Abstract
Programmed death ligand-1 (PD-L1)/PD-1 checkpoint extensively serves as a central mediator of immunosuppression. A tumor-promoting role for abundant PD-L1 in several cancers is revealed. However, the importance of PD-L1 and how the PD-L1 expression is controlled in breast cancer remains obscure. Here, the mechanisms of controlling PD-L1 at the transcription and protein acetylation levels in promoting breast cancer growth are presented. Overexpressed PD-L1 accelerates breast cancer growth in vitro and in vivo. RNA-seq uncovers that PD-L1 can induce some target genes affecting many cellular processes, especially cancer development. In clinical breast cancer tissues and cells, PD-L1 and HBXIP are both increased, and their expressions are positively correlated. Mechanistic exploration identifies that HBXIP stimulates the transcription of PD-L1 through co-activating ETS2. Specifically, HBXIP induces PD-L1 acetylation at K270 site through interacting with acetyltransferase p300, leading to the stability of PD-L1 protein. Functionally, depletion of HBXIP attenuates PD-L1-accelerated breast tumor growth. Aspirin alleviates breast cancer via targeting PD-L1 and HBXIP. Collectively, the findings display new light into the mechanisms of controlling tumor PD-L1 and broaden the utility for PD-L1 as a target in breast cancer therapy.
Similar content being viewed by others
Introduction
Programmed death ligand-1 (PD-L1, B7-H1) as one of the crucial immune checkpoints can manipulate the immune escape of the tumor cells by interacting with programmed death 1 (PD-1), resulting in T-cell apoptosis or exhaustion and immune cell inhibitory signaling [1, 2]. The abundance of PD-L1 is found in several cancers including ovarian, breast, cervical, melanoma, and lung cancer [3,4,5,6,7,8]. Many studies in past years are focused on the effects of PD-L1 as a transmembrane protein, especially on immune evasion. Higher levels of PD-L1 lead to an increased impairment of T-cell activity and tumor immune escape [9]. The adenosine A1 receptor (ADORA1) deficiency markedly inhibits T cell-induced cytotoxicity via the PD-L1 overexpression on tumor cell membrane [10]. Inhibition of PD-L1 stability by epidermal growth factor (EGF) strengthens anti-tumor T-cell immune response [11]. Some evidence reveals the roles of PD-L1 in the development of several specific cancers, such as melanoma, bladder cancer, ovarian cancer, and lung cancer [4, 8, 12]. Knockdown of PD-L1 with a polyethylenimine-dermatan sulfate complex suppresses melanoma B16F10 cell growth both in T-cell dependent and independent mechanisms [13]. High PD-L1 inhibits autophagy and promotes tumor growth via mTOR signals in melanoma and ovarian cancer [8]. Highly expressed tumor PD-L1 can serve as a direct marker for the prediction of anti-PD-L1/PD-1 effect in cancer clinical therapy [14]. However, the factors and the associated mechanisms controlling tumor PD-L1 overexpression are not fully revealed. Some transcriptional factors, such as RORC, ATF3, TEAD1, NF-κB, and MEF2D, function in the transcription regulation of tumor PD-L1 [4, 10, 15,16,17]. As for the regulation of oncogenic protein expression during the progression of cancers, in addition to transcription regulation, the role of posttranslational modifications (PTMs) has attracted the majority of researchers. There exist about 20 different types of PTMs in cells and their deregulations contribute to cancer progression [18, 19]. For the protein stability regulation of PD-L1, glycosylation, phosphorylation, palmitoylation, and ubiquitination play vital roles [3, 11, 20,21,22]. Yet, the function of tumor PD-L1 and related regulatory mechanism in breast cancer need to be further elucidated.
Serving as an 18 kDa well conserved protein, mammalian hepatitis B X-interacting protein (HBXIP), also called LAMTOR5, forms a regulator complex with p18, p14, and MP1, which is essential for Rag GTPases and mTOR lysosomal localization and mTORC1 activation under amino acid stimulation [23]. HBXIP can mediate cell apoptosis in collaboration with survivin [24]. Some studies have pointed out that HBXIP is overexpressed in multiple types of cancers and can function as an oncogenic transcriptional co-activator of many transcriptional factors, such as LXR, c-Myb, STAT4, Sp1, E2F1, and p53, accelerating breast cancer progression [25,26,27,28,29,30]. In addition, HBXIP is involved in regulating PTMs. For example, HBXIP enhances the deacetylation of tumor suppressor MST1 via HDAC6, resulting in MST1 protein degradation in breast cancer [31]. HBXIP-increased abnormal O-glycosylation via GALNT1 facilitates breast cancer metastasis [32]. HBXIP-induced HOXB13 acetylation inhibits its protein degradation, resulting in tamoxifen (TAM) resistance to breast cancer [33]. Recently, our group has revealed that HBXIP-induced the acetylation of HMGA2 via PCAF blocks its ubiquitination during the development of esophageal squamous cell carcinoma (ESCC) [34]. Therefore, it is of great interest to find the role of oncogenic HBXIP in PD-L1-mediated breast cancer progression.
Aspirin (ASA) is a well-known nonsteroidal anti-inflammatory drug (NSAID) and can serve as an effective adjuvant therapy in colorectal cancer [35, 36]. Recent studies have pointed out that ASA plays a beneficial role in many cancers via inhibiting the cyclooxygenase 2 (COX2) and NF-κB pathways [37, 38]. ASA can decrease the growth and metastasis of colorectal cancer via upregulating lncRNA OLA1P2 and then blocking the homodimers formation of phosphorylated STAT3 [39]. ASA also reduces mTOR signaling to activate autophagy in colorectal cancer cells [40]. Long-term use of ASA is beneficial to gastric cancer in the general population of South Korea at a certain dose [41]. In hepatocellular carcinoma, ASA inhibits the collagen deposition to relieve the liver tumor growth via targeting P4HA2 [42]. In breast cancer, ASA is related to lower risk of relapse rate and tumor death [43]. Recently, our group has demonstrated that ASA can reverse TAM resistance in breast cancer by regulating the HBXIP/HOXB13 axis [33]. In ESCC, ASA can reduce tumor growth via targeting HBXIP and HMGA2 [34]. Accordingly, we are interested in whether ASA is effective for HBXIP/PD-L1-induced breast tumor.
In the present study, we clarify that PD-L1 can stimulate the tumor growth and HBXIP serves as a key regulator to activate the transcription and induce the acetylation of PD-L1 in breast cancer. On the one hand, HBXIP is able to co-activate transcriptional factor ETS2 to promote the PD-L1 transcription. On the other hand, HBXIP induces the acetylation of PD-L1 at the lysine 270 residue (K270) via p300 to increase its stability. Functionally, HBXIP is essential for PD-L1-mediated breast tumor growth in breast cancer cell model and mouse model. As a classic anti-inflammation drug, ASA can target PD-L1 and HBXIP to suppress tumor growth. Our finding may identify HBXIP/PD-L1 pathway as a promising target for combating breast cancer.
Materials and methods
Tissue specimens
Paired human clinical adjacent tissues and breast cancer tissues (n = 37) were collected from the patients in Tianjin Tumor Hospital (Tianjin, China). The patient information is displayed in Supplementary Table S1. Two breast tumor tissue microarrays (Catalog No. 1005 and Catalog No. HBreD030PG03) were purchased from Xi’an Aomei Biotechnology (Xi’an, China) and Shanghai Outdo Biotech Co., Ltd (Shanghai, China), respectively, including 131 breast cancer tissues and 15 noncancerous breast tissues. The information is displayed in Supplementary Table S2. A total of 146 cases of breast tumor samples were stained as previously described [44]. All protocols of sample use were agreed by the Research Ethics Committee of Tianjin Tumor Hospital and Nankai University. Written consent was gained from all participants before tissues were collected.
Immunohistochemical staining analysis
Five micrometer of paraffin-embedded mouse tumor tissues were stained for Ac-K270-PD-L1, PD-L1, HBXIP or Ki67. Tissue sections were deparaffinized and rehydrated gradually. Primary antibodies (Supplementary Table S3) were incubated with tissues at 4 °C for overnight, HRP-conjugated secondary antibody was incubated with tissues for 45 min, and then tissues were stained with 3,3′-diaminobenzidine (DAB) (Zhong Shan Jin Qiao, Beijing, China) and hematoxylin. The score of staining was calculated by Image-Pro Plus software. For staining of breast tumor tissue microarrays, we scored the PD-L1 and HBXIP staining levels according to the intensity of staining 0 = negative, 1 = low, 2 = moderate, 3 = high, and distribution 0 (0%–10%), 1 (11%–30%), 2 (31%–70%), and 3 (71%–100%). The score was generated via multiplying intensity and distribution with a range from 1 to 9, 0 and 1 represent negative grade (−), 1, 2, and 3 represent weak grade (+), 4 and 6 represent moderate grade (++), 9 represents intense grade (+++).
Cells culture and treatment
The breast cancer cell lines MCF-7, SK-BR3, and MDA-MB-453 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Grand Island, NY, USA). MDA-MB-231 and BT549 cells were cultured in Roswell Park Memorial Institute 1640 (1640, Gibco). SUM159 cells were cultured in F12 media (Gibco). All cell culture mediums were supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U/mL of penicillin, and 0.1 mg/mL of streptomycin at 37 °C in a humidified incubator with 5% CO2. Cells were transfected with indicated plasmids and/or siRNAs by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Reagents used in this study include Trichostatin A (TSA, MedChem Express, Monmouth Junction, NJ, USA), cycloheximide (CHX, MedChem Express) and aspirin (ASA, Sigma-Aldrich, St. Louis, MO, USA). Cells were negative for mycoplasma.
RNA-sequencing (RNA-seq) and data analysis
MCF-7 cells were transfected with pCMV or pCMV-PD-L1 for 36 h with two biological replicates. Total RNAs were prepared with Trizol (Thermo Fisher Scientific, Waltham, MA, USA) and RNA-seq was performed by Shanghai Majorbio Biotechnology Co., Ltd, using Illumina HiSeq Xten (Illumina, USA). For bioinformatics analyses, fold change ≥1.5 and P value < 0.05 were calculated and visualized to identify significantly regulated genes utilizing edgeR software. Heat map of gene groups was presented by the Majorbio I-Sanger Cloud Platform (www.i-sanger.com). Gene Ontology (GO, http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) analyses were taken to determine functional and biological properties of genes. The RNA-sequencing data are presented at the Gene Expression Omnibus database (GSE158696).
Quantitative real-time PCR (qRT-PCR) and reverse transcription-PCR (RT-PCR)
The total RNAs were extracted with Trizol (Thermo Fisher Scientific). RNAs were reversely transcribed into cDNAs using Superscript III reverse transcription kit (Invitrogen) according to the manufacturer’s instructions. QRT-PCR was performed in three biological replicates per sample using the Platinum® Quantitative PCR System (TransGen Biotech, Beijing, China) and SYBR® Premix Ex Taq™ (TransGen Biotech) following the manufacturer’s instructions. RT-PCR was performed as previously reported [44]. The relative gene expression changes were calculated by the comparative method (2−ΔΔCt). The level of mRNAs was normalized to GAPDH gene in the same samples. ΔCt value means the average of triplicates. Primers used for the indicated gene products are listed in Supplementary Table S4.
Cell proliferation assays
For 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, the cells (2000 cells/well) were treated with indicated plasmids or ASA (0, 2.5, 5 mM). The cell viability was measured at indicated times. The cells were incubated with MTT reagent (Sigma-Aldrich) for 4 h at 37 °C. And the MTT formazan was dissolved with 150 μL DMSO for each well. The absorbance (490 nm) was examined using an absorbance reader. For 5-ethynyl-2-deoxyuridine (EdU) staining, cells (4000 cells/well) treated with indicated plasmids or ASA (5 mM). The cell proliferative capacity was measured with Cell-Light™ EdU kit (RiboBio, China) according to the manufacturer’s instructions. Briefly, EdU reagent was incubated with the indicated cells for 2 h, and fixed with paraformaldehyde for 30 min. The excessive aldehydes were neutralized with Glycine for 5 min. Then the cells underwent Apollo 567 staining for 30 min and Hoechst 33342 staining for 30 min. Cells were finally imaged by fluorescence microscope and calculated.
Colony forming assay
MCF-7, SK-BR3, or MDA-MB-231 cells (1000-3000 cells/well) were treated with indicated plasmids or ASA, and then incubated for 2 weeks. All mediums were replaced every 3 days. Cells were washed three times with PBS, fixed with methanol, and then stained with 0.05% (w/v) crystal violet (25% methanol). Colonies were imaged, and then calculated.
Western blotting analysis
The proteins of cells or tumor tissues were lysed in RIPA buffer (Solarbio, Beijing, China). The protein samples (10-15 μg/lane) were separated by 8%-15% SDS-PAGE and transfected to PVDF membranes (Merck Millipore, Billerica, MA, USA). Membranes were blocked with 5% nonfat dry milk in TBST for 1 h and incubated with primary antibodies (Supplementary Table S3) overnight at 4 °C. The antibody of acetylated PD-L1 at lysine 270 residue (Ac-K270) was generated with a synthetic acetylated human PD-L1 peptide: LRKGRMMDVK(AcK)KCGIQDTNSKKQS (Jia Xuan Zhi Rui, Beijing, China). Membrane was incubated with appropriate secondary antibodies and then the signal was detected with ECL Kit (Millipore). The intensity of Western blotting images was determined by Image J software.
Dual-luciferase reporter assay
The indicated breast cancer cells were co-transfected with luciferase reporter constructs of CD274 (PD-L1) promoter (100 ng/well, pGL3-P1, pGL3-P2, pGL3-P3, pGL3-P4, ETS2-M, and ELF5-M) and internal control Renilla luciferase (pRL-TK) (10 ng/well), and pcDNA, pcDNA-HBXIP, siHBXIP, siETS2, or siControl. The cells were collected, and the luciferase activities were examined with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Each experiment was repeated in triplicates. Fold changes of promoter activities were normalized to the control group.
Immunofluorescence staining
MCF-7 cells were transfected with indicated plasmids. Then cells were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature, permeabilized with 0.1% Triton X-100, and then blocked with 3% BSA in PBS. Cells were incubated with anti-p300 antibody (Santa Cruz, CA, USA) or anti-Flag antibody (Sigma-Aldrich) overnight and incubated with the secondary antibody for 1 h. Nuclei were counterstained with DAPI (Sigma-Aldrich) for 15 min. Images were visualized with a confocal microscopy (LeicaTCSSP5, Wetzlar, Germany).
Plasmids and small interfering RNAs (siRNAs)
The plasmids pCMV-Tag2B, pcDNA3.1(+), pcDNA3.1(+)-HBXIP, and pSilencer-HBXIP were kept in our laboratory [24,25,26, 30, 34]. The promoter (−1227/+243) of CD274 (PD-L1) (NCBI Reference Sequence: NC_000009.12) was obtained by amplification from the MCF-7 genomic DNA, subsequently inserted into the pGL3-Basic vector (Promega) using the KpnI/XhoI site and named pGL3-P1. The truncated promoter regions (−579/+243, −336/+243, and −105/+243) of CD274 (PD-L1) promoter were generated from the pGL3-P1 by the related primers and were cloned into the pGL3-Basic vector, named as pGL3-P2, pGL3-P3, and pGL3-P4, respectively. The mutant sequences of PD-L1 promoter carrying a substitution of nucleotides within the binding sites of ETS2 (AGGAA, named ETS2-M) and ELF5-binding site (TCCTA, named ELF5-M) were generated by PCR from the pGL3-P2. The human PD-L1 cDNA sequence (NCBI Reference Sequence: NM_014143.4) was inserted into pCMV-tag2B plasmid using EcoRI/XhoI restriction enzyme site. The PD-L1 mutations of Lys (K) at 270, 280, or 270/280 site into Arg (R) and Glu (Q) were generated by PCR from the pCMV-PD-L1 plasmid using EcoRI/XhoI restriction enzymes, called 270R, 280R, DM, 270Q, 280Q, or DM, respectively. Small interfering RNAs were synthesized by RiboBio (Guangzhou, China). All siRNA sequences and the related primers used are presented in Supplementary Table S4.
Chromatin immunoprecipitation (ChIP)-PCR
ChIP-PCR assay was conducted by using ChIP Assay Kit (Epigentek, USA) according to manufacturer’s instructions. Briefly, MDA-MB-231 cells were transfected with siControl or siHBXIP#1, and then were cross-linked with 1% formaldehyde for 10 min and lysed with ChIP buffer on ice. DNA from these samples was sheared by sonication. The sheared DNA was incubated with anti-ETS2 antibody (Supplementary Table S3), or with IgG as a negative control. DNA was eluted and amplified with PCR assay using specific primers for the expected ETS2-binding site in CD274 promoter, left: 5′-GATGTAGCTCGGGATGGGAAGTTCT-3′ and right: 5′-ACCCATTTGGCTTTGGTTTTTATTC-3′.
Co-immunoprecipitation (Co-IP) assay
The cell lysates of indicated cells were prepared with the lysis buffer. The cell lysates were centrifuged, and the supernatants were collected and incubated with specific primary antibodies (Supplementary Table S3) followed by incubation with protein G-Sepharose (Santa Cruz) at 4 °C, or with anti-Flag M2 affinity gel (Sigma-Aldrich). The immunocomplexes were washed with IP lysis buffer for three times, resuspended in 5× SDS loading buffer and boiled, and then examined by Western blotting using the indicated primary antibodies (Supplementary Table S3).
In vivo xenograft tumor growth experiments
All animal procedures were approved by the guidelines of National Institutes of Health Guide for the Care and Use of Laboratory Animals. BALB/c athymic nude mice (5-week-old, female) were purchased from Experimental Animal Center of Peking (Beijing, China). MCF-7 cells transfected with pCMV, pCMV-PD-L1, pSilencer-HBXIP or Control were subcutaneously injected into immune-deficient nude mice (3 × 107 cells/mL in 0.2 mL saline, n = 6/group). For ASA treatment experiment, MDA-MB-231 cells were subcutaneously injected into immune-deficient nude mice (3 × 107 cells/mL in 0.2 mL saline, n = 6/group). When tumor size of mouse exceeded 100 mm3, the saline or ASA at 75 mg/kg was initiated daily oral administration. The tumor dimensions and body weight were measured every 3 or 5 days and the tumor volume was calculated by the formula (tumor volume = (length × width × width/2)). When the tumor size reached ~1000 mm3, the mice were sacrificed. Harvested tumors were then used for IHC and Western blotting assays with antibodies against Ac-K270-PD-L1 (Ac-K270), PD-L1, HBXIP, β-actin, or Ki67 (Supplementary Table S3). All animal experiments were performed in accordance with the Research Ethics Committee of Nankai University.
Statistical analysis
All experiments were performed at least three times. Data derived from cell lines and xenografts experiments were assessed by comparing mean values (±SD). Statistical tests including Student’s t test and One-way ANOVA with Dunnett-t test were applied as indicated. Pearson’s correlation analysis was used to evaluate the correlation of the mRNA levels between CD274 (PD-L1) and HBXIP or ETS2 in human clinical breast cancer tissues. GraphPad Prism 5 for Windows (IBM, USA) was used for the above statistical analysis. Pearson chi-square independence test was used to evaluate the relationship of the protein level of HBXIP and PD-L1 in clinical breast cancer tissues using SPSS 22.0 (IBM). P < 0.05 was considered significant (*P < 0.05; **P < 0.01; ***P < 0.001, and not significant (NS)).
Results
PD-L1 accelerates breast cancer growth in vitro and in vivo
Consistent with previous studies, PD-L1 is important in promoting the development of some specific types of cancers including bladder cancer, liver cancer, lung cancer, and melanoma [4, 20, 45, 46]. To clarify the effect of PD-L1 on breast cancer growth, we introduced exogenous PD-L1 into MCF-7 and SK-BR3 breast cancer cells, and then investigated the protein level of PD-L1 in PD-L1 or control-transfected MCF-7 and SK-BR3 cells via Western blotting. Our data displayed that compared to control, the protein level of PD-L1 was significantly higher in the PD-L1-transfected cells, indicating that PD-L1 was overexpressed in MCF-7 and SK-BR3 cells (Supplementary Fig. S1a). Then we analyzed cell proliferation ability using MTT, colony forming, and EdU assays. PD-L1 overexpression markedly enhanced the viability of MCF-7 and SK-BR3 cells (Fig. 1a, b). Furthermore, the colony forming capacity was obviously increased in PD-L1-transfected MCF-7 and SK-BR3 cells (Fig. 1c and Supplementary Fig. S1b). Similarly, PD-L1-overexpressed group showed a higher relative fold fraction of EdU-positive cells (Fig. 1d and Supplementary Fig. S1c). Taken a further step, we identified the function of PD-L1 in tumor growth independent of immune in vivo. MCF-7 or PD-L1-overexpressing MCF-7 cells were subcutaneously injected into immune-deficient nude mice. The results showed that PD-L1 overexpression could remarkably induce the growth of breast tumor (Fig. 1e). Strikingly, tumor volume and weight were markedly increased in the PD-L1-overexpressed group than those in the control mice (Fig. 1f, g). Through immunohistochemical (IHC) staining analysis of mouse tumor tissues, we observed higher expression of Ki67, a marker of cell proliferation, in PD-L1-overexpressed group compared with the control group (Fig. 1h). In mouse tumor samples, we identified high expression of PD-L1 in PD-L1-transfected group (Fig. 1i). Collectively, these findings indicate that PD-L1 can promote the growth of breast cancer.
a, b MCF-7 cells (a) and SK-BR3 cells (b) were transfected with pCMV-PD-L1 or pCMV vectors. The cell viability was measured by MTT assay at indicated times. c MCF-7 cells were transfected with pCMV-PD-L1 or pCMV vectors and then the colony forming efficiency was assessed by crystal violet. d MCF-7 cells were transfected with pCMV-PD-L1 or pCMV vectors. The cell proliferation was tested by EdU incorporation assay. Scale bar = 50 μm. e-g Images of the tumors derived from immune-deficient nude mice transplanted with MCF-7 cells pretreated with indicated plasmids. Tumor photographs at the time endpoint (e), tumor volumes (f), and tumor weights (g) in each group were shown (each group, n = 6). h Tumors tissues were stained for Ki67 expression via immunohistochemical staining. Representative images of Ki67 staining are shown (left) and the percentage of Ki67 positive cells was digitally counted (right). Scale bar = 50 μm. i The protein levels of PD-L1 in treated explant tissue lysates of mice were analyzed by Western blotting. Data are presented as mean ± SD (n = 3). *P < 0.05; ***P < 0.001, NS, not significant, P value was assessed by Student’s t test.
PD-L1 induces cell growth signaling in breast cancer cells
To gain better insight into the molecular basis by which PD-L1 overexpression induces the growth of breast cancer cells, we performed genome-wide RNA-seq to investigate the gene transcription in MCF-7 cells with or without PD-L1 transfection (Fig. 2a). Based on the 29478 mapped genes, we defined 529 differentially expressed genes including 248 upregulated and 281 downregulated genes by setting absolute fold change ≥1.5 and P value < 0.05 as thresholds (Fig. 2b, c and Supplementary Table S5). Subsequently, GO analyses manifested that the differentially expressed genes upon PD-L1 overexpression was involved in several important biological processes, for instance cellular, metabolic, and development process in MCF-7 cells (Fig. 2d). KEGG analyses of the identified differentially expressed genes revealed that some cellular signal pathways were obviously affected by PD-L1, such as cell growth, death, cell motility, translation, endocrine or immune system, and cancers (Fig. 2e). At last, we performed qRT-PCR analysis to confirm the RNA-seq data. Our result revealed that the mRNA levels of several oncogenes were significantly increased, such as TPR, ITIH4, RAC2, TERT, and EGR1 in PD-L1 overexpressing MCF-7 cells compared with the control cells (Fig. 2f). Taken together, these data imply that PD-L1 can regulate the transcription of distinct groups of genes involved in cell growth or motility in breast cancer.
a Experimental scheme of RNA-Seq in MCF-7 cells and subsequent data analysis. b The differentially expressed genes in pCMV-PD-L1-transfected MCF-7 cells compared to pCMV-transfected cells were analyzed by transcriptomics and shown by volcano plots. c Heat map of differentially expressed genes in pCMV-PD-L1 or pCMV-transfected MCF-7 cells from RNA-Seq results. d Gene-Ontology (GO) analysis of RNA-seq data. The y axis displays the GO terms, and the x-axis displays number of genes enriched in GO terms. e KEGG pathway analysis of the discovered differentially expressed genes from RNA-seq data. The x-axis displays different KEGG pathways, and the y axis displays number of genes enriched in KEGG terms. f The level of five representatively upregulating genes in pCMV-PD-L1-transfected MCF-7 cells from RNA-Seq results was analyzed by qRT-PCR assay. GAPDH was used for normalization. Data are presented as mean ± SD (n = 3). **P < 0.01; ***P < 0.001, P value was assessed by Student’s t test.
PD-L1 is positively related to HBXIP in human breast cancer tissues and cells
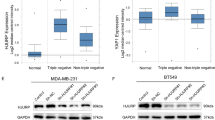
Some evidence has revealed that highly expressed PD-L1 play crucial roles in progression of several types of cancers [4, 6, 8, 47]. HBXIP can function as an oncogenic protein during the development of numbers of cancers, including breast cancer, hepatoma carcinoma, and esophageal squamous cancer [24, 31, 32, 34, 48]. In the present investigation, we are interested in whether there is a relationship between PD-L1 and HBXIP in the progression of breast cancer. Using clinical cancerous and their paired noncancerous tissues from 37 breast cancer patients, we analyzed the mRNA levels of CD274 (PD-L1) and HBXIP through qRT-PCR assay (Supplementary Table S1). The data showed that both CD274 and HBXIP at the mRNA levels were increased in clinical breast cancer tissues compared with the adjacent noncancerous tissues (Fig. 3a, b). Notably, the mRNA level of CD274 was positively related to the HBXIP level (Pearson’s correlation, r = 0.4773, P < 0.01, Fig. 3c). The IHC staining analysis of human breast cancer serial section tissue microarrays containing 131 breast cancer tissues and 15 noncancerous breast tissues indicated that PD-L1 and HBXIP were obviously increased (Fig. 3d, e and Supplementary Table S2). Furthermore, a positive correlation protein expression of HBXIP and PD-L1 in human tissues was revealed (Pearson chi-square independence test, χ2 = 37.96, P < 0.001, Fig. 3f). In addition, using six breast cancer cell lines including MCF-7, SK-BR3, MDA-MB-453, SUM159, BT549, and MDA-MB-231 cells, we further identified that the expression of HBXIP was positively related to the PD-L1 expression (Supplementary Fig. S2). To further evaluate the influence of PD-L1 and HBXIP on the clinical relevance of breast cancer patients, Kaplan–Meier survival analysis was performed by a public database (Kaplan-Meier Plotter, https://kmplot.com/analysis/). The results indicated that high HBXIP or PD-L1 expression was associated with poor relapse free survival in breast cancer patients (Fig. 3g, h). Therefore, we conclude that HBXIP exhibits a positive relationship with PD-L1 in breast cancer tissues and cells.
a, b Relative mRNA levels of CD274 (PD-L1) (a) and HBXIP (b) in 37 cases of human clinical breast tumor tissues and paired peritumor tissues. c The correlation of CD274 and HBXIP mRNA level was analyzed in 37 cases of human clinical breast cancer tissues by Spearman’s correlation analysis. d Representative IHC staining images of HBXIP and PD-L1 in human breast cancer serial section tissue microarrays containing 131 breast cancer samples and 15 noncancerous breast tissues. Scale bar = 100 μm. e Heat map of the correlation between PD-L1 and HBXIP expression in human breast cancer serial section tissue microarrays is shown. f The association between PD-L1 and HBXIP expression in human breast cancer serial section tissue microarrays containing 131 breast cancer samples and 15 noncancerous breast tissues was statistically analyzed by Pearson chi-square independence test, χ2 = 37.96, P < 0.001. g Kaplan-Meier analysis of RFS (n = 3951, P = 0.00097) curve for breast cancer patients with high or low HBXIP expression. h Kaplan-Meier analysis of RFS (n = 204, P = 0.012) curve for breast cancer patients with high or low PD-L1 expression. *P < 0.05; **P < 0.01; ***P < 0.001, P value was assessed by Student’s t test.
PD-L1 is induced by HBXIP in breast cancer cells
To further ascertain the role of HBXIP in the modulation of PD-L1, we transfected full-length HBXIP into MCF-7 cells or siRNAs targeting HBXIP mRNA into MDA-MB-231 cells, and then detected the RNA and protein levels of PD-L1. Firstly, we evaluated RNA interfering efficiency of two different HBXIP siRNAs (siHBXIP#1 or siHBXIP#2) and found that siHBXIP#1 was more effective than siHBXIP#2 in MDA-MB-231 cells (Supplementary Fig. S3a). Using RT-PCR and qRT-PCR assays we found that CD274 were induced by increasingly overexpressed HBXIP at the mRNA level in MCF-7 cells and then silencing of HBXIP could decrease the expression of CD274 at the mRNA level in MDA-MB-231 cells (Supplementary Fig. S3b and Fig. 4a, b). Our results further showed that the protein levels of PD-L1 were significantly upregulated or downregulated in a dose-dependent manner when HBXIP was overexpressed in MCF-7 or depleted in MDA-MB-231 cells (Fig. 4c). In addition, we exogenously overexpressed HBXIP in MCF-7 cells and tested the expression of HBXIP and PD-L1 using Western blotting and immunofluorescence analysis. Our results revealed that HBXIP expression was induced and then the HBXIP overexpression caused a significant increase of cytoplasm staining of PD-L1 (Supplementary Fig. S3c and Fig. 4d). To determine whether PD-L1 is activated by HBXIP, we generated the core promoter region of CD274 (−1227/+243) [4, 10, 15,16,17] into the pGL3-basic luciferase reporter vector. The data revealed that HBXIP overexpression markedly increased the activities of CD274-Luc vector (Fig. 4e), and knockdown of HBXIP hindered the activity of CD274-Luc vector (Fig. 4f), suggesting that HBXIP could induce the activation of CD274. Thus, we find that HBXIP can promote the expression of PD-L1 in breast cancer cells.
a, b MCF-7 cells and MDA-MB-231 cells were transfected with pcDNA or pcDNA-HBXIP vectors (a) or siControl or siHBXIP#1 (b), respectively. The mRNA levels of CD274 and HBXIP by qRT-PCR assay. c MCF-7 cells and MDA-MB-231 cells were transfected with pcDNA or pcDNA-HBXIP vectors or siControl or siHBXIP#1, respectively. The proteins were collected 48 h after transfection and assessed for the protein levels of PD-L1 and HBXIP by Western blotting assay. The intensity of the protein levels of PD-L1 and HBXIP was calculated by Image J. d MCF-7 cells were transfected with pcDNA or pcDNA-HBXIP vectors. Anti-PD-L1 antibody (red) was applied for IF staining. Nuclei were stained with DAPI (blue). Scale bar = 20 μm. e, f Dual-luciferase reporter assay of the CD274 promoter activity in MCF-7 cells transfected with pcDNA or pcDNA-HBXIP vectors (e) and in MDA-MB-231 cells transfected with siControl or siHBXIP#1 (f). Data are presented as mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001, P value was assessed by One-way ANOVA with Dunnett-t test.
HBXIP stimulates the transcription of PD-L1 via co-activating transcription factor ETS2
To investigate the molecular mechanism of HBXIP-mediated CD274 activation, we constructed three truncated promoter regions shorter than −1227/+243 (pGL3-P1), including −579/+243 (pGL3-P2), −336/+243 (pGL3-P3), and −105/+243 (pGL3-P4). Dual-luciferase reporter assays revealed that compared with pGL3-P2 (−579/+243), pGL3-P3 activity showed a collapse under HBXIP transfection, implying that the fragment −579/−336 might be the key regions of the CD274 promoter regulated by HBXIP (Fig. 5a). Using bioinformatics analysis of transcriptional factor binding prediction servers (PROMO, http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3 and JASPAR, http://jaspar.genereg.net/), we found that this promoter region contains ETS2 and ELF5 putative binding sites. To screen the transcriptional factor responsible for CD274 activation-mediated by HBXIP, we generated the promoter region containing the mutated ETS2 or ELF5-binding sites individually (ETS2-M or ELF5-M) and transfected all the constructs into MCF-7 cells. Compared with wild type (WT), HBXIP still can stimulate the luciferase activity of ELF5-M, but not the ETS2-M mutant (Fig. 5b). These results indicated that ETS2 could serve as the transcriptional factor in HBXIP-enhanced CD274 activation. To explore the role of ETS2 in HBXIP-induced CD274 activation, we employed two different siRNAs targeting ETS2 to silence the expression of ETS2 and found that siETST2#1 was more effective than siETST2#2 (Supplementary Fig. S4a). Indeed, knockdown of ETS2 with siETS2#1 sharply hampered the enhancement of CD274-P2 promoter activity induced by HBXIP (Fig. 5c). We next performed Chromatin Immunoprecipitation (ChIP) to examine whether ETS2 is recruited to the CD274 promoter. Through ChIP-PCR assays we observed that ETS2 could occupy the CD274 promoter but knockdown of HBXIP by siRNAs attenuated the occupation in MDA-MB-231 cells (Fig. 5d). In addition, we found that HBXIP was co-immunoprecipitated endogenously by ETS2 in MDA-MB-231 cells. Reciprocally, ETS2 was also co-immunoprecipitated by HBXIP in MDA-MB-231 cells (Fig. 5e). Using cBioportal Web resource online (cBioportal for Cancer Genomic) we analyzed the TCGA database (breast cancer-TCGA, Nature 2012), and found that there was a positive relationship between the mRNA levels of ETS2 and CD274 in 463 cases of clinical breast cancer samples (Pearson’s correlation, r = 0.24, P < 0.001, Fig. 5f). Consistently, the mRNA and protein levels of ETS2 could be upregulated by HBXIP in breast cancer cells using qRT-PCR and Western blotting assays (Supplementary Fig. S4b, c). Together, all these results implicate that HBXIP can activate the PD-L1 promoter via binding to transcriptional factor ETS2 in breast cancer cells.
a Relative luciferase activity of CD274 in MCF-7 cells transfected with pcDNA or pcDNA-HBXIP vectors. b Diagram of CD274 promoter with the predicated ETS2 and ELF5-binding site within −336 to −579 region by the online tool PROMO and JASPAR. The red rectangular box highlights the mutated ETS2 or ELF5-binding sites (ETS2-M or ELF5-M). Luciferase reporter gene assay of ETS2-M or ELF5-M promoter activity was performed in MCF-7 cells transfected with pcDNA or pcDNA-HBXIP vectors. c Luciferase reporter gene analysis of CD274 (PD-L1)-P2 promoter activity in MCF-7 cells co-transfected with pcDNA, pcDNA-HBXIP vectors and ETS2 siRNAs including siETS2#1 and siETS2#2. d ChIP assay of the binding of ETS2 with the CD274 promoter region in MDA-MB-231 cells with or without siHBXIP#1 treatment. e Co-IP analysis of the interaction of endogenous HBXIP with ETS2 in MDA-MB-231 cells and Western blotting assay of HBXIP and ETS2. IgG serves as a negative control. f The correlation analysis of CD274 and ETS2 in 463 cases of clinical breast cancer tissues (Pearson’s correlation, r = 0.24, ***P < 0.001). **P < 0.01; ***P < 0.001, NS, not significant, P value was assessed by Student’s t test (a, b) or One-way ANOVA with Dunnett-t test (c).
HBXIP acetylates PD-L1 and maintains PD-L1 stability
Only recently have the researchers focused on posttranslational modifications (PTMs) of PD-L1, such as phosphorylation, N-linked glycosylation, palmitoylation, and polyubiquitination, which can function as the therapeutic targets for cancers [3, 11, 20,21,22]. Some investigations have implied that HBXIP is important in regulating PTMs of some key factors to affect their stabilities, including MST1, HOXB13, or HMGA2 [31, 33, 34]. Here, we want to explore whether HBXIP engaged in the modulation of PTM and stability of PD-L1 in breast cancer. Firstly, we treated MCF-7 cells and MDA-MB-231 with cycloheximide (CHX), a protein synthesis inhibitor, for the indicated periods to block protein translation, and then the PD-L1 protein degradation rates were examined by Western blotting. Of note, when HBXIP was overexpressed in MCF-7 cells the half-life of endogenous PD-L1 protein was extended, while knockdown of HBXIP accelerated PD-L1 degradation in MDA-MB-231 cells (Fig. 6a and Supplementary Fig. S5a). It has been reported that among numbers of types of PTMs the lysine acetylation plays a key role in modulating the stability and function of proteins [49, 50]. To determine whether HBXIP could increase the PD-L1 protein stability via regulating its acetylation, we treated the indicated cells with trichostatin A (TSA), an inhibitor of histone deacetylase HDACI/II and/or CHX. The data demonstrated that the PD-L1 protein levels were still increased along with the elevated concentration of TSA after CHX prevented the protein synthesis of PD-L1, implying that PD-L1 could be acetylated in breast cancer cells (Fig. 6b). We then transfected Flag-tagged PD-L1 into MCF-7 cells and found that HBXIP overexpression promoted the acetylation levels of exogenous Flag-PD-L1 using immunoprecipitation analysis (Fig. 6c). According to the prediction of PhosphoSitePlus (http://www.phosphosite.org/homeAction.action) and GPS-PAIL 2.0 there are two potential acetylation sites in PD-L1 (K270 and K280). Then, we mutated the 270 and 280 lysine residues to arginine (R, acetylation-deficient mimics), as well as mutated these two lysine residues to glutamine (Q, hyperacetylation mimics). Our findings concluded that compared with the WT group, the acetylation levels of the mutants including K270R, K270R/280R, K270Q, and K270Q/280Q, were not induced by HBXIP, indicating that K270 might be the major acetylation site of PD-L1 mediated by HBXIP (Fig. 6d and Supplementary Fig. S5b). The alignment analysis of amino acid sequences in PD-L1 homologs from different species showed that K270 was conserved across all six species during the evolution (Supplementary Fig. S5c). To further confirm whether K270 of PD-L1 is acetylated, we generated a specific rabbit polyclonal antibody that only recognized the acetylated-K270 (Ac-K270) of PD-L1 from a company. We identified the specificity of the Ac-K270 antibody of PD-L1 for it could recognize the K270-acetylated peptides, but not the unacetylated PD-L1 peptides (Fig. 6e). Moreover, Ac-K270 of PD-L1 antibody could recognize the acetylation of ectopically expressed WT of PD-L1, but not its K270R or K270Q mutant (Fig. 6f), supporting that HBXIP-induced PD-L1 acetylation occurred at the K270 site. Our data further revealed that compared with WT group, the hyperacetylation K270Q mutant was more stable and then the acetylation-deficient K270R mutant degraded faster (Fig. 6g). Acetyltransferase plays a vital role in many processes and is regarded as a potential target for cancer treatment. To further explore the acetyltransferase is responsible for PD-L1 acetylation, we examined different acetyltransferases including CBP, GCN5, p300 or PCAF on PD-L1 acetylation. We firstly screened the effective siRNAs from two different siRNAs targeting CBP (siCBP#1 or siCBP#2), GCN5 (siGCN5#1 or siGCN5#2), p300 (sip300#1 or sip300#2) or PCAF (siPCAF#1 or siPCAF#2), respectively (Supplementary Fig. S5d). The result revealed that only silencing of p300 could significantly abolish HBXIP-induced PD-L1 acetylation at K270 site, but not silencing of GCN5, CBP or PCAF (Fig. 6h). Additionally, the immunoprecipitation analysis showed that p300 could interact with PD-L1 and HBXIP. Confocal microscopy indicated that PD-L1 was co-localized with p300 and HBXIP (Supplementary Fig. S5e and Fig. 6i). In summary, our results imply that HBXIP induces the PD-L1 acetylation at the K270 site via acetyltransferase p300 in breast cancer cells.
a MCF-7 cells were transfected with pCMV or pCMV-HBXIP vectors, and then treated with CHX (100 μg/mL) for the indicated times. The degradation of PD-L1 protein was analyzed by Western blotting assay. Quantification graph is shown. b MCF-7 and MDA-MB-231 cells were treated with CHX (100 μg/mL) and the indicated concentrations of TSA for 18 h, and endogenous PD-L1 protein levels were evaluated by Western blotting. c Exogenous Flag-PD-L1 was immunoprecipitated with Flag-beads in MCF-7 cells, and then the acetylation level of PD-L1 protein was detected in precipitation samples by Western blotting analysis with anti-acetylated-lysine antibody. d MCF-7 cells were transfected with the indicated vectors. Exogenous Flag-tagged PD-L1 was immunoprecipitated with Flag-beads in MCF-7 cells, and then the acetylation level of PD-L1 was detected by Western blotting analysis with anti-acetylated-lysine antibody. The protein expressions of PD-L1 and HBXIP were examined using anti-Flag or anti-HBXIP antibodies, respectively. e The specificity of anti-acetylated-K270-PD-L1 antibody was detected by dot blot assay. Nitrocellulose membrane was spotted with different amounts of acetylated-K270 peptide (LRKGRMMDVK(AcK)KCGIQDTNSKKQS) or unmodified peptide (LRKGRMMDVKKCGIQDTNSKKQS) and probed with anti-AcK270-PD-L1 (Ac-K270) antibody. f MCF-7 cells were transiently transfected with vector, Flag-PD-L1-WT, Flag-PD-L1-K270R, or Flag-PD-L1-K270Q. Flag-tagged PD-L1 was immunoprecipitated with Flag-beads, and then the acetylated-K270 levels of PD-L1 were tested by Western blotting with anti-AcK270-PD-L1 (Ac-K270) antibody. g MCF-7 cells transiently expressing Flag-PD-L1-WT, Flag-PD-L1-K270R, and Flag-PD-L1-K270Q were treated with CHX (100 μg/mL) for the indicated times, and the protein stabilities of PD-L1 were examined by Western blotting (left). Quantified graph of the PD-L1 protein levels is shown (right). h Flag-PD-L1 vectors accompanied by the indicated plasmids or siRNAs (siRNA of si-CBP#2, si-GCN5#2, si-p300#2, or si-PCAF#2) were transfected into MCF-7 cells. The acetylation and the acetylated-K270 level of PD-L1 protein were tested by Western blotting. i MCF-7 cells were transfected with RFP-HBXIP and/or Flag-PD-L1 vectors. Anti-p300 antibody (red), Flag-PD-L1 (green), and RFP-HBXIP (red) were applied for IF staining. Nuclei were stained with DAPI (blue). Scale bar = 25 μm. Data are presented as mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001, P value was assessed by Student’s t test.
Elevated PD-L1 by HBXIP facilitates breast cancer growth
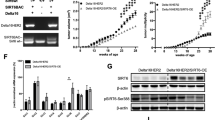
Next, we evaluated the effect of PD-L1 enhancement induced by HBXIP on breast cancer growth in vitro and in vivo. At the first, we transfected pCMV, pCMV-PD-L1 or pCMV-PD-L1/pSilencer-HBXIP plasmids into MCF-7 or SK-BR3 cell lines. Using MTT, colony forming or EdU incorporation assays we found that PD-L1 overexpression dramatically promoted the proliferation of MCF-7 and SK-BR3 cells, and then silence of HBXIP attenuated PD-L1-induced cell proliferation (Fig. 7a-c and Supplementary Fig. S6a-c). Moreover, we explanted MCF-7 cells transfected with Control, PD-L1 overexpression or PD-L1 overexpressed/HBXIP knockdown plasmids into immune-deficient nude mice. We observed that ectopic PD-L1 expression enhanced the growth of breast tumor in mice. However, HBXIP knockdown obviously disrupted PD-L1 droved-tumor growth (Fig. 7d-f). Additionally, the overexpression of PD-L1, acetylated PD-L1, and knockdown of HBXIP in mouse tumors were confirmed by IHC staining and Western blotting analysis (Fig. 7g, h). Ki67, a cell proliferation marker, was elevated in the PD-L1-overexpressed group and then HBXIP silencing abrogated the PD-L1-induced Ki67 in mouse tumors (Fig. 7g and Supplementary Fig. S6d). Together, these findings pinpoint that the enhanced PD-L1 by HBXIP stimulates breast tumor growth.
a MCF-7 cells were co-transfected with pCMV-PD-L1, pSilencer-HBXIP and their controls. The cell viability was measured by MTT analysis at the indicated times. b MCF-7 cells were co-transfected with pCMV-PD-L1 and pSilencer-HBXIP and their controls and then cultured for 2 weeks. Colony forming efficiency was assessed by crystal violet. Cell colonies were quantified. c MCF-7 cells were transfected with the indicated plasmids and their controls. EdU-positive cells were determined by EdU incorporation assays. Scale bar = 50 μm. d-f MCF-7 cells were pretreated with the indicated plasmids and subcutaneously injected into immune-deficient nude mice (3 × 107 cells/mouse). Subcutaneous tumor image (d), tumor growth curves (e), and tumor weights (f) were captured and analyzed. Each bar shows the means ± SD (each group, n = 6). g Representative immunohistochemical staining of AcK270-PD-L1 (Ac-K270), PD-L1, HBXIP, or Ki67 in mice tumor tissues. Scale bar = 50 μm. h Western blotting analysis of AcK270-PD-L1 (Ac-K270), PD-L1, HBXIP, and β-actin in mice tumors. *P < 0.05; **P < 0.01; ***P < 0.001, NS, not significant, P value was assessed by One-way ANOVA with Dunnett-t test.
ASA suppresses breast cancer growth via the HBXIP/PD-L1 axis
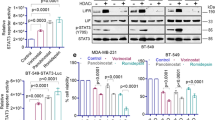
The anticancer function of ASA has been revealed in multiple cancers, which can reduce tumor growth [51]. Regular use of ASA decreases the risk of lethal prostate cancer and is beneficial to gastric cancer therapy in the population of South Korea [38, 41]. ASA treatment inhibits rheumatoid arthritis fibroblast-like synoviocytes proliferation and induces cell apoptosis [52]. In hepatocellular carcinoma, ASA alleviates hepatic immunopathology and severity of liver fibrosis [53]. ASA is able to prevent human colorectal cancer cell growth via lncRNA OLA1P2, mTOR signaling or H3K9 modification [39, 40, 54]. Our group has identified that ASA can block the HBXIP to overcome tamoxifen resistance or inhibit the progression of ESCC [33, 34]. In the present study, we are wondering whether ASA can target PD-L1 or HBXIP to affect the growth of breast tumor. We treated MD-MBA-231 cells with the increased concentration of ASA and the data presented that ASA inhibited the mRNA levels of HBXIP and CD274 in a dose-dependent manner (Fig. 8a). Moreover, ASA could significantly decrease the acetylation of PD-L1 at K270 site and the protein levels of PD-L1 and HBXIP in MD-MBA-231 cells in a dose-dependent manner (Fig. 8b). Besides, we investigated the possibility by which decreasing PD-L1 expression by ASA could inhibit the breast tumor growth in vitro and in vivo. To this end, we treated breast cancer MDA-MB-231 cells with ASA and observed that ASA could decrease the growth of breast cancer cells in vitro using MTT, colony forming, and EdU assays (Fig. 8c–e). Finally, we monitored mouse xenograft tumor growth using MDA-MB-231 cells and the result proved that ASA was able to attenuate the breast tumor growth in vivo (Fig. 8f–h). Expectedly, ASA decreased the mRNA levels of HBXIP, CD274, and ETS2 in mouse tumors (Fig. 8i). The IHC staining of Ki67 confirmed attenuated proliferation under ASA treatment (Fig. 8j and Supplementary Fig. S7). Meanwhile, ASA could restrain the acetylation of PD-L1 at K270 site and the protein levels of PD-L1 and HBXIP in mouse tumor samples through IHC staining and Western blotting analysis (Fig. 8j, k). Collectively, our results imply that ASA is capable of reducing HBXIP and PD-L1 to prevent the growth of breast tumor.
a MDA-MB-231 cells were treated with ASA (0 mM, 2.5 mM, 5 mM), and then examined for the mRNA levels of HBXIP and CD274 by qRT-PCR assay. b MDA-MB-231 cells were treated with ASA (0 mM, 2.5 mM, 5 mM), and then the protein levels of AcK270-PD-L1 (Ac-K270), PD-L1, or HBXIP were examined by Western blotting analysis. c MDA-MB-231 cells were treated with ASA (0 mM, 2.5 mM, 5 mM). The cell viability was measured by MTT assay at the indicated times. d MDA-MB-231 cells were treated with ASA (0 mM, 2.5 mM, 5 mM) for 2 weeks. The cell colonies were quantified. e MDA-MB-231 cells were treated with ASA (0 mM, 2.5 mM, 5 mM) for 24 h. The proliferation ability of cells was examined by EdU incorporation assay. f-h MDA-MB-231 cells (3 × 107 cells/mouse) were subcutaneously injected into immune-deficient nude mice (each group, n = 6). After the tumor size reached ~100 mm3, mice were daily oral administered with saline as the control or ASA (75 mg · kg−1 · d−1). Xenografts were harvested 20 days post ASA treatment. Subcutaneous tumor image (f), tumor growth curves (g), and tumor weight (h) were captured and analyzed. i The mRNA levels of HBXIP, CD274, and ETS2 in mouse tumor tissues were analyzed by qRT-PCR analysis. GAPDH was used as a control. Each bar shows the means ± SD (each group, n = 6). j Representative immunohistochemical staining of AcK270-PD-L1 (Ac-K270), PD-L1, HBXIP, or Ki67 in mouse tumor tissues. Scale bar = 50 μm. k Western blotting analysis of AcK270-PD-L1 (Ac-K270), PD-L1, HBXIP, and β-actin in mouse tumor tissues. **P < 0.01; ***P < 0.001, NS not significant, P value was assessed by One-way ANOVA with Dunnett-t test (a) or Student’s t test (c–i).
Discussion
PD-L1/PD-1 immune checkpoint blockade has presented beneficial therapeutic effect, arousing widely attention. PD-L1 can be expressed in diverse kinds of immune and tumor cells, which plays a major role in inhibiting immune surveillance. After macrophages are treated with PD-L1 antibody, its proliferation, survival, and activation are significantly stimulated [55]. Depleted PD-L1 expression in colorectal cancer cells by targeting PD-L1 palmitoylation significantly stimulates T-cell immune responses against tumors [21]. Most work on the regulations and functions of PD-L1 in cancers has focused on PD-L1 as a transmembrane protein, especially on the surface of immune cells. Despite recent work reveals that PD-L1 has important effects on the development of bladder cancer, cervical cancer, ovarian cancer, lung cancer, and melanoma through regulating some key signaling pathways [4, 6, 8, 12], a comprehensive understanding of the function and regulatory mechanism of highly PD-L1 during the development of breast cancer has been insufficient.
Considering some evidence based on PD-L1 has shown that it plays an oncogenic role in lung, melanoma, ovarian, bladder, and colon cancer [3,4,5,6,7], we firstly tried to explore the effect of PD-L1 on breast cancer growth. Our findings revealed that highly expressed PD-L1 could induce the breast cancer cell proliferation in vitro and the growth of breast tumor in vivo using MTT, colony forming, EdU incorporation, and tumor formation assay. IHC staining revealed an increase of Ki67 in the mouse tumor tissues with PD-L1 overexpression, which further supported our above finding. Next, we are interested in the molecular alterations caused by overexpressed PD-L1 in the promotion of breast tumor growth. Through transcriptome analysis by RNA-seq, we found that there were 529 differentially expressed genes in PD-L1 overexpressed-breast cancer cells. Using GO and KEGG pathway analysis, we observed that PD-L1 could regulate great numbers of target genes involved in tumor growth or taking part in many other signal pathways, such as cell growth, death, cell motility, translation, endocrine or immune system, and cancers. Thus, our findings indicate that PD-L1 can promote the growth of breast tumor through affecting many signaling pathways.
In next investigation, we tried to identify the regulatory mechanisms of PD-L1 overexpression in breast cancer. HBXIP, MAPK, and MTOR activator 5, is highly expressed in many cancers and plays critical roles in keeping cancer proliferation, invasion, and drug resistance as an oncogenic transcriptional co-activator [25, 29, 32, 33]. Accordingly, we are wondering if HBXIP takes part in PD-L1 regulation in the promotion of breast cancer. Our data revealed that both PD-L1 and HBXIP were highly expressed, and the expression of these two proteins was positively correlated in clinical breast cancer samples using qRT-PCR and IHC staining assays. The results from six breast cancer cell lines also confirmed the relationship between PD-L1 and HBXIP expression, and high expression of HBXIP or PD-L1 predicts poor prognosis. Further examination showed that HBXIP was able to induce the mRNA and protein levels and activate the promoter of CD274 in breast cancer cells. PD-L1 is mainly regulated by two signaling pathways including PI3K/AKT and MAPK. PI3K/AKT pathway and NF-κB can not only activate the PD-L1 transcription but also directly increase the PD-L1 protein level [15, 45, 47, 56, 57]. In melanoma cells, MAPK pathway activates PD-L1 and EGFR [58, 59]. Several oncogenic transcriptional factors, such as MEF2D, STAT3, HDAC8, and TEAD1 can also activate the promoter of PD-L1 in many different cancers [15, 17, 60]. Here, we found that there was one binding site of transcriptional factor ETS2 in the promoter of PD-L1. ETS2, as a target gene of MAPK signaling pathway, can be activated in various solid tumors [61]. Our result presented that HBXIP could interact with and co-activate ETS2, resulting in the PD-L1 transcription in breast cancer cells. Furthermore, by analyzing the data from the cBioportal for Cancer Genomic, we demonstrated that ETS2 was positively related to PD-L1 expression in 463 cases of breast cancer samples. Interestingly, we observed that the mRNA and protein levels of ETS2 were upregulated by HBXIP in breast cancer cells. Therefore, our studies reveal that ETS2 functions as a transcriptional factor of PD-L1 induced by HBXIP in breast cancer cells.
PTMs of PD-L1, such as phosphorylation, palmitoylation, glycosylation, and ubiquitination, have deemed as the significant mechanisms that mediate the protein stability of PD-L1 and tumor immunity. The study has shown that PD-L1 is highly N-glycosylated at N192, N219, and N200 [11, 62]. NF-κB p65-induced COP9 signalosome 5 (CSN5) can inhibit the ubiquitination and degradation of PD-L1 [3]. IL-6-activated JAK1 is able to phosphorylate Tyr112 of PD-L1 and then N-glycosyltransferase STT3A is recruited to catalyze the glycosylation and stability of PD-L1 [20]. PD-L1 can be palmitoylated at Cys272 site and subsequently stabilized to protect breast cancer cells from T-cell killing and promote tumor growth [63]. In the present study, in addition to the transcriptional modulation, we are interested in whether PTMs play a role in HBXIP-elevated PD-L1 in breast cancer. Our study has reported that HBXIP can promote the protein stability of HOXB13 via stimulating its acetylation [33]. In addition, HBXIP can induce the deacetylation and degradation of MST1 by upregulating HDAC6 during the development of breast cancer [31]. HBXIP can also promote abnormal O-glycosylation via modulating the expression and dislocation of GALNT1 in facilitating metastasis of breast cancer [32]. Recently, our group has revealed that HBXIP can activate the acetylation of HMGA2 at K26 site by PCAF in the progression of ESCC [34]. Therefore, in next study we analyzed the effect of HBXIP on PTMs of PD-L1. We found that HBXIP could stabilize PD-L1 at the posttranslational level.
Acetylation and methylation are the most widely affected epigenetic pathways in cancer [18]. The protein acetylation is mainly found in histone lysine residues or nonhistone lysine residues. The acetylation of nonhistone protein regulates key cellular processes related to diverse cellular processes including gene transcription, cell cycle, DNA damage repair, and cellular signaling [64,65,66,67]. Acetylation can influence protein functions through many different mechanisms, such as getting command of protein–protein, and protein–DNA connection, cross-regulation of other nonhistone modifications, or regulating protein stability [50, 68,69,70,71,72]. Previously published data has indicated that the EGF treatment might induce acetylation, phosphorylation, and ubiquitination of PD-L1 by a PTM enrichment screening [73]. However, there is no more deeply investigation for the functions of PD-L1 acetylation and its related mechanism.
In next study, we revealed that HBXIP could induce the acetylation of PD-L1 and its subsequent stability in breast cancer cells. According to the website prediction of conserved sequences of PD-L1 of various species, we found that there were two putative acetylation sites within PD-L1. Utilizing mutagenic experiment, we identified the conserved K270 residue as the mainly PD-L1 acetylation site. We then got a rabbit polyclonal antibody specifically recognizing the acetylated PD-L1-K270 to confirm whether K270 of PD-L1 can be acetylated in breast cancer cells. The specificity of AcK270-PD-L1 (Ac-K270) antibody was testified for it could recognize the K270-acetylated peptide and failed to recognize the unacetylated PD-L1 peptide. The AcK270-PD-L1 (Ac-K270) antibody was able to recognize acetylated PD-L1-WT at K270 and failed to recognize K270Q or K270R mutations. Further data showed that the K270R and K270Q mutant PD-L1 were more stable than WT PD-L1 in breast cancer cells. Thus, our data suggest that HBXIP can induce the acetylation of PD-L1 at K270 site and its stability in breast cancer.
CREB-binding protein (CBP), EP300 (p300), general control nonderepressible-5 (GCN5), and p300/CBP-associated factor (PCAF) are main members of acetyltransferase family required for the protein acetylation. To screen the acetyltransferase responsible for PD-L1 acetylation, we designed siRNAs targeting CBP, GCN5, p300, and PCAF. Our data indicated that only the p300 silencing effectively suppressed the HBXIP-mediated increase in PD-L1 acetylation. Given that p300 may be an acetyltransferase of PD-L1, we are wondering if p300 could interact with PD-L1. We observed the p300 interacts with exogenous PD-L1 in breast cancer cells. Further experiments showed that p300, PD-L1, and HBXIP were co-localized in the cells. Thus, our findings imply that HBXIP can recruit p300 to acetylate K270 of PD-L1, resulting in the increase of the protein stability of PD-L1. Except for acetylation, the other PTMs including lactylation or methylation in PD-L1 protein remains an open question. In the future, the screening of novel PTMs on PD-L1 may amplify the understanding of the regulatory network behind PD-L1 level and provide new directions to improve cancer therapy.
Functionally, HBXIP-mediated the acetylation of PD-L1 at K270 site induced its stability, leading to the cell proliferation in vitro and the tumor growth in vivo. Given that HBXIP promoted PD-L1 expression by activating its promoter and acetylation in breast cancer, we speculated that the inhibitor of HBXIP could also inhibit PD-L1 and improve the effect of anti-PD-L1 treatment in breast cancer. ASA, as a classic NSAID, is currently approved by FDA for treating colorectal cancer, which can also affect cell proliferation, apoptosis, migration, or other functions in some other cancers including ovarian cancer, colorectal cancer, breast cancer, or liver cancer [33, 35, 37, 40, 42]. Our study has identified that ASA can inhibit HBXIP/HOXB13 signaling to overcome tamoxifen resistance in breast cancer [33]. In ESCC, ASA is able to decrease the level of HBXIP and HMGA2 to bate the tumor growth [34]. An investigation in colorectal cancer with ASA treatment has shown that there is a better survival rate of the patients with low-level PD-L1 than that with high-level PD-L1, implying that adjuvant ASA treatment might be useful for PD-L1 immunotherapy [74]. Emerging evidence suggested a possible synergism of PD-1 immune checkpoint blockade and ASA-induced prostaglandin inhibition for combination immunotherapy treatment [75, 76]. Therefore, we are interested in the effect of ASA on HBXIP/PD-L1-induced breast cancer. Actually, we observed an obvious inhibition of HBXIP and PD-L1 expression in ASA-treated breast cancer cells. Furthermore, ASA could suppress the growth of breast cancer in vitro and in vivo. Notably, Western blotting and IHC staining manifested that the protein levels of HBXIP and PD-L1, as well as the level of Ac-K270-PD-L1 were markedly reduced in ASA-treated mouse tumors, which further supported our above findings that ASA could reduce breast cancer growth via targeting HBXIP and PD-L1. Our results present a potential therapeutic use of ASA for breast cancer with the high expression of HBXIP and PD-L1.
In summary, our results demonstrate a new mechanism of the activation and acetylation of PD-L1 via HBXIP in breast cancer (Supplementary Fig. S8). On the one hand, oncogenic HBXIP can induce the transcription of CD274 through upregulating and co-activating the transcriptional factor, ETS2. On the other hand, HBXIP can increase the acetylation of PD-L1 at K270 site by binding to the acetyltransferase p300, subsequently resulting in the stability and accumulation of PD-L1 protein in breast cancer cells. PD-L1 is highly expressed and promotes breast cancer growth via regulating multiple signal pathways. ASA prevents the growth of breast cancer through targeting HBXIP and PD-L1. Thus, HBXIP and PD-L1 may serve as a new biomarker for breast cancer and as a potential target for the treatment of breast cancer patients. Therapeutically, ASA can be a promising strategy for breast cancer treatment through targeting HBXIP and PD-L1.
References
Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64.
Cai G, Karni A, Oliveira EM, Weiner HL, Hafler DA, Freeman GJ. PD-1 ligands, negative regulators for activation of naive, memory, and recently activated human CD4+ T cells. Cell Immunol. 2004;230:89–98.
Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, et al. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925–39.
Cao D, Qi Z, Pang Y, Li H, Xie H, Wu J, et al. Retinoic acid-related orphan receptor C regulates proliferation, glycolysis, and chemoresistance via the PD-L1/ITGB6/STAT3 signaling axis in bladder cancer. Cancer Res. 2019;79:2604–18.
Ritprajak P, Azuma M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol. 2015;51:221–8.
Wang S, Li J, Xie J, Liu F, Duan Y, Wu Y, et al. Programmed death ligand 1 promotes lymph node metastasis and glucose metabolism in cervical cancer by activating integrin beta4/SNAI1/SIRT signaling pathway. Oncogene. 2018;37:4164–80.
Hersey P, Gallagher S. A focus on PD-L1 in human melanoma. Clin Cancer Res. 2013;19:514–6.
Clark CA, Gupta HB, Sareddy G, Pandeswara S, Lao S, Yuan B, et al. Tumor-intrinsic PD-L1 signals regulate cell growth, pathogenesis, and autophagy in ovarian cancer and melanoma. Cancer Res. 2016;76:6964–74.
Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, et al. Aberrant PD-L1 expression through 3'-UTR disruption in multiple cancers. Nature. 2016;534:402–6.
Liu H, Kuang X, Zhang Y, Ye Y, Li J, Liang L, et al. ADORA1 inhibition promotes tumor immune evasion by regulating the ATF3-PD-L1 axis. Cancer Cell. 2020;37:324–39.e8.
Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632.
Asgarova A, Asgarov K, Godet Y, Peixoto P, Nadaradjane A, Boyer-Guittaut M, et al. PD-L1 expression is regulated by both DNA methylation and NF-κB during EMT signaling in non-small cell lung carcinoma. Oncoimmunology. 2018;7:e1423170.
Kwak G, Kim D, Nam GH, Wang SY, Kim IS, Kim SH, et al. Programmed cell death protein ligand-1 silencing with polyethylenimine-dermatan sulfate complex for dual inhibition of melanoma growth. ACS Nano. 2017;11:10135–46.
Chen Y, Liu Q, Chen Z, Wang Y, Yang W, Hu Y, et al. PD-L1 expression and tumor mutational burden status for prediction of response to chemotherapy and targeted therapy in non-small cell lung cancer. J Exp Clin Cancer Res. 2019;38:193.
Feng J, Yang H, Zhang Y, Wei H, Zhu Z, Zhu B, et al. Tumor cell-derived lactate induces TAZ-dependent upregulation of PD-L1 through GPR81 in human lung cancer cells. Oncogene. 2017;36:5829–39.
Maeda T, Hiraki M, Jin C, Rajabi H, Tagde A, Alam M, et al. MUC1-C induces PD-L1 and immune evasion in triple-negative breast cancer. Cancer Res. 2018;78:205–15.
Xiang J, Zhang N, Sun H, Su L, Zhang C, Xu H, et al. Disruption of SIRT7 increases the efficacy of checkpoint inhibitor via MEF2D regulation of programmed cell death 1 ligand 1 in hepatocellular carcinoma cells. Gastroenterology. 2020;158:664–78.e24.
Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27.
Wan J, Xu W, Zhan J, Ma J, Li X, Xie Y, et al. PCAF-mediated acetylation of transcriptional factor HOXB9 suppresses lung adenocarcinoma progression by targeting oncogenic protein JMJD6. Nucleic Acids Res. 2016;44:10662–75.
Chan LC, Li CW, Xia W, Hsu JM, Lee HH, Cha JH, et al. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest. 2019;129:3324–38.
Yao H, Lan J, Li C, Shi H, Brosseau JP, Wang H, et al. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat Biomed Eng. 2019;3:306–17.
Hsu JM, Li CW, Lai YJ, Hung MC. Posttranslational modifications of PD-L1 and their applications in cancer therapy. Cancer Res. 2018;78:6349–53.
Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–208.
Marusawa H, Matsuzawa S, Welsh K, Zou H, Armstrong R, Tamm I, et al. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003;22:2729–40.
Zhao Y, Li H, Zhang Y, Li L, Fang R, Li Y, et al. Oncoprotein HBXIP modulates abnormal lipid metabolism and growth of breast cancer cells by activating the LXRs/SREBP-1c/FAS signaling cascade. Cancer Res. 2016;76:4696–707.
Wang Y, Fang R, Cui M, Zhang W, Bai X, Wang H, et al. The oncoprotein HBXIP up-regulates YAP through activation of transcription factor c-Myb to promote growth of liver cancer. Cancer Lett. 2017;385:234–42.
Liu S, Li L, Zhang Y, Zhang Y, Zhao Y, You X, et al. The oncoprotein HBXIP uses two pathways to up-regulate S100A4 in promotion of growth and migration of breast cancer cells. J Biol Chem. 2012;287:30228–39.
Jiang Y, Wang D, Ren H, Shi Y, Gao Y. Oncogenic HBXIP enhances ZEB1 through Sp1 to accelerate breast cancer growth. Thorac Cancer. 2018;9:1664–70.
Li H, Liu Q, Wang Z, Fang R, Shen Y, Cai X, et al. The oncoprotein HBXIP modulates the feedback loop of MDM2/p53 to enhance the growth of breast cancer. J Biol Chem. 2015;290:22649–61.
Xu F, You X, Liu F, Shen X, Yao Y, Ye L, et al. The oncoprotein HBXIP up-regulates Skp2 via activating transcription factor E2F1 to promote proliferation of breast cancer cells. Cancer Lett. 2013;333:124–32.
Li L, Fang R, Liu B, Shi H, Wang Y, Zhang W, et al. Deacetylation of tumor-suppressor MST1 in Hippo pathway induces its degradation through HBXIP-elevated HDAC6 in promotion of breast cancer growth. Oncogene. 2016;35:4048–57.
Fang R, Xu F, Shi H, Wu Y, Cao C, Li H, et al. LAMTOR5 raises abnormal initiation of O-glycosylation in breast cancer metastasis via modulating GALNT1 activity. Oncogene. 2019;39:2290–30.
Liu B, Wang T, Wang H, Zhang L, Xu F, Fang R, et al. Oncoprotein HBXIP enhances HOXB13 acetylation and co-activates HOXB13 to confer tamoxifen resistance in breast cancer. J Hematol Oncol. 2018;11:26.
Wu Y, Wang X, Xu F, Zhang L, Wang T, Fu X, et al. The regulation of acetylation and stability of HMGA2 via the HBXIP-activated Akt-PCAF pathway in promotion of esophageal squamous cell carcinoma growth. Nucleic Acids Res. 2020;48:4858–76.
Chan AT, Ogino S, Fuchs CS. Aspirin use and survival after diagnosis of colorectal cancer. JAMA. 2009;302:649–58.
McCowan C, Munro AJ, Donnan PT, Steele RJ. Use of aspirin post-diagnosis in a cohort of patients with colorectal cancer and its association with all-cause and colorectal cancer specific mortality. Eur J Cancer. 2013;49:1049–57.
Trabert B, Ness RB, Lo-Ciganic WH, Murphy MA, Goode EL, Poole EM, et al. Aspirin, nonaspirin nonsteroidal anti-inflammatory drug, and acetaminophen use and risk of invasive epithelial ovarian cancer: a pooled analysis in the Ovarian Cancer Association Consortium. J Natl Cancer Inst. 2014;106:djt431.
Downer MK, Allard CB, Preston MA, Gaziano JM, Stampfer MJ, Mucci LA, et al. Regular aspirin use and the risk of lethal prostate cancer in the physicians’ health study. Eur Urol. 2017;72:821–7.
Guo H, Liu J, Ben Q, Qu Y, Li M, Wang Y, et al. The aspirin-induced long non-coding RNA OLA1P2 blocks phosphorylated STAT3 homodimer formation. Genome Biol. 2016;17:24.
Din FV, Valanciute A, Houde VP, Zibrova D, Green KA, Sakamoto K, et al. Aspirin inhibits mTOR signaling, activates AMP-activated protein kinase, and induces autophagy in colorectal cancer cells. Gastroenterology. 2012;142:1504–15.e3.
Kim MH, Chang J, Kim WJ, Banerjee S, Park SM. Cumulative dose threshold for the chemopreventive effect of aspirin against gastric cancer. Am J Gastroenterol. 2018;113:845–54.
Wang T, Fu X, Jin T, Zhang L, Liu B, Wu Y, et al. Aspirin targets P4HA2 through inhibiting NF-kappaB and LMCD1-AS1/let-7g to inhibit tumour growth and collagen deposition in hepatocellular carcinoma. EBioMedicine. 2019;45:168–80.
Holmes MD, Chen WY, Li L, Hertzmark E, Spiegelman D, Hankinson SE. Aspirin intake and survival after breast cancer. J Clin Oncol. 2010;28:1467–72.
Hu N, Zhang J, Cui W, Kong G, Zhang S, Yue L, et al. miR-520b regulates migration of breast cancer cells by targeting hepatitis B X-interacting protein and interleukin-8. J Biol Chem. 2011;286:13714–22.
Bouillez A, Rajabi H, Jin C, Samur M, Tagde A, Alam M, et al. MUC1-C integrates PD-L1 induction with repression of immune effectors in non-small-cell lung cancer. Oncogene. 2017;36:4037–46.
Kleffel S, Posch C, Barthel SR, Mueller H, Schlapbach C, Guenova E, et al. Melanoma cell-intrinsic PD-1 receptor functions promote tumor growth. Cell. 2015;162:1242–56.
Lastwika KJ, Wilson W 3rd, Li QK, Norris J, Xu H, Ghazarian SR, et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76:227–38.
Cai X, Cao C, Li J, Chen F, Zhang S, Liu B, et al. Inflammatory factor TNF-α promotes the growth of breast cancer via the positive feedback loop of TNFR1/NF-κB (and/or p38)/p-STAT3/HBXIP/TNFR1. Oncotarget. 2017;8:58338–52.
Huang C, Zhang Z, Chen L, Lee HW, Ayrapetov MK, Zhao TC, et al. Acetylation within the N- and C-terminal domains of Src regulates distinct roles of STAT3-mediated tumorigenesis. Cancer Res. 2018;78:2825–38.
Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–61.
Dai X, Yan J, Fu X, Pan Q, Sun D, Xu Y, et al. Aspirin inhibits cancer metastasis and angiogenesis via targeting heparanase. Clin Cancer Res. 2017;23:6267–78.
Zhang X, Feng H, Du J, Sun J, Li D, Hasegawa T, et al. Aspirin promotes apoptosis and inhibits proliferation by blocking G0/G1 into S phase in rheumatoid arthritis fibroblast-like synoviocytes via downregulation of JAK/STAT3 and NF-kappaB signaling pathway. Int J Mol Med. 2018;42:3135–48.
Sitia G, Aiolfi R, Di Lucia P, Mainetti M, Fiocchi A, Mingozzi F, et al. Antiplatelet therapy prevents hepatocellular carcinoma and improves survival in a mouse model of chronic hepatitis B. Proc Natl Acad Sci USA. 2012;109:E2165–72.
Chen Z, Li W, Qiu F, Huang Q, Jiang Z, Ye J, et al. Aspirin cooperates with p300 to activate the acetylation of H3K9 and promote FasL-mediated apoptosis of cancer stem-like cells in colorectal cancer. Theranostics. 2018;8:4447–61.
Hartley GP, Chow L, Ammons DT, Wheat WH, Dow SW. Programmed cell death ligand 1 (PD-L1) signaling regulates macrophage proliferation and activation. Cancer Immunol Res. 2018;6:1260–73.
Gowrishankar K, Gunatilake D, Gallagher SJ, Tiffen J, Rizos H, Hersey P. Inducible but not constitutive expression of PD-L1 in human melanoma cells is dependent on activation of NF-kappaB. PLoS ONE. 2015;10:e0123410.
Song M, Chen D, Lu B, Wang C, Zhang J, Huang L, et al. PTEN loss increases PD-L1 protein expression and affects the correlation between PD-L1 expression and clinical parameters in colorectal cancer. PLoS ONE. 2013;8:e65821.
Jiang X, Zhou J, Giobbie-Hurder A, Wargo J, Hodi FS. The activation of MAPK in melanoma cells resistant to BRAF inhibition promotes PD-L1 expression that is reversible by MEK and PI3K inhibition. Clin Cancer Res. 2013;19:598–609.
Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-Driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J Thorac Oncol. 2015;10:910–23.
Wang YF, Liu F, Sherwin S, Farrelly M, Yan XG, Croft A, et al. Cooperativity of HOXA5 and STAT3 Is critical for HDAC8 inhibition-mediated transcriptional activation of PD-L1 in human melanoma cells. J Investig Dermatol. 2018;138:922–32.
Han M, Yan W, Guo W, Xi D, Zhou Y, Li W, et al. Hepatitis B virus-induced hFGL2 transcription is dependent on c-Ets-2 and MAPK signal pathway. J Biol Chem. 2008;283:32715–29.
Lee HH, Wang YN, Xia W, Chen CH, Rau KM, Ye L, et al. Removal of N-Linked glycosylation enhances PD-L1 detection and predicts Anti-PD-1/PD-L1 therapeutic efficacy. Cancer Cell. 2019;36:168–78.e4.
Yang Y, Hsu JM, Sun L, Chan LC, Li CW, Hsu JL, et al. Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res. 2019;29:83–6.
Reed SM, Quelle DE. p53 acetylation: regulation and consequences. Cancers. 2014;7:30–69.
Kawasumi R, Abe T, Arakawa H, Garre M, Hirota K, Branzei D. ESCO1/2’s roles in chromosome structure and interphase chromatin organization. Genes Dev. 2017;31:2136–50.
Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–25.
Li Y, Xu W, McBurney MW, Longo VD. SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008;8:38–48.
Fujisawa T, Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat Rev Mol Cell Biol. 2017;18:246–62.
Westerheide SD, Anckar J, Stevens SM Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–6.
Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277:50607–11.
Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43:33–44.
Zhao D, Zou SW, Liu Y, Zhou X, Mo Y, Wang P, et al. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell. 2013;23:464–76.
Horita H, Law A, Hong S, Middleton K. Identifying regulatory posttranslational modifications of PD-L1: a focus on monoubiquitinaton. Neoplasia. 2017;19:346–53.
Hamada T, Cao Y, Qian ZR, Masugi Y, Nowak AJ, Yang JH, et al. Aspirin use and colorectal cancer survival according to tumor CD274 (Programmed Cell Death 1 Ligand 1) expression status. J Clin Oncol. 2017;35:1836–44.
Hutchinson L. Immunotherapy: evading immune escape: synergy of COX and immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2015;12:622.
Wang D, DuBois RN. The role of prostaglandin E2 in tumor-associated immunosuppression. Trends Mol Med. 2016;22:1–3.
Acknowledgements
This work was supported by National Natural Science Foundation of China (82072943, 82072929, and 31870752), and Project of Prevention and Control of Key Chronic Non-Infectious Diseases (No. 2016YFC1303401).
Author information
Authors and Affiliations
Contributions
FFX, WYZ, and LHY designed and conceived experiments. FFX designed the research methods, analyzed the data, and drafted the paper. FFX, HMS, and RPF performed the experiments and analyzed the data. LZ, HS, XW, XLF, XML, XHS, YW, and KY participated in the experiments. FFX, WYZ, and LHY revised the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Xu, Ff., Sun, Hm., Fang, Rp. et al. The modulation of PD-L1 induced by the oncogenic HBXIP for breast cancer growth. Acta Pharmacol Sin 43, 429–445 (2022). https://doi.org/10.1038/s41401-021-00631-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-021-00631-6