Abstract
Phenethyl isothiocyanate (PEITC) is an isothiocyanate that largely exists in cruciferous vegetables and exhibits chemopreventive and chemotherapeutic potential against various cancers. However, it is little known about the molecular mechanisms of its antitumor action against osteosarcoma, which is the second highest cause of cancer-related death in children and adolescents. In this study, we investigated the effects of PEITC on K7M2 murine osteosarcoma both in vitro and in vivo. We found that treatment with PEITC dose-dependently inhibited the viability of K7M2 murine osteosarcoma cells with an IC50 value of 33.49 μM at 24 h. PEITC (1, 15, 30 μM) dose-dependently inhibited the cell proliferation, caused G2/M cell cycle arrest, depleted glutathione (GSH), generated reactive oxygen species (ROS), altered iron metabolism, and triggered multiple forms of cell death, namely ferroptosis, apoptosis, and autophagy in K7M2 cells. We further revealed that PEITC treatment activated MAPK signaling pathway, and ROS generation was a major cause of PEITC-induced cell death. In a syngeneic orthotopic osteosarcoma mouse model, administration of PEITC (30, 60 mg/kg every day, ig, for 24 days) significantly inhibited the tumor growth, but higher dose of PEITC (90 mg/kg every day) compromised its anti-osteosarcoma effect. Histological examination showed that multiple cell death processes were initiated, iron metabolism was altered and MAPK signaling pathway was activated in the tumor tissues. In conclusion, we demonstrate that PEITC induces ferroptosis, autophagy, and apoptosis in K7M2 osteosarcoma cells by activating the ROS-related MAPK signaling pathway. PEITC has promising anti-osteosarcoma activity. This study sheds light on the redox signaling-based chemotherapeutics for cancers.
Similar content being viewed by others
Introduction
Redox metabolism and cell physiology are closely intertwined. The redox system is a molecular target for the development of efficient treatment strategies for cancers [1]. It was reported that reactive oxygen species (ROS) affect cell proliferation and cell survival. Normal and cancer cells present important differences in responding to oxidative stress. Tumor cells normally have persistently high ROS levels and are more vulnerable to ROS due to excessive oxidative stress [2]. Therefore, due to the crucial role of ROS in the initiation and progression of tumors, inducing abnormal ROS generation and accumulation might be a useful antitumor strategy [3].
The glutathione (GSH) system, one of the antioxidant pathways in cells, prevents toxic ROS accumulation; moreover, this system also maintains important redox signaling functions [4]. GSH acts as a core regulator of the GSH antioxidant system and is the most abundant ROS scavenging buffer in living organisms. Cumulative evidence indicates that GSH biosynthesis and homeostasis are poteintial antitumor therapeutic targets [5]. Strategies for depleting cellular GSH to induce oxidative stress include the creation of a shortage of source material for GSH synthesis, inhibition of the GSH synthesis process, direct conjugation of GSH, and the promotion of cellular GSH efflux [6,7,8,9].
β-Phenethyl isothiocyanate (PEITC) is a natural isothiocyanate compound that abundantly exists in cruciferous vegetables and has antitumor therapeutic potential against a variety of cancers [10,11,12,13,14]. PEITC is rapidly absorbed and has high bioavailability [15]. Studies have shown that daily oral administration of PEITC exhibits antitumor activity in both preclinical and clinical trials against lung cancer [14, 16, 17]. One of the mechanisms by which PEITC exerts its antitumor effects is by directly depleting cellular GSH and causing oxidative stress in cancer cells [11].
Osteosarcoma, the most common primary bone malignancy in children and young adults, is characterized by frequent lung metastasis and pathologic bone destruction [18]. Although the treatments (chemotherapy, aggressive surgery, and radiotherapy) for osteosarcomas have improved, people with osteosarcomas still have a very poor prognosis [19]. Therefore, it is of great importance to develop targeted therapeutic strategies for osteosarcoma. The antitumor effects of PEITC have been widely reported, but the molecular mechanisms underlying its actions on osteosarcoma still need to be clarified. In the present study, the antitumor activity of PEITC on murine osteosarcoma was thoroughly studied. We demonstrated that multiple forms of cell death, mainly ferroptosis, autophagy, and apoptosis, were triggered in PEITC-treated K7M2 osteosarcoma cells with decreased GSH, elevated labile iron, increased ROS, and activation of MAPK signaling pathways.
Material and methods
Drugs and chemicals
PEITC was purchased from Shanghai Hushi Laboratorial Equipment Co., Ltd (Shanghai, China). Necrostatin-1 (Nec-1), liproxstatin-1 (Lip-1), bafilomycin A1 (Baf-A1), and 3-methyladenine (3-MA) were purchased from Selleck Chemicals (Houston, TX, USA). Ferrostatin-1 (Fer-1), deferoxamine (DFO), and N-acetyl-L-cysteine (NAC) were purchased from Sigma-Aldrich (Santa Clara, CA, USA). z-VAD-FMK, CCK-8 kit, PI/RNase staining buffer, ROS kit, lipid peroxidation assay kits, GSH/GSSG assay kits, Annexin V-FITC apoptosis detection kit, the JC-1 fluorescent probe, BCA protein assay kits, and antibodies against Caspase3, C-caspase3, Cyclin D1, Cyclin D3, Ki67, and C-PARP were purchased from Beyotime Biotechnology (Shanghai, China). Cell-LightTM EdU® Apollo567 in vitro imaging kit was purchased from RiboBio (Guangzhou, China). Antibodies against β-actin, Beclin1, p62, Cdc2, mTOR, p-mTOR, Stat3, p-Stat3, ERK, p-ERK, p38, p-p38, JNK, and p-JNK were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibodies against Bcl2, Bax, LC3B, FTH1, and GPx4 were purchased from Abcam (Cambridge, MA, USA). Mitotraker TM Green FM and antibodies against TfR1, FPN and DMT1, BODIPY 581/591 C11 and Calcein-AM were purchased from Thermo Fisher Scientific (Waltham, MA, USA). HiScript II Q RT SuperMix and chamQ SYBR qPCR Master Mix were purchased from Vazyme Biotechnology (Nanjing, China). Lysosome staining kit was purchased from Abnova (Taiwan, China).
Cell viability assay
K7M2 murine osteosarcoma cell line used in this study was purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 100 units/mL penicillin–streptomycin at 37 °C in a 5% CO2 incubator. Cell viability was determined by CCK-8 assay. K7M2 cells were seeded in 96-well cell culture plates at a density of 5 × 104 cells/mL and incubated overnight. The cells were treated with different concentrations of PEITC or were treated with PEITC in the presence of 50 μM z-VAD-FMK, 50 μM Nec-1, 10 μM Fer-1, 100 nM Lip-1, 50 μM DFO, 1 μM Baf-A1, 10 mM 3-MA, or 1 mM NAC for 24 h. At the end of the incubation, CCK-8 solution was added to each well, and incubated at 37 °C for 2 h. The plates were read by a Synergy HT multimode microplate reader (BioTek, Winooski, VT, USA) at a wavelength of 450 nm.
Cell proliferation assay
The EdU cell proliferation assay was performed by following the instructions of the Cell-LightTM EdU ® Apollo567 in vitro imaging kit. After PEITC treatment, K7M2 cells were incubated with EdU solution and 1 mL of 4% formaldehyde solution was added and incubated for 10 min. The cells were washed with phosphate-buffered saline (PBS), and then 0.5% Triton X-100 solution was added and incubated at room temperature for 20 min. After removal of the permeabilization buffer, reaction solution was added and the plate was rocked for 30 min at room temperature protected from light. After the reaction, the cells were washed once with PBS, and incubated with Hoechst 33342 solution for 30 min at room temperature protected from light. Hoechst 33342 solution was removed, and the cells were washed with PBS. Finally, the images were captured by an MD IL HC inverted fluorescence microscope (Leica, Wetzlar, Germany).
Clone formation assay
After PEITC treatment, K7M2 cells were detached using trypsin, and an equal number of cells from each treatment were reseeded in six-well plates. The medium was changed every 3 days for 10 consecutive days. The colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 15 min at room temperature. The dye was washed with PBS. The images were photographed by a stereomicroscope (Canon Inc., Tokyo, Japan) and the colonies were counted.
Cell cycle analysis
The cell cycle was analyzed by flow cytometry with PI/RNase staining buffer. After PEITC treatment, K7M2 cells were washed with PBS, trypsinized, and harvested by centrifugation at 1000 × g for 5 min at 4 °C. The cells were once washed with PBS and the pellets were resuspended in 1 mL of 70% ethanol and stored at −4 °C for 24 h. The cells were recentrifuged at 1000 × g for 5 min and washed once with 1 mL cold PBS and resuspended in 500 μL of PI staining solution. The cell suspension was incubated for 30 min at 37 °C in the dark and analyzed on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
Measurement of cytosolic ROS
The generation of intracellular ROS was measured by using ROS kit. After PEITC treatment, K7M2 cells were collected and incubated with DCFH-DA sensor for 30 min at 37 °C protected from light. The stained cells were washed twice with PBS and analyzed by a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
Measurement of lipid ROS
The generation of lipid ROS was evaluated by using BODIPY 581/591 C11. After PEITC treatment, 10 μM BODIPY 581/591 C11 solution was added and K7M2 cells were incubated for 30 min at 37 °C protected from light. Excess BODIPY 581/591 C11 was removed by washing the cells with PBS for three times. Then the cells were imaged by an MD IL HC inverted fluorescence microscope (Leica, Wetzlar, Germany).
Measurement of malondialdehyde
Malondialdehyde (MDA) levels were measured by using a lipid peroxidation MDA assay kit. After PEITC treatment, K7M2 cells were washed with cold PBS, lysed by RIPA lysis buffer, and centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant was collected to determine the MDA level and protein concentration. MDA reacts with thiobarbituric acid (TBA) forming MDA-TBA2 adducts that absorb strongly at 535 nm. MDA was measured by a Synergy HT multimode microplate reader (BioTek, Winooski, Vermont, USA) at 535 nm and the MDA levels were normalized to the protein concentration.
Measurement of GSH/GSSG
The levels of total glutathione and oxidized glutathione were measured by using a GSH/GSSG assay kit. After PEITC treatment, K7M2 cells were washed with PBS, trypsinized, harvested, and lysed by two cycles of freezing and thawing. The samples were then centrifuged at 10,000 × g for 10 min at 4 °C, and the supernatant was collected for determination of total GSH and GSSG. GSH reacts with 5,5′-dithiobis (2-nitrobenzoic acid) to form a stable color with absorbance at 412 nm. Intracellular GSH was determined by using a Synergy HT multimode microplate reader (BioTek, Winooski, VT, USA) at 412 nm. Reduced GSH was determined by subtracting GSSG from the total GSH. Then the ratio of GSH/GSSG was calculated.
Cellular labile iron staining
The relative changes in cellular labile iron were evaluated with calcein-acetoxymethyl ester (calcein-AM). After PEITC treatment, K7M2 cells were washed with PBS and incubated with 1 μM calcein-AM for 15 min. The cells were washed with PBS again and imaged by aN MD IL HC inverted fluorescence microscope (Leica, Wetzlar, Germany).
Iron quantification
The amount of total iron was determined by atomic absorption spectrometer (AAS) (Analytik, Jena, Germany). After PEITC treatment, K7M2 cells were washed with PBS, trypsinized, and harvested by centrifugation at 1000 × g for 5 min at 4 °C. The cells were washed once with PBS and resuspended in PBS for cell counting, protein quantification, and iron quantification. The cell samples for iron quantification were centrifuged and lysed with pure HNO3 at 70 °C for 2 h. Finally, the total iron level was determined by AAS and normalized to the protein concentration and cell number.
Apoptosis assay
Apoptosis was detected by an Annexin V-FITC Apoptosis Detection Kit. After PEITC treatment, K7M2 cells were washed with PBS. Then, 195 μL of binding buffer was added, and the cells were stained with 5 μL of FITC-Annexin V for 10 min at room temperature. The cells were incubated with 10 μL of PI for 10 min in the dark and imaged by an MD IL HC inverted fluorescence microscope (Leica, Wetzlar, Germany).
Morphological observation of mitochondria and nuclei
The mitochondria and nuclei were labeled by MitoTraker TM Green FM and Hoechst 33342, respectively. After PEITC treatment, K7M2 cells were washed with PBS and stained with 20 nM MitoTraker TM Green and Hoechst 33342 for 30 min at 37 °C. Cells were washed with PBS again and imaged by an MD IL HC inverted fluorescence microscope (Leica, Wetzlar, Germany).
Measurement of mitochondrial transmembrane potential
The mitochondrial transmembrane potential was measured with a JC-1 fluorescent probe. After PEITC treatment, K7M2 cells were collected, washed with PBS, and incubated with JC-1 for 20 min at 37 °C. The stained cells were washed with PBS and analyzed by a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
Detection of acidic vesicular organelles
To detect the formation of acidic vesicular organelles, cells were stained with acridine orange (AO). After PEITC treatment, K7M2 cells were washed with PBS and incubated with 10 μM AO solution for 15 min at 37 °C. Cells were washed with PBS three times and then imaged by an MD IL HC inverted fluorescence microscope (Leica, Wetzlar, Germany).
Lysosome staining
Lysosomes were observed by lysosome staining kit (green fluorescence). After PEITC treatment, K7M2 cells were washed with PBS and incubated with lysosome staining solution for 30 min at 37 °C. The cells were washed with PBS and imaged on an MD IL HC inverted fluorescence microscope (Leica, Wetzlar, Germany).
LC3 fluorescence imaging
After PEITC treatment, K7M2 cells were washed with PBS, fixed with 4% paraformaldehyde for 30 min, and permeabilized with 0.1% Triton X-100 for 10 min. Then, the cells were blocked with 5% BAS solution for 30 min and incubated with anti-LC3B primary antibody overnight. Finally, the cells were incubated with FITC-conjugated secondary antibody for 2 h, stained with DAPI for 5 min, and washed with PBS. Images were captured by an Eclipse 80i fluorescence microscope (Nikon, Tokyo, Japan).
Western blotting analysis
Protein samples from K7M2 cells were lysed in RIPA lysis buffer containing 1% protease inhibitor cocktail and 1% phosphatase inhibitor for 20 min on ice. Lysates were centrifuged at 12,000 × g for 10 min and the supernatant was collected. The protein concentration was determined using a BCA protein assay kit according to the manufacturer’s instructions. Equal amounts of proteins were separated by SDS-PAGE. Then, the proteins were transferred onto PVDF membranes. The membranes were blocked with 5% skimmed milk at room temperature for 3 h and then incubated with primary antibodies at 4 °C for overnight. After washing with TBST solution, the membranes were incubated with an HRP-conjugated secondary antibody at room temperature for 2 h. Each band was visualized by BeyoECL Moon on a T5200 multi automatic fluorescence/chemiluminescence imaging system (BioTanon, Shanghai, China).
RNA isolation and qRT-PCR analysis
Total RNA from K7M2 cells was extracted by TRIizol. mRNA was reverse-transcribed into cDNA using HiScript II Q RT SuperMix. The relative mRNA expression levels of TFRC, SLC11A2, FTH1, and SLC40A1 were determined by using qPCR with the cDNA template and ChamQ SYBR qPCR master mix in a CFX96 Touch qPCR System (BioRad, Hercules, CA, USA). The sequences of the forward and reverse primers used are as follows: GADPH forward: 5′-TGC-ACC-ACC-AAC-TGC-TTA-G-3′, reverse: 5′-GGA-TGC-AGG-GAT-GAT-GTT-C-3′; TFRC, forward: 5′-TGG-GCA-GAC-CTC-AAA-ACA-CT, reverse: 5′-TTC-TGG-CTC-AGC-TGC-TTG-AT-3′; SLC11A2, forward: 5′-CAA-TGT-CTT-TGT-CGT-GTC-CGT-3′, reverse: 5′-GCG-ACC- ATT-TTA-GGT-TCA-GGA-AT-3′; FTH1, forward: 5′-CAA-GTG-CGC-CAG-AAC-TAC-CA-3′, reverse: 5′-GCC-ACA-TCA-TCT-CGG-TCA-AAA-3′; SLC40A1, forward: 5′-CTA-TGG-ACT-GGT-GGT-GGC-AG-3′, reverse: 5′-GTG-ACG-TCT-GGG-CCA-CTT-TA-3′. The primers for these genes were synthesized by Sangon Biotech Co., Ltd (Shanghai, China). The data were calibrated to GAPDH and analyzed via the 2−ΔΔCt method.
Establishment of the syngeneic orthotopic osteosarcoma mouse model and PEITC treatment
Male BALB/c mice, aged 4 weeks (14–16 g), were purchased from SPF (Beijing) Biotechnology Co., Ltd (Beijing, China, No. 11401500037845). The mice were provided sterilized food and water. The mice were given at least 1 week for adaptation before starting any experiments. All experimental procedures in the in vivo studies were conducted in accordance with protocols approved by the Northwestern Polytechnical University Animal Care and Use Committee. A syngeneic orthotopic osteosarcoma mouse model was established. Briefly, K7M2 osteosarcoma cells were harvested and suspended in PBS at 4 °C. Mice were anesthetized with isoflurane. The left hindlimbs were shaved and cleaned with 75% ethanol. The knees of the mice were flexed beyond 90° and the cortex of the proximal tibial crest was penetrated using a 25 gauge needle by a rotating action. Once the tibial bone cortex was penetrated, the needles were further inserted 2 mm along the diaphysis, followed by injection of 10 μL of K7M2 osteosarcoma cells (5 × 105 cells) into the tibia. To assess the effect of PEITC on tumor growth, mice with established tumors (mean starting tumor volume: 400 mm3) were randomly divided into four groups (n = 7) that received vehicle (10% sesame-seed oil with saline), 30 mg/kg PEITC (PEITC in 10% sesame-seed oil with saline), 60 mg/kg PEITC, or 90 mg/kg PEITC. All treatments were intragastrically administered once daily for 24 consecutive days. Tumor volumes were calculated from caliper measurements using the standard formula of volume = length × width2 × 0.5. Upon terminal sacrifice, the heart, liver, spleen, lung, kidney, and tumor tissues were harvested, snap-frozen in liquid nitrogen, and stored at −80 °C.
Histopathology and immunohistochemistry
The heart, liver, spleen, lung, kidney, and tumor tissues were collected and placed in 4% paraformaldehyde overnight. Tissue samples were embedded in paraffin and 4 μm sections were cut. Heart, liver, spleen, lung, kidney, and tumor tissue sections were stained with hematoxylin and eosin (H&E) for routine histological examinations and morphometric analysis. For immunohistochemical staining, tumor slides were deparaffinized in xylene, rehydrated with graded alcohol, and incubated in 3% H2O2 to block endogenous peroxidase activity. Antigen retrieval was performed by boiling the slides in 10 mM sodium citrate (pH 6.0) for 30 min. Then, the slides were blocked in 10% normal goat serum for 15 min, followed by incubation with primary antibodies against Ki67, GPx4, C-caspase3, and LC3B at 4 °C overnight. On the next day, the slides were washed with PBS and incubated with secondary antibody for 1 h at room temperature. Diaminobenzidine (DAB) was used as the chromogenic substrate, and nuclei were counterstained by using Harris hematoxylin. The sections were visualized using an Eclipse 80i fluorescence microscope (Nikon, Tokyo, Japan).
Statistical analysis
Statistical analysis was performed using GraphPad Prism software and all data are displayed as the mean ± SD. Student’s t-test or one-way analyses of variance was used for statistical analysis. Significance was defined at *P < 0.05, **P < 0.01, and ***P < 0.001 vs. the indicated control group.
Results
PEITC reduced cell viability, inhibited cell proliferation, and triggered cell death in K7M2 murine osteosarcoma cells
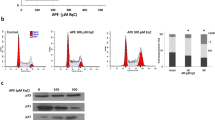
To evaluate the effect of PEITC on K7M2 osteosarcoma cells, we first observed cell morphological changes by using a phase-contrast microscope. Compared with the morphology of cells in the control group, PEITC-treated K7M2 osteosarcoma cells became round and the cell number was reduced as the concentration increased (Fig. 1a). Then, a CCK-8 assay was used to assess the cell viability of K7M2 osteosarcoma cells after treatment with different concentrations of PEITC for different time periods. The results showed that PEITC reduced the viability of K7M2 osteosarcoma cells (Fig. 1b). The IC50 values of PEITC on K7M2 osteosarcoma cells were 33.49, 38.23, and 42.06 μM for 24, 48, and 72 h, respectively.
PEITC reduced cell viability, inhibited cell proliferation, and triggered cell death in K7M2 osteosarcoma cells. a Representative cellular morphological images of K7M2 osteosarcoma cells treated with indicated concentrations of PEITC for 24 h. b Cell viability of K7M2 osteosarcoma cells treated with series concentrations of PEITC for 24, 48, and 72 h. c Cell viability of K7M2 osteosarcoma cells treated with PEITC in the presence of 50 μM z-VAD-FMK or 50 μM necrostatin-1 (Nec-1) or 10 μM ferrostatin-1 (Fer-1) or 100 nM liproxstatin-1 (Lip-1) or 50 μM DFO or 1 μM bafilomycin A1 (Baf-A1) or 10 mM 3-methyladenine (3-MA) or 1 mM N-acetyl-L-cysteine (NAC) for 24 h. d EdU staining assay of K7M2 osteosarcoma cells treated by PEITC. e, f Colony formation assay of K7M2 osteosarcoma cells treated by PEITC. All data were presented as mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group. #P < 0.05, ##P < 0.01, ###P < 0.001 vs. PEITC group
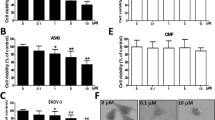
To explore the mechanism by which PEITC induced cell death in K7M2 cells, we selected multiple inhibitors of cell death pathways, including an apoptosis inhibitor (z-VAD-FMK), autophagy inhibitors (Baf-A1 and 3-MA), a necroptosis inhibitor (Nec-1), and ferroptosis inhibitors (Lip-1, Fer-1, and DFO). Unexpectedly, we found that inhibitors of apoptosis, autophagy, necroptosis, and ferroptosis increased cell viability of PEITC-treated K7M2 osteosarcoma cells (Fig. 1c). These results suggest that multiple forms of cell death mechanism are initiated in PEITC-treated K7M2 osteosarcoma cells. Moreover, the viability of K7M2 osteosarcoma cells treated with PEITC in the presence of the antioxidant NAC was also examined. The viability of K7M2 osteosarcoma cells was unaffected by cotreatment with NAC and PEITC. Therefore, we wondered whether PEITC triggers multiple forms of cell death, including apoptosis, autophagy, necroptosis, and ferroptosis, in K7M2 osteosarcoma cells, which are due, at least in part, to ROS generation and accumulation.
The effects of PEITC on the proliferative potential of K7M2 osteosarcoma cells were investigated by conducting both EdU proliferation and colony formation assays. In EdU proliferation assay, the number of living K7M2 osteosarcoma cells decreased, and fewer cells exhibited proliferation potential (Fig. 1d). Moreover, a similar result was observed in the colony formation assay. After PEITC treatment, K7M2 osteosarcoma cells exhibited weak proliferative capacity with fewer colony counts after PEITC treatment in a concentration-dependent manner (Fig. 1e, f). These results suggest that PEITC exerts an anti-proliferative effect on K7M2 osteosarcoma cells.
PEITC induced G2/M cell cycle arrest in K7M2 murine osteosarcoma cells
Since PEITC inhibited viability and proliferative ability of K7M2 osteosarcoma cells, we determined whether PEITC affected the cell cycle. The effect of PEITC on the cell cycle in K7M2 osteosarcoma cells was investigated by PI staining. As shown in Fig. 2a, b, PEITC increased the fraction of K7M2 osteosarcoma cells in the G2/M phase after treatment for 24 h. Likewise, cell cycle-related regulatory proteins were also detected by Western blotting. Decreased expressions of Cdc2, Cyclin D1, and Cyclin D3 were observed in PEITC-treated K7M2 osteosarcoma cells (Fig. 2c–f). Taken together, PEITC induces G2/M arrest in K7M2 osteosarcoma cells.
PEITC induced G2/M cell cycle arrest in K7M2 osteosarcoma cells. a, b The cell cycle distribution of K7M2 osteosarcoma cells after indicated concentrations of PEITC treatment for 24 h by PI staining. c, d Protein expression levels Cdc2, Cyclin D1, and Cyclin D3 in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. e, f Quantitative analysis of Cdc2, Cyclin D1, and Cyclin D3 in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. All data were presented as the mean ± SD, n = 3, *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group
PEITC induced the redox imbalance in K7M2 murine osteosarcoma cells
To evaluate the redox state in K7M2 osteosarcoma cells treated with PEITC, we first measured cellular ROS levels by DCFH-DA sensor. The results indicated that PEITC dramatically increased cellular ROS levels over 12 h of treatment in K7M2 osteosarcoma cells in a concentration-dependent manner (Fig. 3a). Then, lipid ROS were measured by a lipid peroxidation sensor. PEITC treatment for 24 h increased the level of oxidized lipid ROS in K7M2 osteosarcoma cells, as indicated by an intensified green fluorescence (Fig. 3b). To confirm whether ROS induced by PEITC exerted oxidative stress in K7M2 osteosarcoma cells, we measured the level of MDA, an end product that is generated by lipid peroxidation. MDA generation was significantly enhanced by PEITC treatment in K7M2 osteosarcoma cells (Fig. 3c).
PEITC disrupted the redox balance in K7M2 osteosarcoma cells. a ROS level in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 12 and 24 h by DCFH-DA sensor. b Lipid peroxidation level in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 24 h by BODIPY 581/591 C11 sensor. c The MDA level in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 24 h. The data were presented as the mean ± SD, n = 3. *P < 0.05, ***P < 0.001 vs. control group. d GSH/GSSG ratio in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 24 h. **P < 0.01, ***P < 0.001 vs. control group. e, f Expression and quantitative analysis of GPx4 in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h. g, h Expression and quantitative analysis of GPx4 in K7M2 osteosarcoma cells treated by 30 μM PEITC for 4, 12, 24, and 48 h. All the Western blotting data were presented as mean ± SD, n = 3. **P < 0.01, ***P < 0.001 vs. control group
GSH is a major cellular redox buffer. To clarify whether the mechanism by which PEITC induces oxidative stress in K7M2 osteosarcoma cells originates from the depletion of GSH, we further detected the ratio of reduced GSH and GSSG in K7M2 osteosarcoma cells treated with PEITC for 24 h. PEITC significantly decreased the GSH/GSSG ratio in K7M2 osteosarcoma cells at increased concentrations of PEITC (Fig. 3d). GSH is an essential component of the lipid repair function, and glutathione peroxidase 4 (GPx4) utilizes it as substrate. In addition to GSH, we also measured the expression of GPx4 in the cell lysates of K7M2 osteosarcoma cells treated with different concentrations of PEITC or the indicated concentration of PEITC for different times. PEITC inhibited GPx4 expression as concentration and treatment time increased (Fig. 3e–h). Taken together, PEITC treatment disturbed the redox balance in K7M2 osteosarcoma cells by decreasing GSH levels and increasing ROS levels.
PEITC altered iron metabolism in K7M2 murine osteosarcoma cells
Iron metabolism is closely related to tumor initiation, progression, and metastasis. Labile iron directly catalyzes ROS generation through Fenton reaction. First, AAS was used to quantify the total iron content in PEITC-treated K7M2 osteosarcoma cells. Total iron refers to protein-bound iron and labile iron. The level of total iron was significantly increased after PEITC treatment (Fig. 4a, b). Next, the labile iron in K7M2 osteosarcoma cells was evaluated by using calcein-AM. Accordingly, an increase in labile iron was found in PEITC-treated K7M2 osteosarcoma cells as the intensity of green fluorescence decreased (Fig. 4c). Then, the expression of iron metabolism regulatory proteins was detected by Western blotting. PEITC treatment enhanced the expression of the iron-uptake protein transferrin receptor-1 (TfR1) and suppressed the expression of iron-efflux protein ferroportin (FPN), iron storage protein ferritin (FTH1), and divalent metal transporter (DMT1) with increasing concentrations and elongated treatment time, respectively (Fig. 4d–g). These data suggest that the overall outcome of these alterations in iron metabolism regulators increase labile iron after PEITC treatment in K7M2 osteosarcoma cells. The mRNA expression of iron metabolism-related genes was also measured by qRT-PCR. PEITC treatment increased mRNA expression of FTH1 and SLC40A1, and reduced the expression of TFRC, while SLC11A2 expression decreased as the PEITC concentration increased after 12 h of treatment and was unaffected by 24 h PEITC treatment (Fig. 4h–k). In addition to GSH depletion and GPx4 inhibition, an increase in labile iron further increased oxidative stress.
PEITC altered iron metabolism in K7M2 osteosarcoma cells. a, b Total iron content in K7M2 osteosarcoma cells after indicated concentrations of PEITC treatment for 24 h by AAS. Total iron level was normalized to the cell number and protein concentration. The data were presented as mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group. c Labile iron staining in K7M2 osteosarcoma cells after indicated concentrations of PEITC treatment for 24 h by Calcein-AM. d, e Protein expression levels of TfR1, DMT1, FTH1, and FPN in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. f, g Quantitative analysis of TfR1, DMT1, FTH1, and FPN in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. All the Western blotting data were presented as mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group. h–k mRNA expression levels of TFRC, SLC11A2, FTH1, and SLC40A1 in K7M2 osteosarcoma cells after PEITC treatment for 12 and 24 h. The data were presented as the means ± SD, n = 3. *P < 0.05, **P < 0.01 vs. control group of 12 h PEITC treatment; #P < 0.05, ##P < 0.01, ###P < 0.001 vs. control group of 24 h PEITC treatment
PEITC induced apoptosis in K7M2 murine osteosarcoma cells
Because cotreatment with the apoptosis inhibitor z-VAD-FMK increased the viability in PEITC-treated K7M2 osteosarcoma cells, we further explored PEITC-induced apoptosis. First, mitochondrial transmembrane potential was assessed by staining K7M2 cells with JC-1. K7M2 osteosarcoma cells displayed depolarization in mitochondrial transmembrane potential as the concentration of PEITC increased (Fig. 5a). In addition, a MitoTraker sensor was used to label mitochondria and Hoechst 33342 was used to label nuclei. Cells in the PEITC-treated group exhibited condensed chromatin with deeper staining and increased nuclear volume compared to those of the control group. Mitochondria in the control group were uniformly maintained as small dots but appeared swollen and clustered as aggregates in the PEITC treatment groups for 24 h (Fig. 5b). Annexin V-FITC/PI apoptosis assay was performed, and the results showed that PEITC induced a concentration-dependent increase in the number of apoptotic cells (Fig. 5c). Furthermore, factors in the apoptotic cascade pathway were also detected. PEITC-induced apoptosis in K7M2 osteosarcoma cells was associated with decreased levels of Bcl-2 and increased Bax (Fig. 5d–g). To determine whether PEITC-induced apoptosis was associated with caspase activation in K7M2 osteosarcoma cells, C-caspase3 was detected. The results indicated that PEITC promoted Caspase3 activation and significantly increased the level of its substrate C-PARP (Fig. 5d–g). These results suggest that K7M2 osteosarcoma cells undergo mitochondria-mediated apoptosis following PEITC treatment.
PEITC induced apoptosis in K7M2 osteosarcoma cells. a Mitochondrial transmembrane potential in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 12 and 24 h by JC-1. b Mitochondrial and nuclear morphological changes in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 24 h by MitoTraker Green/Hoechst 33342. c Apoptosis and necrosis analysis of K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 24 h by Annexin V-FITC/PI. d, e Protein expression levels of Caspase3, C-caspase3, Bcl-2, Bax, and C-PARP in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. f, g Quantitative analysis of Caspase3, C-caspase3, Bcl-2, Bax, and C-PARP in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. All data were presented as mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group
PEITC induced autophagy in K7M2 murine osteosarcoma cells
The cell viability assay indicated that autophagy may be involved in PEITC-induced cell death in K7M2 osteosarcoma cells. Therefore, autophagic vesicles were examined, and autophagy-related markers were detected. AO staining was used to investigate the changes in acidic vesicles. As shown in Fig. 6a, significant bright red fluorescence appeared in K7M2 osteosarcoma cells as the PEITC concentration increased. This demonstrated that there were more acid vesicles after PEITC treatment. Additionally, lysosome labeling also showed that more autophagic vesicles appeared, and the intensified green fluorescence indicated that autophagy was enhanced after PEITC treatment (Fig. 6b). To further confirm PEITC-induced autophagy, three typical autophagy markers, Beclin1, LC3B, and p62, were detected. As shown in Fig. 6c–f, Beclin1 expression increased in the PEITC treatment group. Treatment of K7M2 osteosarcoma cells with PEITC induced a conversion of LC3I to LC3II over time. p62 levels decreased as PEITC concentration increased. The immunofluorescence results further indicated that PIETC treatment increased LC3 expression (Fig. 6g). These data suggest that PEITC stimulates autophagy in K7M2 osteosarcoma cells through the formation of autophagosomes and the regulation of the autophagy-related proteins.
PEITC induced autophagy in K7M2 osteosarcoma cells. a Acidic vacuole organelles in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 24 h by AO staining analysis. b Lysosomes in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 24 h by LysoTraker Green sensor. c, d Protein expression levels of Beclin1, LC3B, and p62 in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. e, f Quantitative analysis of Beclin1, LC3B, and p62 in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. g Immunofluorescence staining of LC3B in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 24 h. h, i Phosphorylation level of mTOR in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. j, k Quantitative analysis of mTOR in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. l, m Phosphorylation level of Stat3 in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. n, o Quantitative analysis of Stat3 in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. All data were presented as mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group
Mammalian target of rapamycin (mTOR) is the main regulator of autophagy in cancer cells. To better understand the molecular mechanisms by which PEITC induces autophagy in K7M2 osteosarcoma cells, we detected the possible involvement of mTOR. As shown in Fig. 6h–k, phosphorylation of mTOR was inhibited by PEITC treatment. Then, signal transducer and activator of transcription 3 (Stat3), which is the downstream of mTOR, was also detected. Phosphorylation of Stat3 was inhibited in K7M2 osteosarcoma cells treated with PEITC (Fig. 6l–o). Therefore, PEITC stimulates autophagy in K7M2 osteosarcoma cells via the mTOR/Stat3 pathway.
Activation of the MAPK signaling pathway was involved in PEITC-induced cell death in K7M2 murine osteosarcoma cells
Mitogen-activated protein kinases (MAPKs), including extracellular signal-regulated kinase (ERK), p38 kinase, and c-Jun NH2-terminal kinase (JNK), play a key role in the response to extracellular stimuli. Previous studies showed that PEITC activates the MAPK signaling pathway in many types of cancer cells. Therefore, we examined whether MAPKs were activated in PEITC-treated K7M2 osteosarcoma cells. Fig. 7a–d shows that PEITC activated the MAPK signaling pathway in K7M2 osteosarcoma cells by phosphorylating ERK, p38, and JNK. PEITC activated ERK in concentration- and time-dependent manners; p-p38 reached a peak at 4 h after PEITC treatment, and the phosphorylation level declined thereafter; phosphorylation of JNK was enhanced as the PEITC concentration increased and peaked at 12 h of PEITC treatment. These data suggest that the MAPK signaling pathway is activated in PEITC-treated K7M2 osteosarcoma cells.
PEITC activated MAPK signaling pathway in K7M2 osteosarcoma cells. a, b Phosphorylation levels of ERK, p38, and JNK in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. c, d Quantitative analysis of ERK, p38, and JNK in K7M2 osteosarcoma cells treated by indicated concentrations of PEITC for 20 h or 30 μM PEITC for 4, 12, 24, and 48 h. All data were presented as mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group
PEITC-induced cell death in K7M2 murine osteosarcoma cells was dependent on ROS generation
Inhibitors of different cell death pathways partly rescued the reduced cell viability while NAC almost completely blocked the PEITC-induced decrease in viability in K7M2 osteosarcoma cells. Therefore, we examined whether PEITC-induced cell death was due to ROS generation. First, we investigated the effect of PEITC-induced ROS generation on the proliferation of K7M2 osteosarcoma cells. Notably, NAC attenuated the PEITC-induced decrease in cell number and cell proliferation in both the colony formation and EdU assays (Fig. 8a–c). Next, we explored whether the generation of ROS plays a role in the regulatory of iron metabolism. NAC altered the PEITC-induced change in iron metabolism-related proteins (Fig. 8d, e). Moreover, we investigated the role of ROS in PEITC-induced forms of cell death in K7M2 osteosarcoma cells. The Western blotting showed that NAC abrogated the PEITC-induced decrease in the expression of GPx4, Bcl-2 and Cdc2 and the PEITC-induced increase in C-caspase3 and LC3II levels (Fig. 8f, g). The participation of ROS in activating the MAPK signaling pathway was also evaluated. The results supported the hypothesis that ROS played a significant role in the phosphorylation of ERK, p38, and JNK in PEITC-treated K7M2 osteosarcoma cells (Fig. 8h, i). Taken together, these results indicate that PEITC induces cell death in K7M2 osteosarcoma cells predominantly by increasing cellular ROS generation.
PEITC induced cell death via the generation of ROS in K7M2 osteosarcoma cell. a, b Colony formation assay of K7M2 osteosarcoma cells treated by 30 μM PEITC in the presence of NAC or not for 24 h. c EdU staining assay of the proliferation ability of K7M2 osteosarcoma cells treated by 30 μM PEITC in the presence of NAC or not for 24 h. d, e Protein expression levels and quantitative analysis of TfR1, DMT1, FTH1, and FPN in K7M2 osteosarcoma cells treated by 30 μM PEITC in the presence of NAC or not for 24 h. f, g Protein expression levels and quantitative analysis of LC3B, GPx4, C-caspase3, Bcl-2, and Cdc2 in K7M2 osteosarcoma cells treated by 30 μM PEITC in the presence of NAC or not for 24 h. h, i Phosphorylation levels and quantitative analysis of ERK, p38, and JNK in K7M2 osteosarcoma cells treated by 30 μM PEITC in the presence of NAC or not for 24 h. All data were presented as mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group
The effects of PEITC on osteosarcoma in vivo
The in vivo antitumor effect of PEITC was evaluated in a syngeneic orthotopic osteosarcoma mouse model. Fig. 9a shows that there were no significant differences in the average body weights of the mice between the four groups throughout the experiment. The 30 and 60 mg/kg PEITC-treated groups exhibited significantly lower tumor volume and tumor weight compared with those of the control group, but the 90 mg/kg PEITC-treated group showed no differences in tumor volume and tumor weight compared to those of the control group (Fig. 9b–d).
PEITC inhibited osteosarcoma growth in vivo. K7M2 cells were injected into the tibia of BALB/c mice. Two weeks after the syngeneic orthotopic osteosarcoma mouse model was established, the mice were randomly divided into four groups and respectively intragastrically administered 10% sesame-seed oil, 30 mg/kg PEITC, 60 mg/kg PEITC, and 90 mg/kg PEITC once daily for consecutive 24 days. a Body weight change of the four groups. b Volume change of osteosarcoma tissues of the four groups. Data were calculated by the formula: volume = length × width2 × 0.5. The data were presented as the means ± SD, n = 7. *P < 0.05, ***P < 0.001 vs. control group. c Weights of osteosarcoma tissues from the four groups. The data were presented as the means ± SD, n = 7. *P < 0.05, ***P < 0.001 vs. control group. d Macroscopic view of the xenografted tumor of different treatments from the four groups. Scale bar, 1 cm. e H&E staining analysis of tumor tissues. f H&E staining analysis of heart, liver, spleen, lung, and kidney of each group
In addition, we performed histological studies on the main organs and tumor tissues from mice after administration of PEITC for 24 days. H&E staining of tumor sections showed that the control group exhibited a highly cancerous and high mortality bone tumor, while the PEITC-treated groups still had osteoid deposition as the dose increased (Fig. 9e). H&E staining of the main organs revealed that administration of PEITC did not exhibit significant organ-related toxicity compared to that of the control group (Fig. 9f).
PEITC altered iron metabolism, induced cell death, and activated the MAPK signaling pathway in osteosarcoma in vivo
As shown in Fig. 10a, Ki67 staining showed reduced proliferation in osteosarcoma cells after PEITC treatment. To determine whether PEITC induced apoptosis, autophagy, and ferroptosis in osteosarcoma in vivo, immunohistochemistry analysis of tumor sections was also performed. As shown in Fig. 10a, the immunohistochemical staining results showed increased levels of C-caspase3 and LC3II, whereas GPx4 was decreased following PEITC treatment. Then Western blotting was used to confirm the inhibitory effect of PEITC on osteosarcoma in vivo. Accordingly, expression of the cell cycle regulatory d protein Cdc2 was decreased and GPx4 was inhibited as the dose of PEITC increased; C-caspase3 and LC3II were significantly increased in the 60 mg/kg PEITC treatment group (Fig. 10b, c). Moreover, iron metabolism regulatory proteins were also detected. PEITC administration altered iron metabolism in tumor sections with increased expression of the iron import protein TfR1 and decreased expression of the iron export protein FPN and iron storage protein FTH1 (Fig. 10d, e). Finally, the MAPK signaling pathway was detected in tumor sections. Three MAPKs, ERK, p38, and JNK were all activated after PEITC administration in osteosarcoma tissues in vivo (Fig. 10f, g). In conclusion, PEITC delays tumor growth and induces cell death in osteosarcoma in syngeneic orthotopic osteosarcoma mice.
Immunohistochemical analysis of tumor tissue from osteosarcoma mice treated by PEITC. a Immunohistochemical staining analysis of Ki67 (a marker of proliferation), C-caspase3 (a marker of apoptosis), LC3B (a marker of autophagy), and GPx4 (a marker of ferroptosis) in tumor tissues. b, c Protein expression levels and quantitative analysis of Cdc2, GPx4, C-caspase3, and LC3B in tumor tissues. d, e Protein expression levels and quantitative analysis of TfR1, DMT1, FPN, and FTH1 in tumor tissues. f, g Phosphorylation levels and quantitative analysis of ERK, p38, and JNK in tumor tissues. All data were presented as mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group
Discussion
It is widely accepted that a moderate increase in ROS promotes cancer cell proliferation, but excessive ROS generation causes irreversible oxidative damage and induces cell death [20]. Cancer cells develop a set of adaptive antioxidant mechanisms to fight oxidative stress. GSH is the major redox buffer in mammalian cells and under tight homeostatic control both intracellularly and extracellularly. Depletion of GSH leads to cell death in cancer cells [21]. Of note, a large body of evidence suggests that PEITC-induced GSH depletion and ROS generation are associated with cell death in cancer cells [8, 22]. Previous studies have shown that three isothiocyanates, SFN, benzyl isothiocyanate, and PEITC, inhibit U-2 OS osteosarcoma cells, and PEITC had a stronger effect in abolishing tumor metastasis than the other isothiocyanates [23]. PEITC also enhances the inhibitory effects of the first-line antitumor agent adriamycin on U-2 OS cells [24]. The present study demonstrated that PEITC induced multiple forms of cell death, including ferroptosis, autophagy, and apoptosis, in K7M2 osteosarcoma cells, and the mechanism involved GSH depletion, iron metabolism alteration, ROS generation, and the MAPK signaling pathway activation.
Ferroptosis is a form of cell death that is morphologically, biochemically, and genetically distinct from apoptosis, necroptosis, and autophagy. The three biological hallmarks of ferroptosis are iron dependence, GPx4 inactivation, and lipid ROS accumulation [25]. The PEITC-induced reduction in viability in K7M2 osteosarcoma cells was alleviated by DFO. It was proposed that iron was required for PEITC-induced cell death. Iron quantification showed that PEITC caused increased amounts of both total iron and labile iron in K7M2 osteosarcoma cells. Excess labile iron exerted more potential to induce oxidative stress. Moreover, iron metabolism genes and proteins were altered in PEITC-treated K7M2 osteosarcoma cells. There is coordinated feedback control between the labile iron and iron metabolism genes [26, 27]. An increase in labile iron leads to increased translation of FTH1 and SLC40A1 mRNAs and degradation of TFRC mRNA. GPx4 is an essential regulator of ferroptosis which is the only enzyme that can reduce lipid hydroperoxides within biological membranes [28]. GSH is required for lipid repair function, and GPx4 uses it as a substrate. Depletion of GSH indirectly leads to the loss of GPx4 activity, which results in the accumulation of unrepaired lipid peroxides. PEITC treatment depleted GSH levels, further inactivating GPx4 and causing the accumulation of lipid peroxidation in K7M2 osteosarcoma cells. Fer-1 and Lip-1 are two lipid ROS scavengers with robust radical-trapping capacity and that can efficiently inhibit ferroptosis [29]. Both compounds partly protected K7M2 osteosarcoma cells from PEITC-induced cell death. Importantly, the lipid peroxidation level also increased, and the presence of abundant lipid peroxidation products, such as MDA, was also observed in K7M2 osteosarcoma cells treated with PEITC. Therefore, ferroptosis was involved in the PEITC-induced death of K7M2 osteosarcoma cells, and this process was dependent on iron, GPx4 downregulation, and lipid ROS accumulation.
Apoptosis is defined as a cell death process that is driven by a set of molecular mechanisms. z-VAD-FMK, a pan caspase inhibitor of apoptosis, ameliorated PEITC-induced cell death in K7M2 osteosarcoma cells. These findings suggested that apoptosis may be initiated in K7M2 cells by PEITC. Altered mitochondrial and nuclear morphologies which were the visible changes characterizing PEITC-induced apoptosis were observed. Mitochondria play important roles in programmed cell death. In apoptotic cell death, the loss of mitochondrial integrity precedes the release of proapoptotic molecules. Disruption of mitochondrial membrane potential has been regarded as an early event in apoptosis. To elucidate the underlying mechanism of PEITC-induced apoptosis in K7M2 osteosarcoma cells, we found that PEITC treatment decreased mitochondrial transmembrane potential. Reduced expression of the antiapoptotic protein Bcl-2 and increased expression of the proapoptotic protein Bax were also observed. Apoptotic cell death involves the engagement of pathways that result in the activation of caspase proteases, and caspases-mediated cleavage of special substrates, such as poly (ADP ribose) polymerase (PARP), which is also the characteristic feature of apoptosis. PEITC activated Caspase3 and induced C-PARP accumulation in K7M2 osteosarcoma cells. As a result, PEITC treatment caused K7M2 osteosarcoma cells to undergo mitochondria-mediated apoptosis.
In cancer cells, autophagy usually acts as a prosurvival pathway for tumor progression. However, excessive and persistent autophagy in response to anticancer therapies can cause irreversible destruction of cellular contents and eventually trigger cell death [30, 31]. 3-MA is an autophagy inhibitor because of its inhibitory effect on phosphoinositide 3-kinase (PI3K), which is essential for inducing autophagy [32]. Baf-A1, an inhibitor of vacuolar H+-ATPase, inhibits lysosomal acidification and blocks autophagy [33]. We first observed that both 3-MA and Baf-A1 decreased the effects of PEITC on the cell viability of K7M2 osteosarcoma cells. Furthermore, PEITC increased the number of acidic vesicles, and the lysosomes in K7M2 osteosarcoma cells became increasingly evident after PEITC treatment. These findings suggest that the formation of acidified vesicles and activated lysosomes are involved in the PEITC-induced cell death in K7M2 osteosarcoma cells. The following three molecules are autophagy markers: Beclin1 initiates the autophagy pathway; LC3 is a key regulator of autophagy during the formation of autophagosomes; and p62 binds to LC3 and is selectively degraded in autolysosomes as a marker of autophagy promotion. In our study, enhanced expression of Beclin1, increased conversion of LC3I to LC3II, and degradation of p62 were found in PEITC-treated K7M2 osteosarcoma cells. These findings suggest that PEITC induce autophagy in K7M2 osteosarcoma cells. mTOR plays a vital role in the initiation and regulation of autophagy [34]. mTOR inhibition enhances autophagy and suppresses cell proliferation. Stat3, the downstream signal of mTOR, has been implicated in the assembly of autophagosomes and their maturation [35]. PEITC treatment suppressed mTOR phosphorylation and stimulated autophagy. Accordingly, the level of p-STAT3 was reduced by PEITC treatment. Inhibition of Stat3 causes LC3B conversion, p62 degradation and autophagosome formation [36]. This suggests that autophagy process is involved in the PEITC-induced death of K7M2 osteosarcoma cells, possibly through the mTOR/Stat3 pathway.
ROS regulate a number of cellular pathways and play critical roles in determining cell fate [37, 38]. Notably, cancer cells display increased ROS demand and production than their normal counterparts. Moreover, cancer cells also develop increased levels of antioxidant systems to cope with potential oxidative stress. PEITC depleted GSH levels and then dampened the antioxidant capacity. The PEITC-induced decrease in viability of K7M2 osteosarcoma cells was completely prevented by NAC, suggesting that ROS generation is responsible for the induction of cell death. Cotreatment with NAC dramatically altered the changes in proliferation of PEITC-treated K7M2 osteosarcoma cells. Mounting evidence has proposed that ROS participate in regulating signaling pathways that lead to the initiation of multiple forms of cell death. For apoptosis, ROS generation and accumulation preceded mitochondrial transmembrane depolarization, caspase activation, and nuclear condensation. The apoptosis marker (C-caspase3) and the conversion of the autophagic marker (LC3B) were suppressed by NAC in PEITC-treated K7M2 osteosarcoma cells, while the ferroptosis marker (GPx4) was increased with NAC cotreatment. These results indicate a close relationship between ROS generation and cell death in PEITC-treated K7M2 osteosarcoma cells.
MAPK signaling pathways proceed through a phosphorylation cascade creating sequential cascade activation [39, 40]. Mechanistically, previous studies have demonstrated that PEITC triggers activation of the MAPK signaling pathway in many cancer cell types, such as multiple myeloma, ovarian cancer, and prostate cancer [41,42,43,44,45]. In the present study, three regulators in MAPK signaling pathway were all activated in K7M2 osteosarcoma cells treated with PEITC. Activation of MAPKs by ROS and the duration of their activation are recognized as keynote for cell death [46,47,48]. The activation of ERK, p38, and JNK kinases by PEITC in K7M2 osteosarcoma cells was inhibited after NAC cotreatment. ROS may be an upstream signal involved in PEITC-mediated activation of the MAPK signaling pathway. These findings suggest that treatment with PEITC induces multiple forms of cell death accompanied by ROS generation, as well as activation of MAPK signaling pathways.
Administration of 60 mg/kg PEITC significantly suppressed the growth of osteosarcoma in vivo compared to that of the control group, but the group with a higher dose showed indistinguishable tumor volumes and tumor weights. In contrast, H&E staining of tumor slices and Western blotting analysis of tumor lysates indicated that 90 mg/kg PEITC treatment actually exhibited osteoid deposition, LC3B conversion, Caspase3 activation, and GPx4 inhibition. At higher dose, the metabolism of PEITC may be saturated, resulting in increased PEITC tissue distribution. To our knowledge, a higher dose of PEITC causes a transient drop in GSH levels that may leave not only tumor cells but also normal cells vulnerable to oxidative stress [49]. At a lower dose, PEITC had a significantly positive influence on osteosarcoma in mice; when the dose was increased, however, the inhibitory effect of PEITC on the osteosarcoma mice was compromised.
Conclusions
In summary, the present study demonstrated the promising therapeutic effect of PEITC in osteosarcoma both in vitro and in vivo. PEITC triggered multiple forms of programmed cell death, including ferroptosis, apoptosis, and autophagy, by activating the ROS-related MAPK signaling pathway. How the different forms of cell death induced by PEITC in K7M2 osteosarcoma cells connect and switch still needs to be clarified in future research. Undoubtedly, targeting the biosynthesis and homeostasis of the GSH system is a potential antitumor therapeutic strategy.
References
Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–47.
Moloney JN, Cotter TG. ROS signalling in the biology of cancer. Semin Cell Dev Biol. 2018;80:50–64.
de Sa Junior PL, Camara DAD, Porcacchia AS, Fonseca PMM, Jorge SD, Araldi RP, et al. The roles of ROS in cancer heterogeneity and therapy. Oxid Med Cell Longev. 2017;2017:2467940.
Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27:211–22.
Hatem E, El Banna N, Huang ME. Multifaceted roles of glutathione and glutathione-based systems in carcinogenesis and anticancer drug resistance. Antioxid Redox Signal. 2017;27:1217–34.
Habermann KJ, Grünewald L, van Wijk S, Fulda S. Targeting redox homeostasis in rhabdomyosarcoma cells: GSH-depleting agents enhance auranofin-induced cell death. Cell Death Dis. 2017;8:e3067.
Lo M, Wang YZ, Gout PW. The x(c)-cystine/glutamate antiporter: a potential target for therapy of cancer and other diseases. J Cell Physiol. 2008;215:593–602.
Wang J, Luo B, Li X, Lu W, Yang J, Hu Y, et al. Inhibition of cancer growth in vitro and in vivo by a novel ROS-modulating agent with ability to eliminate stem-like cancer cells. Cell Death Dis. 2017;8:e2887.
Barattin R, Perrotton T, Trompier D, Lorendeau D, Di Pietro A, d’Hardemare Adu M, et al. Iodination of verapamil for a stronger induction of death, through GSH efflux, of cancer cells overexpressing MRP1. Bioorg Med Chem. 2010;18:6265–74.
Cheung KL, Kong AN. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J. 2010;12:87–97.
Chen G, Chen Z, Hu Y, Huang P. Inhibition of mitochondrial respiration and rapid depletion of mitochondrial glutathione by beta-phenethyl isothiocyanate: mechanisms for anti-leukemia activity. Antioxid Redox Signal. 2011;15:2911–21.
Conaway CC, Wang CX, Pittman B, Yang YM, Schwartz JE, Tian D, et al. Phenethyl isothiocyanate and sulforaphane and their N-acetylcysteine conjugates inhibit malignant progression of lung adenomas induced by tobacco carcinogens in A/J mice. Cancer Res. 2005;65:8548–57.
Morris ME, Dave RA. Pharmacokinetics and pharmacodynamics of phenethyl isothiocyanate: implications in breast cancer prevention. AAPS J. 2014;16:705–13.
Palliyaguru DL, Yuan JM, Kensler TW, Fahey JW. Isothiocyanates: translating the power of plants to people. Mol Nutr Food Res. 2018;62:e1700965.
Ji Y, Kuo Y, Morris ME. Pharmacokinetics of dietary phenethyl isothiocyanate in rats. Pharm Res. 2005;22:1658–66.
Yuan JM, Stepanov I, Murphy SE, Wang R, Allen S, Jensen J, et al. Clinical trial of 2-phenethyl isothiocyanate as an inhibitor of metabolic activation of a tobacco-specific lung carcinogen in cigarette smokers. Cancer Prev Res. 2016;9:396–405.
De Flora S, Ganchev G, Iltcheva M, La Maestra S, Micale RT, Steele VE, et al. Pharmacological modulation of lung carcinogenesis in smokers: preclinical and clinical evidence. Trends Pharmacol Sci. 2016;37:120–42.
Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer. 2009;125:229–34.
Botter SM, Neri D, Fuchs B. Recent advances in osteosarcoma. Curr Opin Pharm. 2014;16:15–23.
Galadari S, Rahman A, Pallichankandy S, Thayyullathil F. Reactive oxygen species and cancer paradox: to promote or to suppress? Free Radic Biol Med. 2017;104:144–64.
Yu X, Long YC. Crosstalk between cystine and glutathione is critical for the regulation of amino acid signaling pathways and ferroptosis. Sci Rep. 2016;6:30033.
Xiao D, Powolny AA, Moura MB, Kelley EE, Bommareddy A, Kim SH, et al. Phenethyl isothiocyanate inhibits oxidative phosphorylation to trigger reactive oxygen species-mediated death of human prostate cancer cells. J Biol Chem. 2010;285:26558–69.
Jeong YJ, Cho HJ, Chung FL, Wang X, Hoe HS, Park KK, et al. Isothiocyanates suppress the invasion and metastasis of tumors by targeting FAK/MMP-9 activity. Oncotarget. 2017;8:63949–62.
Fan Q, Zhan X, Xiao Z, Liu C. Phenethyl isothiocyanate enhances adriamycin-induced apoptosis in osteosarcoma cells. Mol Med Rep. 2015;12:5945–50.
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72.
Kuhn LC. Iron regulatory proteins and their role in controlling iron metabolism. Metallomics. 2015;7:232–43.
Bogdan AR, Miyazawa M, Hashimoto K, Tsuji Y. Regulators of iron homeostasis: new players in metabolism, cell death, and disease. Trends Biochem Sci. 2016;41:274–86.
Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31.
Angeli JPF, Shah R, Pratt DA, Conrad M. Ferroptosis inhibition: mechanisms and opportunities. Trends Pharmacol Sci. 2017;38:489–98.
Hippert MM, O’Toole PS, Thorburn A. Autophagy in cancer: good, bad, or both? Cancer Res. 2006;66:9349–51.
Chen N, Karantza V. Autophagy as a therapeutic target in cancer. Cancer Biol Ther. 2014;11:157–68.
Wu Y, Wang X, Guo H, Zhang B, Zhang XB, Shi ZJ, et al. Synthesis and screening of 3-MA derivatives for autophagy inhibitors. Autophagy. 2013;9:595–603.
Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1block the fusion of autophagosomes with lysosomes? Autophagy. 2014;4:849–50.
Zou M, Hu C, You Q, Zhang A, Wang X, Guo Q, et al. Oroxylin A induces autophagy in human malignant glioma cells via the mTOR-STAT3-Notch signaling pathway. Mol Carcinog. 2015;54:1363–75.
You L, Wang Z, Li H, Shou J, Jing Z, Xie J, et al. The role of STAT3 in autophagy. Autophagy. 2015;11:729–39.
Jonchère B, Bélanger A, Guette C, Barré B, Coqueret O. STAT3 as a new autophagy regulator. JAKSTAT. 2013;2:e24353.
Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–21.
Circu ML, Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic Biol Med. 2010;48:749–62.
Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. 2010;1802:396–405.
Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18.
Jakubikova J, Cervi D, Ooi M, Kim K, Nahar S, Klippel S, et al. Anti-tumor activity and signaling events triggered by the isothiocyanates, sulforaphane and phenethyl isothiocyanate, in multiple myeloma. Haematologica. 2011;96:1170–9.
Qin CZ, Zhang X, Wu LX, Wen CJ, Hu L, Lv QL, et al. Advances in molecular signaling mechanisms of beta-phenethyl isothiocyanate antitumor effects. J Agric Food Chem. 2015;63:3311–22.
Satyan KS, Swamy N, Dizon DS, Singh R, Granai CO, Brard L, et al. Phenethyl isothiocyanate (PEITC) inhibits growth of ovarian cancer cells by inducing apoptosis: role of caspase and MAPK activation. Gynecol Oncol. 2006;103:261–70.
Xiao D, Singh S. Phenethyl isothiocyanate-induced apoptosis in p53-deficient PC-3 human prostate cancer cell line is mediated by extracellular signal-regulated Kinases. Cancer Res. 2002;62:3615–9.
Hu R, Kim BR, Chen C, Hebbar V, Kong AN. The roles of JNK and apoptotic signaling pathways in PEITC-mediated responses in human HT-29 colon adenocarcinoma cells. Carcinogenesis. 2003;24:1361–67.
Son Y, Cheong YK, Kim NH, Chung HT, Kang DG, Pae HO. Mitogen-activated protein kinases and reactive oxygen species: how can ROS activate MAPK pathways? J Signal Transduct. 2011;2011:792639.
Liu J, Chang F, Li F, Fu H, Wang J, Zhang S, et al. Palmitate promotes autophagy and apoptosis through ROS-dependent JNK and p38 MAPK. Biochem Bioph Res Commun. 2015;463:262–7.
Benhar M, Engelberg D, Levitzki A. ROS, stress-activated kinases and stress signaling in cancer. EMBO J. 2002;3:420–5.
Ioannides C, Konsue N. A principal mechanism for the cancer chemopreventive activity of phenethyl isothiocyanate is modulation of carcinogen metabolism. Drug Metab Rev. 2015;47:356–73.
Acknowledgements
We would like to thank Li Xie, Zhe Wang, Zhouqi Yang, and Yi Lyu for technical assistance. This work was carried out with the support of the National Natural Science Foundation of China [No. 81803032], the Natural Science Foundation of Shaanxi Province [No. 2019JQ632], the Zhejiang Postdoctoral Science Fund, and the Fundamental Research Funds for the Central Universities [No. 3102017OQD111].
Author information
Authors and Affiliations
Contributions
HHL, CXZ, and JYL performed the in vitro experiments and analyzed the data. HHL and CXZ did the in vivo experiments and analyzed the data. HHL designed the research, wrote the article, and titled the manuscript. PS supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Lv, Hh., Zhen, Cx., Liu, Jy. et al. PEITC triggers multiple forms of cell death by GSH-iron-ROS regulation in K7M2 murine osteosarcoma cells. Acta Pharmacol Sin 41, 1119–1132 (2020). https://doi.org/10.1038/s41401-020-0376-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-020-0376-8
Keywords
This article is cited by
-
Harnessing ferroptosis for enhanced sarcoma treatment: mechanisms, progress and prospects
Experimental Hematology & Oncology (2024)
-
Clinical significance and immune landscape of a novel ferroptosis-related prognosis signature in osteosarcoma
BMC Cancer (2023)
-
Autophagy deficiency exacerbates iron overload induced reactive oxygen species production and apoptotic cell death in skeletal muscle cells
Cell Death & Disease (2023)
-
Targeting iron metabolism in osteosarcoma
Discover Oncology (2023)
-
Natural products and the balancing act of autophagy-dependent/independent ferroptosis in cancer therapy
Naunyn-Schmiedeberg's Archives of Pharmacology (2023)