Abstract
Hepatocellular carcinoma (HCC) is one of the most lethal tumours worldwide. However, the effects of first-line sorafenib treatment in advanced HCC fail to prolong patients’ survival due to the highly heterogeneous characteristics of HCC etiology. Cyclin-dependent kinase 9 (CDK9) is an important target in the continuous development of cancer therapy. Here, we demonstrate that CDK9 is closely associated with the progression of HCC and can serve as an HCC therapeutic target by modulating the recovery of wild-type p53 (wt-p53) function. We prove that mouse double minute 2 homologue (MDM2) and Sirtuin 1 (SIRT1) are phosphorylated by CDK9 at Ser166 and Ser47, respectively. Inhibition of CDK9 not only reduces the MDM2-mediated ubiquitination and degradation of wt-p53 but also increases wt-p53 stability by suppressing deacetylase activity of SIRT1. Thus, inhibition of CDK9 promotes the wt-p53 stabilization and prevents HCC progression. However, excessive inhibition by high concentrations of specific CDK9 inhibitors counteracts the promotion of p53 stability and reduces their anti-HCC activity because of extreme general transcription repression. The effects of a novel CDK9 inhibitor named oroxylin A (OA) from Scutellaria baicalensis are explored, with the results indicating that OA shows moderate and controlled inhibition of CDK9 activity and expression, and stabilizes wt-p53 by inhibiting CDK9-regulated MDM2 and SIRT1 signaling. These outcomes indicate the high therapeutic potential of OA against HCC and its low toxicity in normal tissue. This study demonstrates a novel mechanism for the regulation of wt-p53 by CDK9 and indicates that OA is a potential candidate for HCC therapy.
Similar content being viewed by others
Introduction
Primary liver cancer is a serious life-threatening disease and is the sixth most common cancer in the world. Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, and the mortality rate is constantly rising worldwide [1, 2]. In 2020, approximately 905,000 new cases and 830,000 deaths from liver cancer were reported, and more than 80% of these cases were observed in Asia and Africa [3]. The incidence of liver cancer, including HCC, has also increased in Western Europe, North America and parts of Oceania, where it was previously relatively rare [4, 5]. Even though some patients can be diagnosed early and treated with liver transplantation or surgical resection, the recurrence rate is still high, and the prognosis offers little hope for patients with advanced disease. Currently, systematic targeted therapies for liver cancer can be divided into two categories: multiple kinase inhibitors (MKIs) and immunotherapy. MKIs mainly include sorafenib and regorafenib [6]. However, these drugs carry a black box warning about the potential for inducing severe toxicity, and they have not significantly prolonged the survival period of patients [6, 7]. In addition, liver cancer shows heterogeneity, posing difficulties for new drug development. Therefore, the identification of new targets and molecular pathways is urgently needed for drug development in the treatment of HCC.
Cyclin-dependent kinases (CDKs) play key roles in cell cycle regulation progression and RNA transcription [8]. According to different mechanisms of action, CDKs are generally categorized into cell cycle-related subfamilies (CDK1, 2, 4 and 6) and transcriptional subfamilies (CDK7–13) [9]. Cyclin-dependent kinase 9 (CDK9), the catalytic subunit of positive transcription elongation factor b (P-TEFb), is a transcriptional activator recruited to promote the release of paused RNA polymerase II (RNAPII) at the proximal promoter by phosphorylating negative elongation factors (DSIF and NELF) [10, 11]. The case made for CDK9 as a target in oncology is usually based on the regulatory effect of CDK9 on the RNA transcription of short-lived antiapoptotic proteins such as Mcl-1 and XIAP [12]. Many small-molecule CDK9 inhibitors have entered into human clinical trials, such as flavopiridol, dinaciclib, seliciclib, SNS-032 and RGB-286638, and have been shown to be particularly efficient when used in multiple myeloma and chronic myelogenous leukaemia treatment [13,14,15,16]. However, the therapeutic potential of CDK9 inhibitors has rarely been reported in solid tumours such as HCC.
The p53 protein is one of the most widely studied tumour suppressors and is considered a genome guardian, coordinating a series of antiproliferative responses and inhibiting the expansion of malignant cells. In cancer cells harbouring wild-type p53 (wt-p53), recovery of wt-p53 function has become a crucial goal in the development of anticancer strategies [17]. Genomic DNA sequencing of human cancers has indicated that the p53 gene (TP53) is somatically mutated in more than 50% of all human cancers [18]. However, the frequency of the p53 mutation remains low in certain types of human cancers, such as HCC [19]. Therefore, exploration into the approach to and correlation mechanism for the recovery of wt-p53 function in HCC is of great significance. Wt-p53 can be phosphorylated on many serine and threonine residues and thus regulated by different cellular kinases [20]. Previous studies have shown that CDK9 regulates p53 in a cell type-dependent manner. CDK9 phosphorylates Ser392, Ser33 and Ser315 of wt-p53 and enhances its DNA-binding and subsequent transactivation ability [20, 21]. It has also been reported that CDK9 phosphorylates the Pirh2 protein and prevents the degradation of the p53 protein in CNS-derived cells [22]. In contrast, in colon cancer cells, inhibition of CDK9 has been shown to reactivate p53 by downregulating iASPP [23]. MDM2 is a principal regulator of p53 and has been validated as an oncotarget. It has been reported that inhibition of CDK9 suppresses MDM2-mediated p53 degradation by increasing the binding of free ribosomal proteins to the central regulatory domain of the MDM2 protein [24]. In addition to the effect of MDM2, the acetylation of p53 is critical for regulating the stability of wt-p53. Sirtuin 1 (SIRT1), an important NAD-dependent deacetylase, is frequently overexpressed in many malignant tumours and associated with the rapid progression of cancer and poor prognosis for patients [25, 26]. SIRT1 can repress p300/CBP transcription, and its absence leads to enhanced p300-mediated p53 activation and transcription of p53 target genes [27].
In this paper, we reveal a novel mechanism of regulation of p53 stabilization via inhibition of CDK9 in HCC. Recovery of wt-p53 function is critical in the anti-HCC effect of CDK9 inhibitors. More importantly, a novel CDK9 inhibitor named oroxylin A (OA), which is a natural flavonoid extracted from Scutellaria baicalensis (S. baicalensis), was obtained by chemical synthesis and developed. Previous studies have shown that OA has strong anti-hepatoma activity that is mediated via multiple pathways, including through the induction of differentiation and autophagy, regulation of glucose metabolism and reversal of multidrug resistance [28,29,30,31,32]. Here, we show the potent anti-hepatoma activity of OA realized by inhibiting CDK9 and restoring p53 function. Compared with the CDK9 inhibitors recently entered into clinical trials and preclinical tests, OA has greater concentration-dependent efficacy and lower toxicity. Our results indicate a possible therapeutic application of CDK9 targeting as an innovative way to reconstitute the tumour suppressor function of wt-p53 in HCC.
Materials and methods
Reagents
Oroxylin A (C16H12O5, MW 284.26, purity ≥99%) was synthesized by Prof. Zhi-yu Li at China Pharmaceutical University. Synthesized OA powder was dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich, St Louis, MO, USA), and a stock solution of 0.2 M OA was mixed and stored at −80 °C. LDC067 was purchased from Selleck Chemicals (C18H18N4O3S, MW 370.43; purity 99.51%; S7461), dissolved in DMSO and prepared as a 0.05 M stock solution. PHA767491 was purchased from MedChemExpress (C12H12ClN3O, MW 249.70; purity 99.91%; HY-13461), dissolved in DMSO and prepared as a 0.05 M stock solution and stored at –80 °C. For animal studies, the solvent consisted of 30% PEG400, 2% Tween 80 and 5% DMSO was dissolved in physiological saline.
Cell cultures
The human HCC cell lines HepG2, MHCC-97H, HLE, Huh7 and Hep3B, the human liver cell line L02, and the non-small-cell lung cancer cell line H1299 were originally obtained from the Cell Bank of Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). The HepG2, HLE and Huh7 cells were cultured in Dulbecco’s MEM (DMEM, Invitrogen Corporation, Carlsbad, CA, USA), the MHCC-97H, L02 and H1299 cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA), and the Hep3B cells were cultured in MEM (Invitrogen Corporation, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA), 100 U/mL penicillin and 100 μg/mL streptomycin. The cells were cultured in a humidified environment with 5% CO2 at 37 °C.
Patient samples
Two primary human hepatoma biopsy samples were obtained from patients at Jiangsu Cancer Hospital. Approval of the local ethical committees was given and informed consent was obtained from all patients prior to sample acquisition and experimentation. All patient data were used in an anonymized fashion according to the ethical guidelines.
Human hepatocellular carcinoma xenograft tumour models
Animals were cared for according to the Guide for the Care and Use of Laboratory Animals published by the National Institute of Health, USA. Athymic nude female mice and NOD/SCID female mice (4–6 weeks old) weighing 18–22 g were purchased from the Academy of Military Medical Sciences of the Chinese People’s Liberation Army (Certificate No. SCXK(ZHE)2018–0001). The mice were placed in IVC cages with adequate high-temperature sterilized water, sterile feed and regular light. The room temperature was controlled at 24–26 °C, and the humidity was 40%–60%.
The HepG2 cell line is utilized to create the cell Line Derived Xenograft (CDX) HepG2 xenograft mouse model. HepG2 cells (2 × 106) were subcutaneously (s.c.) injected into each experimental mouse. These tumour-bearing mice were grouped according to the tumour volume after 1 week.
The solid tumours from primary hepatoma sample #4 is utilized to create the patient-derived tumour xenograft (PDX) model. Fresh tumour fragments were transplanted subcutaneously (s.c.) into the left flank of anaesthetized NOD/SCID mice. Tumour growth was measured twice weekly using an electronic Vernier calliper. The established PDX model was called passage 1 (P1). When the tumour size of P1 reached approximately 750 mm3, the tumour was separated and sliced into small fragments (approximately 3 × 3 × 3 mm3/fragment) and re-inoculated into new mice as described above. Engrafted tumours were passaged for at least three generations (called P2, P3, and so on). These tumour-bearing mice were grouped according to the tumour volume.
Then, the corresponding drug was administered according to the specific conditions of each experiment. Tumour volume and weight were recorded twice a week using an electronic Vernier callipers. Tumour volume was calculated using the following formula: tumour volume (mm3) = d2 × D/2, in which d and D were the shortest and the longest diameters, respectively. At 3–4 weeks, the tumours were removed and the tumour weight was recorded. The animal medication doses were as follows: 5-FU was administered at a dose of 15 mg/kg to the HepG2 xenograft model and 22.5 mg/kg to the PDX model via intraperitoneal injection (i.p.) once every 2 days, and the mice were sacrificed at 26 and 15 days after treatment, respectively. PHA767491 was administered at a dose of 20 mg/kg via tail vein injection (i.v.) once per day. After 10 days, the treatment was stopped, and the mice continued to be fed and were sacrificed at the same time as the 5-FU administration group. OA was administered at 300 mg/kg via intragastric administration (i.g.) once every 2 days, and these mice were sacrificed after 22 days of treatment.
Western blot (WB) assay
Protein samples were isolated from cells treated with lysis buffer, eluted with SDS buffer, separated by SDS-polyacrylamide gels, and electroblotted onto PVDF membranes. The specific protein bands were stained with High-sig ECL Western Blotting Substrate (Tanon, Shanghai, China), and imaging was performed using an Amersham Imager 600 (GE Healthcare, Little Chalfont, UK). Primary antibodies included antibodies against β-Actin (ABclonal Technology, AC026, 1:200,000), POLR2A (ABclonal, A11181, 1:2000), phospho-POLR2A-S2 (ABclonal, AP0749, 1:2000), CDK9 (Cell Signaling Technology, C12F7, 1:2000), SIRT1 (CST, D739, 1:2000), phospho-MDM2(S166) (Abcam, ab170880, 1:20,000), phospho-SIRT1(S47) (Abcam, ab76039, 1:2000), acetyl-p53(K382) (Abcam, ab75754, 1:3000; this antibody can identify both acetyl-wt-p53 and acetyl-mut-p53, T-47D cells carried mutant p53 [33]), p53 (Proteintech, 10442–1-AP, 1:2000; this antibody can identify both wt-p53 and mut-p53; A431 cells carried mutant p53 [34]), MDM2 (Santa Cruz Biotechnology, SC-965, 1:1000, this antibody was developed against AA 154–167 in MDM2 of human origin and can identify both wt-MDM2 and mut-MDM2 (Ser166)), phospho-CDK Substrate Motif (CST, #9477, 1:2000), recombinant mut-p53 (Abcam, ab32049, 1:2000); phospho-p53(S33) (Abcam, ab75867, 1:2000); phospho-p53(S392) (Abcam, ab33889, 1:2000). Every assay was repeated three times.
Immunohistochemical (IHC) staining
Tumour tissue sections were heated, fixed, deparaffinized and rehydrated in a gradient of decreasing amounts of alcohol and, finally, distilled water. The slides were heated in citric acid buffer for antigen repair. Then, tissue section permeabilization was performed with 0.5% Triton-X/PBS. The slides were treated with 5% goat serum and 0.5% BSA at room temperature for 1 h and incubated overnight with primary antibodies at 4 °C. The primary antibodies included antibodies against CDK9 (CST, C12F7, 1:400), phospho-MDM2(S166) (Abcam, ab170880, 1:500), phospho-SIRT1(S47) (Abcam, ab76039, 1:400), p53 (Proteintech, 10442–1-AP, 1:400), and acetyl-p53(K382) (Abcam, ab75754, 1:200). Finally, the sections were stained with DAB dye, sealed with neutral resin, observed under a microscope and imaged.
Quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR) assay
Total RNA was extracted using a total RNA extraction reagent (Vazyme Biotech, Nanjing, China, R401–01) and then amplified by polymerase chain reaction (PCR). An aliquot of 1 μg of total RNA was used to transcribe first-strand cDNA with a HiScript III 1st Strand cDNA synthesis kit (Vazyme Biotech, R312–01/02). Real-time PCR was completed on an ABI PRISM Sequence Detector 7500 (PerkinElmer, Branchburg, NJ) using Sequence Detector version 1.7 software (Applied Biosystems, Foster City, CA, USA). SYBR Green PCR Master Mix was purchased from Vazyme Biotech (Q131–02/03). The following primer sets were used in the PCR amplification:
p53-sense: 5ʹ-CAGGTAGCTGCTGGGCTC-3ʹ
p53-antisense: 5ʹ-GCTCGACGCTAGGATCTGAC-3ʹ
GAPDH -sense: 5ʹ-TAGTGGAAGGACTCATGACC-3ʹ
GAPDH-antisense: 5ʹ-TCCACCACCCTGTTGCTGTA-3ʹ
P53 gene expression was analysed using quantitative RT-PCR with GAPDH as an internal control.
Immunoprecipitation (IP)
Cell lysates were incubated overnight with the corresponding antibodies at 4 °C with gentle rotation. The primary antibodies included antibodies against CDK9 and SIRT1 (CST, 1:250), MDM2 (Santa, 1:50), and p53 (Proteintech, 1:250). The next day, immunogen-antibody complexes were captured by incubation for 1 h with protein A/G magnetic beads (MedChemExpress, Shanghai, China) at 4 °C with gentle rotation. Then, beads were washed three times with wash buffer. The immunocomplexes were analysed by WB.
Small interfering RNA (siRNAs), plasmid extraction and transient transfection
The CDK9 plasmid was obtained from Addgene (Cambridge, MA, Plasmid #14640). An EndoFree Plasmid Midi kit (CWBIO, Beijing, China, CW2015S) was used to extract plasmids, which were stored at −80 °C. CDK9 siRNA was designed and provided by GenePharma (Shanghai, China). The following siRNA sequences were used:
siRNA-CDK9–315 sense: 5ʹ-GGGAGAUCAAGAUCCUUCATT-3ʹ
siRNA-CDK9–315 antisense: 5ʹ-UGAAGGAUCUUGAUCUCCCTT-3ʹ
siRNA-CDK9–881 sense: 5ʹ-GGCCAAACGUGGACAACUATT-3ʹ
siRNA-CDK9–881 antisense: 5ʹ-UAGUUGUCCACGUUUGGCCTT-3ʹ
For transfection, cells were plated in 6-well plates at 60% density and allowed to attach overnight. One hour before transfection, the medium was replaced with fresh medium. Then, the CDK9 plasmids (3 μg per well) or siRNA (400 pmol per well) was introduced into the cells with Lipofectamine 2000 (Invitrogen, 11668019) according to the manufacturer’s instructions. All steps were performed using antibiotic-free medium.
Primary human liver cancer cells
Tumour biopsy samples were dissected, minced, and digested with 1 mg/mL collagenase I (Invitrogen Life Technologies, San Giuliano Milanese, Milan, Italy) and 1 mg/mL dispase at 37 °C overnight with intermittent shaking. Then, the cells were washed twice with PBS and filtered through a 40 μm filter to generate primary and stabilized cell lines. The cells were cultured in DMEM supplemented with 2 mM glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin and 20% heat-inactivated FBS (Invitrogen Life Technologies) at 37 °C in a humidified atmosphere with 5% CO2. The medium was changed twice per week. Samples capable of growing after only 1–8 culture passages were considered to be primary cell lines. Primary human liver cells were purchased from iCell (iCell Bioscience Inc., Shanghai, China).
Docking study
A molecular docking study was performed using the Discovery Studio 3.0/CDOCKER protocol. The crystal structure of human CDK9 in complex with ATP was available in the Protein Databank, PDB ID: 3BLQ. The structure of 3BLQ was treated by the following procedures: The duplicate B chain was removed, water was removed, and the protein was prepared in DS 3.0 software, the docking sphere was generated using the Define Site with the ATP-binding site. Then, the radius of the SphereObject Attributes was adjusted to 9.0 Å with the other parameters set to default. The OA structure was drawn by ChemBioDraw Ultra 14.0 software and then saved in MOL format. Then, a ligand preparation was introduced to the OA structure, and minimization models in Discovery Studio 3.0 were used to investigate the spatial binding pattern of OA with CDK9. The docking was performed using the CDOCKER protocol with prepared 3BLQ set as the input receptor. In addition, the prepared OA structure was set as the input ligand, and the other parameter values were set to default. The docking result showing the conformation with the lowest docking energy was selected for further analysis. By using the View Interactions module in DS 3.0, the binding mode of OA in complex with CDK9 was obtained: the benzene ring formed hydrophobic interactions with LEU156 and HIS108, and the 6O and 7O atoms of the chromene derivative ring formed hydrogen bonds and electrostatic interactions with LYS48. The proteins in binding mode were visualized by PyMOL.
Cellular thermal shift assay (CETSA)
To determine the direct binding between OA and CDK9 protein in HepG2 cells, we performed a CETSA [35]. First, we treated HepG2 cells with 50 μM OA for 3 h, harvested the cells, resuspended them in phosphate-buffered saline (PBS) containing protease inhibitor cocktail and transferred them to 200 µL tubes. The cells were heat shocked in a T960 thermal cycler (Heal Force, Hangzhou, China) at 37.0–62.3 °C for 3 min to denature the proteins, and then, all the samples were subjected to three freeze-thaw cycles with liquid nitrogen to lyse cells. The samples were centrifuged at 14,000 rpm for 20 min at 4 °C. Loading buffer was added, and the proteins were analysed by WB.
In vitro kinase assays
To determine the effect of OA on CDK9 activity, the cells were examined after incubation with recombinant CDK9/cyclinT (ProQinase) GmbH, Freiburg, Germany) and the recombinant substrate RBERCHKtide (ProQinase) in the presence of different concentrations of OA in 60 mM HEPES-NaOH (pH 7.5), 3 mM MgCl2, 3 mM MnCl2, 3 μM Na-orthovanadate, 1.2 mM DTT, 50 μg/mL PEG20000 and 1 μM ATP for 2 h at 30 °C. The amounts of phosphorylated substrates were determined by WB using specific antibodies against the phosphorylated substrates.
Statistical evaluation
The data are presented as mean ± SD from triplicate parallel experiments unless otherwise indicated. Statistical analyses were performed using one-way ANOVA.
Results
CDK9 is a potential target for HCC therapy
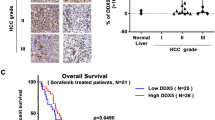
In previous studies, researchers demonstrated that inhibition of CDK9 leads to reduced RNA synthesis, resulting in the apoptosis of leukaemia cells [36]. However, the role of CDK9 in HCC has rarely been studied. Here, we first investigated the clinical relevance of CDK9 RNA expression in liver cancer. We used clinically annotated RNA data from The Cancer Genome Atlas (TCGA) database. The clinicopathological and platform information of these samples is provided in the supporting information (Supplementary Table 1). The rates of liver cancer in men are typically 2- to 4-fold higher than those in women [37]. Quantitative analysis of Kaplan-Meier overall survival curves of male liver cancer cases with low and high CDK9 RNA expression (n = 246) showed a significant correlation between high CDK9 RNA expression and poor overall survival (Fig. 1a). To determine the role of CDK9 protein expression in liver cancer, we assessed the expression of CDK9 protein in a liver cancer tissue microarray consisting of 32 HCC patient tissues and relevant paracancerous tissues by IHC analysis (Fig. 1b). According to the level of CDK9 expression, we divided the tissue samples into four levels and scored the CDK9 expression in each sample (Supplementary Table 2). We found that CDK9 was expressed at low levels (negative and level 1 groups) in 62.5% of the paracancerous tissues (n = 10/16) and in 25.0% of the HCC tissues (n = 4/16). In addition, 75.0% of the HCC tissues (n = 12/16) showed high CDK9 expression (level 2 and 3 groups). Then, we sought to determine the expression of CDK9 in normal versus liver cancer cell lines by immunoblotting. As shown in Fig. 1c, the human hepatoma cell lines HepG2, MHCC-97H and Huh7 showed high CDK9 protein expression; in contrast, the human hepatoma cell line HLE and the human liver cell line L02 showed much lower CDK9 protein expression. Thus, CDK9 was overexpressed in most HCC cell lines and human HCC tissues, and its overexpression was correlated with poor survival in patients with HCC.
a Kaplan-Meier overall survival curves in male liver cancer cases with CDK9 RNA low and high expression (FPKM expression data were used) obtained from the TCGA database (downloaded from https://www.proteinatlas.org). The patient survival proportion is plotted versus time since diagnosis in months. In analyses of Kaplan-Meier curves with a log-rank test, a P value < 0.05 was considered significant. b Immunohistochemistry (IHC) staining for the CDK9 protein in a tissue microarray of HCC samples. Representative images of CDK9 IHC staining of HCC tissues and paracancerous liver tissues showing negative, weak (1), moderate (2) and high (3) intensity stain. The results of the tissue microarray were quantified. c Whole-cell lysates were prepared and subjected to WB assay for CDK9 in human hepatoma cell lines and a normal liver cell line. d Survival rates of human hepatoma cell lines treated with LDC067 for 24 h were measured by MTT assay. e Primary human hepatoma cells and hepatocytes were subjected to WB assay to measure CDK9 levels. f Survival rates of primary human hepatoma cells treated with LDC067 for 24 h were measured by MTT assay. “None” indicated that the absolute IC50 cannot be calculated because the inhibitory effect of LDC067 failed to reach 50% in the concentration range of MTT assay. g, h HepG2 cells were transfected with CDK9 siRNA-315. Cell proliferation rates were measured by MTT assay. Bar, SD. *P < 0.05 or **P < 0.01 versus the untreated control.
To investigate the functional significance of CDK9 on the growth of HCC, HepG2 cells with high CDK9 expression and HLE cells with low CDK9 expression were treated with the CDK9-specific inhibitor LDC067. The results showed that LDC067 significantly inhibited the survival of the HepG2 cells and showed a lesser effect on the HLE cells (Fig. 1d). Thus, HepG2 cells were used for our subsequent studies. The influence of CDK9 on the growth of human primary hepatoma cells was also investigated. LDC067 showed a strong growth inhibitory effect on primary hepatoma cells from a hepatoma sample with high CDK9 expression (#4) but little effect on cells from a hepatoma sample with low CDK9 expression (#2) (Fig. 1e, f). After HepG2 cells were transfected with CDK9-targeted siRNAs (siCDK9–315 and siCDK9–881), cell growth was suppressed (Fig. 1g, h and Supplementary Fig. 1a, b).
In further studies, we constructed a HepG2 cell xenograft model and HCC PDX models and investigated the influence of CDK9 on tumour growth in vivo. Because the toxicity and lethality of LDC067 in vivo is particularly high (the early lethality was greater than 50%), we used another CDK9 inhibitor, PHA767491, for the in vivo studies. PHA767491 inhibited the growth of HepG2 cells in vitro but showed a weaker effect than LDC067 (Supplementary Fig. 2a). As shown in Fig. 2a–f, PHA767491 and the positive control drug 5-FU had obvious inhibitory effects on the proliferation of xenograft tumours. The tumour growth inhibition rates of PHA767491 in the HepG2 xenograft model and PDX model were 41.4% and 38.12%, respectively, which were equivalent to the effect of 5-FU. These results demonstrated that CDK9 is a potential anti-hepatoma target. In a disappointing result, PHA767491 decreased the mouse weight during administration and led to greater mortality than 5-FU (Fig. 2g, h). Therefore, CDK9 is a potential target for HCC therapy, but more CDK9 inhibitors with lower toxicity need to be explored.
HepG2 cell xenograft mouse models and the human hepatoma sample (#4) PDX mouse models were treated with physiological saline, PHA767491 and 5-FU. The HepG2 xenograft model mice were sacrificed 26 days after drug administration, the PDX model mice were sacrificed 15 days after drug administration, and the tumour tissues were analysed by IHC. a–c Tumour volumes (a), inhibition rates of tumour weight (b) and a tumour picture of (c) HepG2 xenograft model mice. d–f The tumour volumes (d), inhibition rates of tumour weight (e) and a tumour picture (f) of PDX model mice. g, h Changes in mouse weight (g) and survival rates (h) of the HepG2 xenograft model mice. Bar, SD. *P < 0.05 or **P < 0.01 versus the untreated control.
Controlled inhibition of CDK9 suppressed MDM2-mediated wt-p53 degradation
Previous studies have demonstrated that restoration of wt-p53 function can reverse the tumour growth of human cancers with a low frequency of p53 mutations, such as HCC. Developing molecules that reactivate the wt-p53 pathway is a paramount challenge in human cancers [38]. Thus, to investigate whether the inhibition of CDK9 disrupted cell growth by modulating wt-p53, we first assayed the influence of CDK9 on the growth of p53-null Hep3B cells (an HCC cell line) and H1299 cells (a human non-small-cell lung cancer cell line) [39, 40]. The results showed that the inhibitory effect of the CDK9 inhibitor LDC067 on the p53-null Hep3B and H1299 cells was weaker than that on the HepG2 cells with wt-p53 (Fig. 3a, b and Supplementary Fig. 3a, b). Interestingly, the protein expression of CDK9 in the Hep3B cells was lower than that in the HepG2 cells; in contrast, the H1299 cells showed CDK9 levels similar to those of the HepG2 cells. In addition, the HCC cell line HLE expressed mutant p53 (p53 codon 249 position 3G → C) [39]. We also identified the status of p53 in the HLE cells using an antibody specific to mutant p53 (Supplementary Fig. 3c). The HLE cells showed low CDK9 protein expression and hyposensitivity to LDC067 (Fig. 1c, d). Therefore, wt-p53 might be critical to the growth inhibitory effect of CDK9 inhibitors, but there was no regulatory genetic association between CDK9 and p53. Then, we explored the influences of CDK9 on wt-p53 expression in HepG2 cells. Strangely, the protein level of wt-p53 was significantly increased with low concentrations of LDC067 but gradually decreased with increasing LDC067 concentration (Fig. 3c). This, to some degree, explained our confusion over the results showing that selective CDK9 inhibitors, such as LDC067 and PHA767491, led to strong extracellular inhibitory effects on CDK9 activity but did not inhibit the high growth rate of HepG2 cells, especially at high concentrations. It has been reported that transactivation by p53 prevails only over generic repression caused by incomplete CDK9 blockade [41]. Thus, only controlled inhibition of CDK9 had the potential to recover wt-p53 function, and therefore, we chose to use lower concentrations of LDC067 (<20 μM) in subsequent experiments.
a Whole-cell lysates of HepG2 and Hep3B cells were prepared and subjected to WB assay for wt-p53 and CDK9 level assessment. b Survival rates of the HepG2 and Hep3B cells treated with LDC067 for 24 h. “None” indicated that the absolute IC50 cannot be calculated because the inhibitory effect of LDC067 failed to reach 50% in the concentration range of MTT assay. c WB assay was performed to assess the wt-p53 level in HepG2 cells treated with LDC067 for 24 h. d–f WB assays were performed to assess the POLR2A, p-POLR2A-S2, wt-p53 and CDK9 levels in HepG2 cells treated with LDC067 (9, 12 and 15 μM) for 24 h (d), in HepG2 cells treated with LDC067 (15 μM) for different times (e) and in HepG2 cells transfected with CDK9 siRNA-315 (f). g Quantitative RT-PCR was performed to detect p53 mRNA expression after HepG2 cells were treated with LDC067 for 24 h. Bar, SD. **P < 0.01 versus the untreated control.
RNAPII is the specific substrate of CDK9, which phosphorylates the Ser2 site in the C-terminal repeat domain (CTD) of RNAPII [42]. The activity of CDK9 is demonstrated by the level of Ser2 phosphorylation of RNA polymerase II subunit A (POLR2A). Upon treatment with lower concentrations of LDC067, the protein level of phospho-POLR2A-Ser2 (p-POLR2A-S2) was decreased in a concentration- and time-dependent manner, suggesting that CDK9 activity was suppressed (Fig. 3d). The wt-p53 protein level was upregulated by LDC067 exposure in a concentration-dependent manner (Fig. 3e). Knockdown of CDK9 expression by siRNAs (siCDK9–315 and siCDK9–881) in HepG2 cells decreased CDK9 activity and increased wt-p53 protein levels (Fig. 3f and Supplementary Fig. 1c). In further studies, a qRT-PCR analysis revealed that the trend of p53 mRNA levels also increased early but decreased later with increasing LDC067 concentration (Fig. 3g). These findings further supported the speculation that high concentrations of LDC067 cause high general transcription inhibition via the suppression of CDK9-regulated RNAPII. The opposite trend between CDK9 protein and mRNA levels upon treatment with 15 μM LDC067 indicated that CDK9 modulates the stability of the wt-p53 protein.
Mouse double minute 2 homologue (MDM2) is an E3 ubiquitin ligase that mediates the ubiquitination degradation of wt-p53 [43]. Inhibition of CDK9 by treatment of HepG2 cells with low concentrations (<20 μM) of LDC067 reduced the level of phospho-MDM2 (p-MDM2 Ser166) but increased the protein level of MDM2 (Fig. 4a). Upon exposure to high concentrations (>20 μM) of LDC067, the expression of the MDM2 protein was decreased gradually, similar to wt-p53 expression (Supplementary Fig. 4a). We speculated that the upregulation of MDM2 induced by exposure to low concentrations of LDC067 might have been the result of feedback regulation of p53 because the upregulation of wt-p53 induced by LDC067 was evident before changes to the MDM2 level were observed (Supplementary Fig. 4b). Knockdown of CDK9 also decreased the level of p-MDM2 (Ser166) (Fig. 4b). Thus, inhibition of CDK9 by exposure to lower concentrations of LDC067 suppressed the phosphorylation of MDM2 at Ser166 to promote wt-p53 stability. The reduction in p-MDM2 (Ser166) level and the increase in wt-p53 expression induced by 12 μM LDC067 in HepG2 cells were weakened by CDK9 overexpression (Fig. 4c). In human primary hepatoma cells (#4), 12 μM LDC067 also decreased the p-MDM2 (Ser166) and upregulated the wt-p53 protein levels (Fig. 4d). At the same concentration, the influences of PHA767491 on p-POLR2A-S2, p-MDM2 (Ser166) and wt-p53 in HepG2 cells were the same as those of LDC067 (Supplementary Fig. 2b). Observations of tumour tissues after IHC staining also showed that the CDK9 expression was changed a little in the PHA767491-treated group; however, p-MDM (Ser166) expression was significantly decreased and wt-p53 expression was increased (Fig. 4e, f).
a, b WB assays were performed to assess the p-MDM2 (Ser166) and MDM2 levels in HepG2 cells treated with LDC067 for 24 h (a) or in HepG2 cells transfected with CDK9 siRNA-315 (b). c HepG2 cells were transfected with a HA-labelled CDK9 plasmid and then treated with 12 μM LDC067 for 24 h. WB assays were performed to assess the p-POLR2A-S2, p-MDM2 (Ser166), wt-p53 and CDK9 levels. d Human primary hepatocarcinoma #4 cells were treated with LDC067 (12 μM) for 24 h. The expression of CDK9, wt-p53, p-POLR2A-S2 and p-MDM2 (Ser166) in the #4 cells was analysed by WB. e, f Xenograft models with HepG2 cells (e) and PDX models with human hepatoma samples (#4) (f) treated with physiological saline control or PHA767491. IHC detection of CDK9, wt-p53 and p-MDM2 (Ser166) in tumour tissues (400×; scale bar, 50 μm). g HepG2 cells were treated with 12 μM LDC067 for 24 h. The binding of CDK9 with MDM2 was assayed by immunoprecipitation (IP). h HepG2 cells were transfected with a wt-MDM2 plasmid. Cell lysates were incubated with MDM2-conjugated beads for IP. Immunoprecipitates and whole-cell lysates were assayed by WB. The MDM2 signal was visualized with an anti-p-CDK substrate motif antibody. β-Actin was used as the loading control. i CDK9-overexpressing HepG2 cells were transfected with wt-MDM2 and mut-MDM2 (Ser166) plasmids. Cell lysates were incubated with CDK9-conjugated beads for IP assay. Immunoprecipitates and whole-cell lysates were assayed by IB. j HepG2 cells were treated with 12 μM LDC067 for 24 h. The binding of wt-p53 with MDM2 was assayed by IP.
Then, we further investigated the modulation of the phosphorylation of MDM2 as induced by CDK9. The IP results showed that a low concentration (12 μM) of LDC067 inhibited the interaction between CDK9 and MDM2 (Fig. 4g). To determine whether MDM2 was the substrate of CDK9, a specific antibody against the key phospho-CDK substrate motif was used. As shown in Fig. 4h, MDM2 contains a consensus amino acid sequence for the phospho-CDK9 substrate. Since LDC067 decreased the level of p-MDM2 (Ser166) expression, we mutated the amino acid at site 166 from serine to alanine in MDM2 and transfected HepG2 cells expressing high levels of CDK9 with the wt-MDM2 plasmid and Ser166-mutated MDM2 plasmid. The results showed that the mutation at Ser166 of MDM2 disrupted the binding of MDM2 with CDK9 (Fig. 4i). Thus, MDM2 is the substrate of CDK9 and is phosphorylated by CDK9 at the Ser166 site. LDC067 (12 μM) decreased the phosphorylation of MDM2 at Ser166, interfering with the interaction between MDM2 and wt-p53 (Fig. 4j). In conclusion, lower concentrations of LDC067 reduced wt-p53 degradation by preventing the phosphorylation of MDM2 at Ser166 and interfering with the interaction between MDM2 and wt-p53.
Lower concentrations of CDK9 inhibitor increased the acetylation of wt-p53 by preventing the phosphorylation of SIRT1 at the Ser47 site
The acetylation of p53 is also critical for regulating the stability of wt-p53. IP assays showed that the NAD-dependent deacetylase SIRT1 also contains a consensus amino acid sequence for binding the phospho-CDK substrate (Fig. 5a), and inhibition of CDK9 expression by exposure to a lower concentration (12 μM) of LDC067 disrupted the binding of SIRT1 with CDK9 (Fig. 5b). Phosphorylation of SIRT1 at Ser47 had been previously shown to activate the SIRT1 deacetylase [44]. In the present study, IP assays showed that the mutation at Ser47 of SIRT1 disrupted the interaction between the CDK9 and SIRT1 proteins (Fig. 5c). Therefore, we concluded that SIRT1 is a substrate of CDK9 and that it can be phosphorylated at the Ser47 site by CDK9.
a HepG2 cells were transfected with wt-SIRT1 plasmid. Cell lysates were incubated with SIRT1-conjugated beads for IP assay. Immunoprecipitates and whole-cell lysates were assessed by WB. The SIRT1 signal was visualized with an anti-p-CDK substrate motif antibody. β-Actin was used as the loading control. b HepG2 cells were treated with LDC067 for 24 h, and the lysates were immunoprecipitated with anti-CDK9 antibody. WB assay was performed for assessing CDK9 and SIRT1 levels. c CDK9-overexpressing HepG2 cells were transfected with wt-SIRT1 and mut-SIRT1 (Ser47) plasmids in combination with a CDK9 plasmid, and immunoprecipitates and whole-cell lysates were assessed by WB. The CDK9 plasmid was HA-labelled. d WB assays were performed to assess the SIRT1, p-SIRT1 (Ser47) and Ac-p53 (Lys382) levels in HepG2 cells treated with LDC067 for 24 h. e IP assay showing the interaction between p53 and SIRT1 in HepG2 cells treated with 12 μM LDC067 for 24 h, and whole-cell lysates were subjected to WB assay. f WB assays were performed to determine the p-SIRT1 (Ser47), Ac-p53 (Lys382) and CDK9 levels in HepG2 cells transfected with CDK9 siRNA-315. g HepG2 cells were transfected with a HA-labelled CDK9 plasmid and then treated with 12 µM LDC067 for 24 h. WB analysis was performed for assessing p-SIRT1, Ac-p53 and CDK9 levels. h Human primary hepatocarcinoma (#4) cells were treated with LDC067 (12 μM) for 24 h. The expression of p-SIRT1 and Ac-p53 in the #4 cells was analysed by WB. i, j Protein expression of p-SIRT1 and Ac-p53 in tumour tissues obtained from HepG2 cell xenograft model mice (i) and PDX model mice (j) as detected by IHC (400×; scale bar, 50 μm). The tumour tissue samples used for IHC assay of (i, j) were obtained from the same samples for those shown in Fig. 4e, f, respectively.
P53 interacts directly with SIRT1 and is deacetylated at carboxy-terminal lysine 382, resulting in instability and attenuated antiproliferative effects [45, 46]. Inhibition of CDK9 in HepG2 cells treated with lower concentrations of LDC067 reduced phospho-SIRT1 (p-SIRT1 Ser47) levels, upregulated acetylated p53 (Ac-p53 Lys382) levels (Fig. 5d) and suppressed the binding between SIRT1 and wt-p53 (Fig. 5e). Knockdown of CDK9 also decreased the phosphorylation of SIRT1 and deacetylation of wt-p53 (Fig. 5f and Supplementary Fig. 1c). The effects of 12 µM LDC067 were attenuated by the overexpression of CDK9 (Fig. 5g). In human primary hepatoma cells (#4), 12 μM LDC067 decreased p-SIRT1 (Ser47) levels and increased Ac-p53 (Lys382) levels (Fig. 5h). The influences of 9 μM PHA767491 on p-SIRT1 (Ser47) and Ac-p53 (Lys382) were the same as those on LDC067 (Supplementary Fig. 2b). After the mice were treated with the CDK9 inhibitor PHA767491, the changes in the protein levels of p-SIRT1 (Ser47) and Ac-p53 (Lys382) in xenograft tumour tissue showed changes consistent with those obtained in vitro (Fig. 5i, j).
These results indicated that the inhibition induced by treatments with lower concentrations of CDK9 inhibitor downregulated the phosphorylation and activity of SIRT1, enhancing the acetylation and stability of wt-p53 in HCC cells.
Oroxylin A is a novel CDK9 inhibitor with potent anti-hepatoma activity
Naturally occurring and synthesized flavonoids have been identified as protein kinase (PK) inhibitors, targets associated with many of the processes related to cancer, diabetes and osteoporosis [47,48,49]. Flavopiridol, a semisynthetic flavonoid derivative from rohitukine, was the first CDK inhibitor to enter clinical trials [50]. Despite the promising preclinical efficacy of flavopiridol against various tumour types, the results of early clinical trials were disappointing [50, 51]. OA is a natural flavonoid obtained from S. baicalensis and has potent anti-hepatoma activity [28]. Cell-free kinase activity assay results showed that OA inhibited CDK9 activity in a concentration-dependent manner (Fig. 6a). Compared with the highly specific inhibitor LDC067 with an IC50 value of 44 ± 10 nM for CDK9 extracellular activity [52], the EC50 value of OA for CDK9 was 8.022 ± 1.00 μM. This finding suggested that CDK9 inhibition by OA was more moderate than that of LDC067. A CETSA was performed to study the thermal stabilization of proteins upon ligand binding. We applied this method to further confirm the interaction between OA and CDK9. As shown in Fig. 6b, the thermal stabilization of CDK9 protein in the OA treatment group of in HepG2 cells was stronger than that in the negative control group. The binding between OA and CDK9 was also assayed through a molecular docking study. As shown in Fig. 6c, at the lowest energy conformation, OA could bind with Lys48 of CDK9, forming strong hydrogen bonding (green) as well as salt bridges and ion interactions (orange). In addition, OA formed intramolecular hydrogen bonds (green) and hydrogen bonds with CDK9 Leu156 (pink). In HepG2 cells, OA reduced the level of p-POLR2A-S2 in a time- and concentration-dependent manner (Fig. 6d, e). In contrast to LDC067, OA decreased CDK9 protein levels, suggesting that OA might have a CDK9-independent regulatory mechanism. OA showed a remarkable in vitro growth inhibitory effect on HepG2 cells with high CDK9 expression and wt-p53, and the effects of high concentrations of OA was much better than those of the CDK9 inhibitor LDC067 (Fig. 6f). In p53-mut HLE cells and p53-null Hep3B cells, OA showed a weak growth inhibitory effect (Fig. 6g). After CDK9 was overexpressed in HepG2 cells, the growth inhibitory effect of OA was reduced (Fig. 6h). Thus, the growth inhibitory effect of OA on HCC cells was associated with the recovery of wt-p53 function and suppression of CDK9. In the HepG2 cell xenograft model, the tumour growth inhibitory effect of OA (300 mg/kg, i.g.) was similar to the effect of the CDK9-specific inhibitor PHA767491 (20 mg/kg, i.v.) (Fig. 6i, j and Supplementary Fig. 5a–c). However, OA had no significant effect on the body weight or survival rate of mice (Supplementary Fig. 5d, e). OA also inhibited the growth of human primary hepatoma cells and xenografts in PDX models (Supplementary Fig. 6a, c–e). Overall, the novel CDK9 inhibitor OA showed potent anti-hepatoma activity.
a CDK9 was incubated with substrate peptide and [33P]-ATP in the presence of different concentrations of OA. The kinase activity is described as % of [33P]-phosphorylated substrate peptide. The EC50 values are indicated. b HepG2 cells were treated with OA (50 µM) for 3 h. The temperatures were increased from 37.0 to 62.3 °C during the CETSA assay directed towards CDK9. The results from the quantitative analysis of CDK9 protein expression are presented as mean ± SD of three independent experiments. c Computer docking simulation of the crystal structure of human CDK9 in complex with OA. d, e WB assays were performed to assess the POLR2A, p-POLR2A-S2 and CDK9 levels in HepG2 cells treated with 15 μM OA for different times (d) and HepG2 cells treated with OA for 48 h (e). f Survival rates of the HepG2 cells treated with LDC067 (24 h) and OA (48 h). g Survival rates of different hepatoma cell lines treated with OA for 48 h. h Survival rates of CDK9-overexpressing HepG2 cells treated with OA for 48 h. “None” indicated that the absolute IC50 cannot be calculated because the inhibitory effect of OA failed to reach 50% in the concentration range of MTT assay. i, j Tumour volumes and weights of the HepG2 xenograft model mice treated with physiological saline, PHA767491 (20 mg/kg, i.v.) or OA (300 mg/kg, i.g.). The in vivo experiments based on OA treatment were performed in parallel with those based on PHA767491 and 5-FU treatments, as shown in Fig. 2b. Bar, SD. **P < 0.01 versus the untreated control.
Oroxylin A inhibited HCC and enhanced the stability of wt-p53 by suppressing the CDK9-regulated function of MDM2 and SIRT1
In our previous studies, we showed that recovery of wt-p53 function via negative regulation of MDM2 transcription is important for the anticancer activity of OA [53]. Here, we found that OA significantly decreased p-MDM2 (Ser166) and p-SIRT1 (Ser47) levels, increased wt-p53 protein levels and acetylated wt-p53 at Lys382 in HCC cells (Fig. 7a and Supplementary Fig. 6b). Moreover, OA led to MDM2 expression maintained at a low level by other CDK9-independent mechanisms. The p53 mRNA level was slightly downregulated (Fig. 7b), suggesting that OA restored wt-p53 function by regulating its protein stability. The interaction of wt-p53 with SIRT1 and MDM2 in HepG2 cells was interrupted by OA treatment (Fig. 7c, d). OA also inhibited the binding of CDK9 with SIRT1 and MDM2 (Fig. 7e, f). The overexpression of CDK9 in HepG2 cells weakened the regulatory effects of OA on wt-p53 stability (Fig. 7g), suggesting that the recovery of wt-p53 by OA was closely associated with its inhibition of CDK9. Consistent with the results in vitro, IHC experiments showed that CDK9, p-SIRT1 (Ser47) and p-MDM2 (Ser166) protein expression was decreased by OA treatment, and the levels of Ac-p53 (Lys382) and wt-p53 protein were significantly increased in the tumour tissues of the OA-administered group (Fig. 7h and Supplementary Fig. 6f). Therefore, OA was shown to be a potential candidate for hepatoma treatment by inhibiting CDK9-regulated MDM2 and SIRT1 signaling and enhancing the stability of wt-p53.
a WB detection of CDK9, wt-p53 and related protein levels in HepG2 cells treated with OA for 48 h. b Quantitative RT-PCR was performed to detect the p53 mRNA expression in HepG2 cells treated with OA for 48 h. c, d HepG2 cells treated with 12 μM OA for 48 h were immunoprecipitated against wt-p53. Then, WB assays were performed to assess the MDM2, SIRT1 and wt-p53 levels. e, f HepG2 cells treated with 12 μM OA for 48 h were immunoprecipitated with anti-CDK9 and anti-MDM2 antibodies. Then, WB assays were performed to assess the SIRT1, MDM2 and CDK9 levels. g WB assays were performed to assess the CDK9, wt-p53 and related protein levels in CDK9-overexpressing HepG2 cells treated with 12 µM OA for 48 h. h Images of IHC-stained CDK9, wt-p53 and related proteins (400×; scale bar, 50 μm). The in vivo experiments based on OA treatment were performed in parallel with those based on PHA767491 and 5-FU treatments, as shown in Fig. 2b. The tumour tissue slices of “Control” group used for IHC assay of (h) were the same one for that shown in Fig. 4e, 5i.
Discussion
HCC is a highly heterogeneous disease at both the clinical and molecular levels. Currently, sorafenib is the only approved standard first-line systemic therapy for advanced HCC, but the therapeutic effect has failed to prolong patient survival. Regorafenib has shown a survival benefit in HCC patients as a second-line therapy in combination with prior sorafenib treatment. However, there was no obvious therapeutic effect for many patients in phase III clinical trials [54]. The proposed reason for this outcome is the molecular heterogeneity of HCC aetiology.
CDK9 is required for the efficient expression of most genes, except certain short genes with no introns, and at first glance, it did not appear to be an ideal target for any disease [55]. Previous studies have shown that transient inhibition of RNAPII phosphorylation at similar levels in both transformed and non-transformed cells induced caspase-dependent apoptosis only for transformed cells because, in contrast to non-transformed cells, transformed cells are addicted to the sustained expression of antiapoptotic proteins [56]. Thus, inhibition of CDK9 has an ideal therapeutic effect on tumour tissues and a certain safety level in normal tissues. In recent years, some CDK9 inhibitors have been developed for cancer treatment and entered clinical trials mainly as leukaemia therapeutics, but has rarely been studied as solid tumour treatments. In this work, we found a novel CDK9 inhibitor obtained from S. baicalensis named OA and showed that OA had potent anti-hepatoma activity by inhibiting CDK9-regulated MDM2 and SIRT1 signaling and enhancing wt-p53 stability (Fig. 8). Both MDM2 and SIRT1 were found to be substrates of phospho-CDK9. Inhibition of CDK9 by OA suppressed the phosphorylation of MDM2 at Ser166 and SIRT1 at Ser47, leading to the suppression of the ubiquitination and deacetylation of p53 and resulting in the recovery of wt-p53 function. Importantly, OA inhibited CDK9 moderately and controllably and did not induce strong repression generally, in contrast to the effect of high concentrations of selective CDK9 inhibitors. Thus, the anticancer effect of OA is more stable and concentration-dependent, and the toxicity to normal tissue is low.
CDK9 is highly expressed in HCC. OA shows potent anti-hepatoma activity by inhibiting the activity and expression of CDK9 and enhancing p53 stability. Inhibition of CDK9 by OA led to dephosphorylation of MDM2 and SIRT1 and stabilization of the p53 protein by suppressing MDM2-mediated ubiquitination-triggered degradation and SIRT1-mediated deacetylation of p53. Thus, p53-mediated promotion of apoptosis, inhibition of DNA repair, metabolic disorders and so on suppressed HCC. Importantly, OA inhibited CDK9 moderately and controllably and did not induce strong general transcription repression, in contrast to high concentrations of selective CDK9 inhibitors, such as LDC067. Thus, the novel CDK9 inhibitor OA shows great and concentration-dependent anticancer effects.
P53 is a key regulatory protein in cell growth, survival and metabolism [57]. Exploration of the mechanism for the restoration of wt-p53 in HCC is of great significance, because the frequency of the p53 mutation in HCC is much lower than in many other types of cancer [19]. Previous studies have reported some inconsistent connections between the CDK9 and p53 proteins in different cells or tissues. CDK9 not only phosphorylates p53 through physical interaction [20] but also prevents degradation of the p53 protein in CNS-derived cells by phosphorylating the Pirh2 protein [22]. In addition, activation of p53 reduced CDK9 recruitment [58]. We also investigated the effects of the CDK9 inhibitors LDC067 and OA on the phosphorylation of wt-p53 in HCC and found that LDC067 and OA did not downregulate the phosphorylation of wt-p53 at Ser33 or Ser392 but increased the phosphorylation of these proteins (Supplementary Fig. 7). This result suggested that the phosphorylation of wt-p53 at Ser33 and Ser392 in HCC was not CDK9-dependent. It has been reported that casein kinase 2 (CK2) also phosphorylates wt-p53 at Ser392 [59]. Thus, OA not only enhanced wt-p53 stability in a CDK9-dependent manner but also increased wt-p53 activity in a CDK9-independent manner in HCC. Our previous studies showed that OA also stabilized wt-p53 by promoting sirtuin3-mediated deacetylation of PTEN (phosphatase and tensin homologue) and PTEN-mediated negative regulation of MDM2 transcription [53]. Akt has been reported to be a negative regulator of p-MDM2 Ser166 [60]. Both the CDK9 inhibitors LDC067 and OA had previously been shown to attenuate the activation of Akt signaling pathways [61, 62]. Therefore, OA might also inhibit the phosphorylation of MDM2 and regulate the function of wt-p53 by interrupting Akt signaling. The recovery of wt-p53 function through multiple paths indicates that OA is more stable and has wider anticancer effects than highly specific inhibitors, since liver cancer has strong heterogeneity such that a small proportion of patients are sensitive to a single kind of highly specific inhibitor.
Recently, the CDK inhibitors SNS-032 and dinaciclib (SCH 727965), which inhibit CDK2, 7, 9 and CDK1, 2, 5, 9 enzyme activities, respectively, have been evaluated in clinical trials for cancer treatment [63]. Flavopiridol was the first CDK inhibitor to enter clinical trials. However, the main form of toxicity and side effects of flavopiridol treatment included secretory diarrhoea and tumour lysis syndrome (TLS), which correlated with the area under the curve (AUC) of plasma concentration-time for the flavopiridol glucuronide metabolite [51]. The CDK9 inhibitor OA showed much less toxicity and better druggability than the specific CDK9 inhibitor in vivo. Toxicological studies in vivo showed that the no-observed-adverse-effect level (NOAEL) of OA administered orally and measured by acute cytotoxicity tests was 3000 mg/kg, and the NOAEL of OA on the central nervous system function and respiratory system function of mice subjected to general pharmacology experiments was 2400 and 1200 mg/kg, respectively (Supplementary Table 3). The NOAEL dose of OA was much higher than the effective dose in vivo; thus, the drug safety window of OA was wide, and its safety was high.
In summary, our work led to the identification of CDK9 as an important regulator critical for the anticancer function of wt-p53. The CDK9 inhibitor OA stabilized the wt-p53 level by inhibiting CDK9-modulated activation of SIRT1 and MDM2. Recovery of wt-p53 function led to the promotion of apoptosis, inhibition of DNA repair, disrupted metabolism and so on and suppressed the progression of HCC. This study demonstrates a new mechanism for the recovery of wt-p53 function by CDK9 and provides a potential drug candidate for use in HCC therapy.
References
Connor F, Rayner TF, Aitken SJ, Feig C, Lukk M, Santoyo-Lopez J, et al. Mutational landscape of a chemically-induced mouse model of liver cancer. J Hepatol. 2018;69:840–50.
Villanueva A, Hernandez-Gea V, Llovet JM. Medical therapies for hepatocellular carcinoma: a critical view of the evidence. Nat Rev Gastroenterol Hepatol. 2013;10:34–42.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Wu J, Zhu P, Lu T, Du Y, Wang Y, He L, et al. The long noncoding RNA LncHDAC2 drives the self-renewal of liver cancer stem cells via activation of Hedgehog Signaling. J Hepatol. 2018;70:918–29.
Williams R, Alexander G, Aspinall R, Batterham R, Bhala N, Bosanquet N, et al. Gathering momentum for the way ahead: fifth report of the Lancet Standing Commission on Liver Disease in the UK. Lancet. 2018;392:2398–412.
Vecsei L. [Nature Reviews Drug Discovery: editorial article of neuroscientists from Szeged about kynurenine (IF: 33.078)]. Ideggyogy Sz. 2014;67:70.
Bayard Q, Meunier L, Peneau C, Renault V, Shinde J, Nault JC, et al. Cyclin A2/E1 activation defines a hepatocellular carcinoma subclass with a rearrangement signature of replication stress. Nat Commun. 2018;9:5235.
Whittaker SR, Mallinger A, Workman P, Clarke PA. Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharmacol Ther. 2017;173:83–105.
Olson CM, Jiang B, Erb MA, Liang Y, Doctor ZM, Zhang Z, et al. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol. 2018;14:163–70.
Garriga J, Grana X. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene. 2004;337:15–23.
Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–31.
Bose P, Simmons GL, Grant S. Cyclin-dependent kinase inhibitor therapy for hematologic malignancies. Expert Opin Investig Drugs. 2013;22:723–38.
Morales F, Giordano A. Overview of CDK9 as a target in cancer research. Cell Cycle. 2016;15:519–27.
Wei L, Zhou Y, Qiao C, Ni T, Li Z, You Q, et al. Oroxylin A inhibits glycolysis-dependent proliferation of human breast cancer via promoting SIRT3-mediated SOD2 transcription and HIF1alpha destabilization. Cell Death Dis. 2015;6:e1714.
Chen R, Wierda WG, Chubb S, Hawtin RE, Fox JA, Keating MJ, et al. Mechanism of action of SNS-032, a novel cyclin-dependent kinase inhibitor, in chronic lymphocytic leukemia. Blood. 2009;113:4637–45.
Gojo I, Zhang B, Fenton RG. The cyclin-dependent kinase inhibitor flavopiridol induces apoptosis in multiple myeloma cells through transcriptional repression and down-regulation of Mcl-1. Clin Cancer Res. 2002;8:3527–38.
Kunst C, Haderer M, Heckel S, Schlosser S, Muller M. The p53 family in hepatocellular carcinoma. Transl Cancer Res. 2016;5:632–8.
Kim J, Yu L, Chen W, Xu Y, Wu M, Todorova D, et al. Wild-type p53 promotes cancer metabolic switch by inducing PUMA-dependent suppression of oxidative phosphorylation. Cancer Cell. 2019;35:191–203 e8.
Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12:303–12.
Claudio PP, Cui J, Ghafouri M, Mariano C, White MK, Safak M, et al. Cdk9 phosphorylates p53 on serine 392 independently of CKII. J Cell Physiol. 2006;208:602–12.
Radhakrishnan SK, Gartel AL. CDK9 phosphorylates p53 on serine residues 33, 315 and 392. Cell Cycle. 2006;5:519–21.
Bagashev A, Fan S, Mukerjee R, Claudio PP, Chabrashvili T, Leng RP, et al. Cdk9 phosphorylates Pirh2 protein and prevents degradation of p53 protein. Cell Cycle. 2013;12:1569–77.
Wu J, Liang Y, Tan Y, Tang Y, Song H, Wang Z, et al. CDK9 inhibitors reactivate p53 by downregulating iASPP. Cell Signal. 2020;67:109508.
Cirstea D, Hideshima T, Santo L, Eda H, Mishima Y, Nemani N, et al. Small-molecule multi-targeted kinase inhibitor RGB-286638 triggers P53-dependent and -independent anti-multiple myeloma activity through inhibition of transcriptional CDKs. Leukemia. 2013;27:2366–75.
Yuan H, Su L, Chen WY. The emerging and diverse roles of sirtuins in cancer: a clinical perspective. Onco Targets Ther. 2013;6:1399–416.
Wei Z, Jia J, Heng G, Xu H, Shan J, Wang G, et al. Sirtuin-1/mitochondrial ribosomal protein S5 axis enhances the metabolic flexibility of liver cancer stem cells. Hepatology. 2019;70:1197–213.
Ong ALC, Ramasamy TS. Role of Sirtuin1-p53 regulatory axis in aging, cancer and cellular reprogramming. Ageing Res Rev. 2018;43:64–80.
Wei L, Dai Y, Zhou Y, He Z, Yao J, Zhao L, et al. Oroxylin A activates PKM1/HNF4 alpha to induce hepatoma differentiation and block cancer progression. Cell Death Dis. 2017;8:e2944.
Dai Q, Yin Q, Wei L, Zhou Y, Qiao C, Guo Y, et al. Oroxylin A regulates glucose metabolism in response to hypoxic stress with the involvement of hypoxia-inducible factor-1 in human hepatoma HepG2 cells. Mol Carcinog. 2016;55:1275–89.
Dai Q, Yin Y, Liu W, Wei L, Zhou Y, Li Z, et al. Two p53-related metabolic regulators, TIGAR and SCO2, contribute to oroxylin A-mediated glucose metabolism in human hepatoma HepG2 cells. Int J Biochem Cell Biol. 2013;45:1468–78.
Zou M, Lu N, Hu C, Liu W, Sun Y, Wang X, et al. Beclin 1-mediated autophagy in hepatocellular carcinoma cells: implication in anticancer efficiency of oroxylin A via inhibition of mTOR signaling. Cell Signal. 2012;24:1722–32.
Zhu B, Zhao L, Zhu L, Wang H, Sha Y, Yao J, et al. Oroxylin A reverses CAM-DR of HepG2 cells by suppressing Integrinbeta1 and its related pathway. Toxicol Appl Pharmacol. 2012;259:387–94.
Lu X, Errington J, Curtin NJ, Lunec J, Newell DR. The impact of p53 status on cellular sensitivity to antifolate drugs. Clin Cancer Res. 2001;7:2114–23.
Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–17.
Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundback T, Nordlund P, et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9:2100–22.
Bazarbachi A. CDK9 inhibition for ATL therapy. Blood. 2017;130:1074–75.
Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16.
Nguyen D, Liao WJ, Zeng SX, Lu H. Reviving the guardian of the genome: small molecule activators of p53. Pharmacol Ther. 2017;178:92–108.
Hsu IC, Tokiwa T, Bennett W, Metcalf RA, Welsh JA, Sun T, et al. p53 gene mutation and integrated hepatitis B viral DNA sequences in human liver cancer cell lines. Carcinogenesis. 1993;14:987–92.
Vaughan CA, Singh S, Windle B, Sankala HM, Graves PR, Andrew Yeudall W, et al. p53 mutants induce transcription of NF-kappaB2 in H1299 cells through CBP and STAT binding on the NF-kappaB2 promoter and gain of function activity. Arch Biochem Biophys. 2012;518:79–88.
Albert TK, Antrecht C, Kremmer E, Meisterernst M. The establishment of a hyperactive structure allows the tumour suppressor protein p53 to function through P-TEFb during limited CDK9 kinase inhibition. PLoS One. 2016;11:e0146648.
Zhang H, Pandey S, Travers M, Sun H, Morton G, Madzo J, et al. Targeting CDK9 reactivates epigenetically silenced genes in cancer. Cell. 2018;175:1244–58 e26.
Dhar D, Antonucci L, Nakagawa H, Kim JY, Glitzner E, Caruso S, et al. Liver cancer initiation requires p53 inhibition by CD44-enhanced growth factor signaling. Cancer Cell. 2018;33:1061–77 e6.
Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, et al. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One. 2009;4:e8414.
Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–48.
Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–59.
Yokoyama T, Kosaka Y, Mizuguchi M. Structural insight into the interactions between death-associated protein kinase 1 and natural flavonoids. J Med Chem. 2015;58:7400–8.
Kim K, Choe H, Jeong Y, Lee JH, Hong S. Ru(II)-catalyzed site-selective hydroxylation of flavone and chromone derivatives: the importance of the 5-hydroxyl motif for the inhibition of Aurora kinases. Org Lett. 2015;17:2550–3.
Chin YW, Kong JY, Han SY. Flavonoids as receptor tyrosine kinase FLT3 inhibitors. Bioorg Med Chem Lett. 2013;23:1768–70.
Zhao L, Yuan X, Wang J, Feng Y, Ji F, Li Z, et al. A review on flavones targeting serine/threonine protein kinases for potential anticancer drugs. Bioorg Med Chem. 2019;27:677–85.
Blachly JS, Byrd JC, Grever M. Cyclin-dependent kinase inhibitors for the treatment of chronic lymphocytic leukemia. Semin Oncol. 2016;43:265–73.
Albert TK, Rigault C, Eickhoff J, Baumgart K, Antrecht C, Klebl B, et al. Characterization of molecular and cellular functions of the cyclin-dependent kinase CDK9 using a novel specific inhibitor. Br J Pharmacol. 2014;171:55–68.
Zhao K, Zhou Y, Qiao C, Ni T, Li Z, Wang X, et al. Oroxylin A promotes PTEN-mediated negative regulation of MDM2 transcription via SIRT3-mediated deacetylation to stabilize p53 and inhibit glycolysis in wt-p53 cancer cells. J Hematol Oncol. 2015;8:41.
Rebouissou S, La Bella T, Rekik S, Imbeaud S, Calatayud AL, Rohr-Udilova N, et al. Proliferation markers are associated with MET expression in hepatocellular carcinoma and predict tivantinib sensitivity in vitro. Clin Cancer Res. 2017;23:4364–75.
Wang S, Fischer PM. Cyclin-dependent kinase 9: a key transcriptional regulator and potential drug target in oncology, virology and cardiology. Trends Pharmacol Sci. 2008;29:302–13.
Sonawane YA, Taylor MA, Napoleon JV, Rana S, Contreras JI, Natarajan A. Cyclin dependent kinase 9 inhibitors for cancer therapy. J Med Chem. 2016;59:8667–84.
Bieging KT, Attardi LD. Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol. 2012;22:97–106.
Pirngruber J, Johnsen S. Induced G1 cell-cycle arrest controls replication-dependent histone mRNA 3ʹ end processing through p21, NPAT and CDK9. Oncogene. 2010;29:2853–63.
Lain S, Xirodimas D, Lane DP. Accumulating active p53 in the nucleus by inhibition of nuclear export: a novel strategy to promote the p53 tumor suppressor function. Exp Cell Res. 1999;253:315–24.
Malmlof M, Roudier E, Hogberg J, Stenius U. MEK-ERK-mediated phosphorylation of Mdm2 at Ser-166 in hepatocytes. Mdm2 is activated in response to inhibited Akt signaling. J Biol Chem. 2007;282:2288–96.
Xue S, Shao Q, Zhu LB, Jiang YF, Wang C, Xue B, et al. LDC000067 suppresses RANKL-induced osteoclastogenesis in vitro and prevents LPS-induced osteolysis in vivo. Int Immunopharmacol. 2019;75:105826.
Xu M, Lu N, Sun Z, Zhang H, Dai Q, Wei L, et al. Activation of the unfolded protein response contributed to the selective cytotoxicity of oroxylin A in human hepatocellular carcinoma HepG2 cells. Toxicol Lett. 2012;212:113–25.
Polier G, Giaisi M, Kohler R, Muller WW, Lutz C, Buss EC, et al. Targeting CDK9 by wogonin and related natural flavones potentiates the anti-cancer efficacy of the Bcl-2 family inhibitor ABT-263. Int J Cancer. 2015;136:688–98.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 81873051 and 81830105), the National Science & Technology Major Project (Nos. 2017ZX09301014 and 2018ZX09711001–003–007), Social Development Project of Jiangsu Provincial Science and Technology Department (BE2018711), “Double First-Class” University project (CPU 2018GF11 and CPU2018GF05), and Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX20_0657).
Author information
Authors and Affiliations
Contributions
JYY, SX, QLG, and LBW conceptualized and designed the study. JYY, SX, YNS, YX, QLG, and LBW. contributed to the acquired data. JYY, SX, YNS, and LBW contributed to the in vitro study. JYY and YX contributed to the in vivo study. All authors prepared, reviewed, and edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Yao, Jy., Xu, S., Sun, Yn. et al. Novel CDK9 inhibitor oroxylin A promotes wild-type P53 stability and prevents hepatocellular carcinoma progression by disrupting both MDM2 and SIRT1 signaling. Acta Pharmacol Sin 43, 1033–1045 (2022). https://doi.org/10.1038/s41401-021-00708-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-021-00708-2
Keywords
This article is cited by
-
Selective autophagy in cancer: mechanisms, therapeutic implications, and future perspectives
Molecular Cancer (2024)
-
CDK9 inhibitors downregulate DKK1 expression to suppress the metastatic potential of HCC cells
Genes & Genomics (2023)
-
Anticancer potential of oroxylin A: from mechanistic insight to synergistic perspectives
Naunyn-Schmiedeberg's Archives of Pharmacology (2023)
-
Targeting cyclin-dependent kinase 9 in cancer therapy
Acta Pharmacologica Sinica (2022)
-
MicroRNA-874 targets phosphomevalonate kinase and inhibits cancer cell growth via the mevalonate pathway
Scientific Reports (2022)