (2022).
Kumari, R., Rathi, R., Pathak, S. R. & Dalal, V. Computational investigation of potent inhibitors against YsxC: Structure-based pharmacophore modeling, molecular docking, molecular dynamics, and binding free energy. J. Biomol. Struct. Dyn. 41(3), 930–941 (2021).
Singh, V. et al. Drug-repurposing approach to combat Staphylococcus aureus: Biomolecular and binding interaction study. ACS Omega 7, 38448–38458 (2022).
Petersen, H. G. Accuracy and efficiency of the particle mesh Ewald method. J. Chem. Phys. 103, 3668–3679 (1995).
Kräutler, V., Van Gunsteren, W. F. & Hünenberger, P. H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 22, 501–508 (2001).
Izaguirre, J. A., Catarello, D. P., Wozniak, J. M. & Skeel, R. D. Langevin stabilization of molecular dynamics. J. Chem. Phys. 114, 2090–2098 (2001).
Roe, D. R. & Cheatham, T. E. III. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013).
Turner, P. J. XMGRACE, Version 5.1. 19. Cent. Coast. Land-Margin Res. Oregon Grad. Inst. Sci. Technol. Beaverton, OR (2005).
Hou, T., Wang, J., Li, Y. & Wang, W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 51, 69–82 (2011).
Genheden, S., Kuhn, O., Mikulskis, P., Hoffmann, D. & Ryde, U. The normal-mode entropy in the MM/GBSA method: Effect of system truncation, buffer region, and dielectric constant. J. Chem. Inf. Model. 52, 2079–2088 (2012).
Miller, B. R. et al. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 8, 3314–3321 (2012).
Woods, C. J. et al. Rapid decomposition and visualisation of protein-ligand binding free energies by residue and by water. Faraday Discuss. 169, 477–499 (2014).
Ismail, S., Ahmad, S. & Azam, S. S. Immunoinformatics characterization of SARS-CoV-2 spike glycoprotein for prioritization of epitope based multivalent peptide vaccine. J. Mol. Liq. 314, 113612 (2020).
Alamri, M. A. et al. Screening of marine natural products for potential inhibitors targeting biotin biosynthesis pathway in Mycobacterium tuberculosis. J. Biomol. Struct. Dyn. https://doi.org/10.1080/07391102.2022.2135596 (2022).
Altharawi, A., Ahmad, S., Alamri, M. A. & ulQamar, M. T. Structural insight into the binding pattern and interaction mechanism of chemotherapeutic agents with Sorcin by docking and molecular dynamic simulation. Colloids Surfaces B Biointerfaces 208, 112098 (2021).
Van De Waterbeemd, H. & Gifford, E. ADMET in silico modelling: towards prediction paradise?. Nat. Rev. Drug Discov. 2, 192–204 (2003).
Pires, D. E. V., Blundell, T. L. & Ascher, D. B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 58, 4066–4072 (2015).
Gupta, D. N. et al. Biochemical characterization and structure-based in silico screening of potent inhibitor molecules against the 1 cys peroxiredoxin of bacterioferritin comigratory protein family from Candidatus Liberibacter asiaticus. J. Biomol. Struct. Dyn. https://doi.org/10.1080/07391102.2022.2096118 (2022).
Rathi, R., Kumari, R., Pathak, S. R. & Dalal, V. Promising antibacterials for LLM of Staphylococcus aureus using virtual screening, molecular docking, dynamics, and MMPBSA. J. Biomol. Struct. Dyn. https://doi.org/10.1080/07391102.2022.2119278 (2022).
Kumari, R., Rathi, R., Pathak, S. R. & Dalal, V. Structural-based virtual screening and identification of novel potent antimicrobial compounds against YsxC of Staphylococcus aureus. J. Mol. Struct. 1255, 132476 (2022).
Lun, S. et al. Therapeutic potential of coumestan Pks13 inhibitors for tuberculosis. Antimicrob. Agents Chemother. 65, e02190-e2220 (2021).
Dalal, V. et al. Structure-based identification of potential drugs against FmtA of Staphylococcus aureus: Virtual screening, molecular dynamics, MM-GBSA, and QM/MM. Protein J. 40, 148–165 (2021).
Dhankhar, P., Dalal, V., Golemi-Kotra, D. & Kumar, P. In-silico approach to identify novel potent inhibitors against GraR of S. aureus. Front. Biosci. 25, 1337–1360 (2020).
Dalal, V. et al. Repurposing an ancient protein core structure: Structural studies on FmtA, a novel esterase of Staphylococcus aureus. J. Mol. Biol. 431, 3107–3123 (2019).
Malik, A., Dalal, V., Ankri, S. & Tomar, S. Structural insights into Entamoeba histolytica arginase and structure-based identification of novel non-amino acid based inhibitors as potential antiamoebic molecules. FEBS J. 286, 4135–4155 (2019).
Dhankhar, P., Dalal, V., Singh, V., Tomar, S. & Kumar, P. Computational guided identification of novel potent inhibitors of N-terminal domain of nucleocapsid protein of severe acute respiratory syndrome coronavirus 2. J. Biomol. Struct. Dyn. 40, 4084–4099 (2022).
Acknowledgements
This study is supported via funding from Prince Sattam Bin Abdulaziz University project number (PSAU/2023/R/1444).
Author information
Authors and Affiliations
Contributions
A.A. and M.A.A. performed the experiments and wrote the main text. M.M.A. and S.M.A. validated the findings and revised the manuscript. M.T.Q. designed this study and finalized the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Altharawi, A., Alossaimi, M.A., Alanazi, M.M. et al. An integrated computational approach towards novel drugs discovery against polyketide synthase 13 thioesterase domain of Mycobacterium tuberculosis. Sci Rep 13, 7014 (2023). https://doi.org/10.1038/s41598-023-34222-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-34222-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.