Abstract
Chinese herbal medicine (CHM) addresses complex diseases through polypharmacological interactions. However, systematic studies of herbal medicine pharmacology remain challenging due to the complexity of CHM ingredients and their interactions with various targets. In this study, we aim to address this challenge with computational approaches. We investigated the herb-target-disease associations of 197 commonly prescribed CHMs using the similarity ensemble approach and DisGeNET database. We demonstrated that this method can be applied to associate herbs with their putative targets. In the case study of three well-known herbs, Radix Glycyrrhizae, Flos Lonicerae, and Rhizoma Coptidis, approximately 70% of the predicted targets were supported by scientific literature. By linking 406 targets to 2439 annotated diseases, we further analyzed the pharmacological functions of 197 herbs. Finally, we proposed a strategy of target-oriented herbal formula design and illustrated the target profiles for four common chronic diseases, namely, Alzheimer’s disease, depressive disorder, hypertensive disease, and non-insulin-dependent diabetes mellitus. This computational approach holds great potential in the target identification of herbs, understanding the molecular mechanisms of CHM, and designing novel herbal formulas.
Similar content being viewed by others
Introduction
Chinese herbal medicine (CHM) has been intensively used in China for more than 2000 years. As an essential branch of traditional Chinese medicine (TCM), CHM has influenced health practices in East Asia and has become a worldwide alternative medicine. Starting from the first herbal medical literature, Shen Nong’s Materia Medica (Shen Nong Ben Cao Jing, ~220 CE), to date, TCM doctors have collected thousands of herbal materials for the treatment of diseases. Among them, approximately 200 herbs are frequently prescribed [1, 2].
Despite a holistic practice in CHM that addresses one’s health from a systematic point of view, reductionist studies of each herb to the molecular level are becoming inevitable if one aims to understand the rationale behind the narratives of ancient medical classics [3]. Consequently, one can demystify the power of a curative herb with its biochemical nature. During the past century, scientists have been working in this direction on many herbs with fruitful outcomes; the most successful story was that of Artemisia annua. A natural compound derived from this herb and its derivatives saved millions of people from malaria. One of the inventors, Youyou Tu, was rewarded the 2015 Nobel Prize in Physiology or Medicine for her tremendous contributions [4, 5].
With computational chemistry and biology becoming a cutting-edge technology in biomedical research, many studies have demonstrated its power in the field of herbal medicine as well [6]. Such approaches offer us not only rapid and accurate predictions but also, and more importantly, a comprehensive understanding at different levels. For example, TCM databases provide us with chemical representations of each herb for knowledge discovery [7, 8]. Docking methods offer suggestions of binding affinities for herbal compounds and targets [9, 10]. Herb-based docking analysis can further suggest herb-target associations [11,12,13]. Systems biology, e.g., ordinary differential equations, can simulate the dynamics of a biological network with the treatment of herbal medicine [11,12,13]. Taken together, CHM can be reviewed and designed similarly to the development of new chemical entities for a certain disease with specific targets.
In recent decades, ligand-based techniques have been developed and applied to reveal the possible associations between CHM and drug targets. For instance, Zobir et al. studied the mode of action of 45 TCM therapeutic action classes by in silico target prediction algorithms, of which the targets were annotated with the Kyoto Encyclopedia of Genes and Genomes pathway [14]. Huang et al. used a most-similar ligand-based approach to predict the mechanism of action targets of aloe-emodin discovered from phenotypic screening and traditional medicine [15]. However, the systems pharmacology of individual herbs or herbal formulas remains largely elusive, which to some extent has hindered modern herbal drug development.
The similarity ensemble approach (SEA) is one of the pioneering ligand-based methods in computational systems pharmacology [16,17,18]. In SEA, molecules are expressed in topological fingerprints as bit strings [19]. Given two strings of fingerprints, one can calculate the overlapping bits divided by the total number of nonoverlapping bits, termed the Tanimoto coefficient (TC) [20], which is a common index to quantify the similarity between two compounds and ranges from 0 to 1. SEA leverages thousands of pairwise TC calculations between the two compound sets and adopts a BLAST-like model to remove the biases of ligand size and chemical composition [21].
SEA has been successfully applied to some interesting questions related to known compounds. For example, SEA was used to compare 3665 FDA approved and investigational drugs against 246 sets of ligands from known targets. As a result, 23 new drug-target associations were confirmed with experiments [22]. In another study to predict side effect targets, SEA was applied to investigate 656 marketed drugs on 73 unintended targets, and approximately half of the predictions were confirmed [23]. However, no study using SEA to understand herbal medicine has been reported thus far. In this study, we first confirmed that SEA can be reliably applied to study CHM and then used SEA to build the associations of 197 commonly prescribed herbs with their potential targets and corresponding diseases. Finally, we proposed a computational strategy for target-oriented herbal formula design.
Materials and methods
The successful representation of herbs in chemical space is the foundation of CHM research. Thanks to the currently available herbal databases, such as the Traditional Chinese Medicine Database (TCMD) [24, 25], the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) [26], and the TCM Database@Taiwan [27], in silico studies were made possible. The TCMD was used in this study because it is of high quality and offers detailed information on more than 20 000 natural compounds. We selected 197 commonly prescribed herbal materials (Supplementary Table S1). Each herb was labeled with information on its Latin name, origin, Chinese name, pinyin (Chinese Romanization) and the number of compounds retrieved from the TCMD.
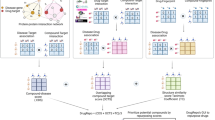
An overview of the computational workflow and scheme are depicted in Fig. 1. The natural compounds collected from the TCMD were converted into SMILES (simplified molecular input line entry system) format by Open Babel (http://openbabel.org/wiki/Main_Page) [28]. SMILES is a line notation that represents molecules and is unique for each compound. With this format, one can obtain topological information for different purposes [29]. Then, the SEA was applied to associate compound sets of an herb and a target (e.g., S1 and S2) [16]. The algorithm first sums the pairwise TCs above a threshold as the raw score (Eq. 1). Then, by taking the difference between the raw score expected at random and dividing by the standard deviation, the raw score is converted into Z-score (Eq. 2). The Z-score is finally transformed into an E-value based on an extreme value distribution and the number of set comparisons (Ndb) made in the database search (Eq. 3) [21].
An overview of the (a) computational workflow and (b) scheme. A total of 197 herbs were associated with 2439 diseases via 406 targets by the similarity ensemble approach and DisGeNET platform. All the information constitutes the strategy of target-oriented herbal formula design, which replaces the traditional narratives of herbal healing
On the SEA Search Server (http://sea.bkslab.org), we used ChEMBL (EBI medicinal chemistry database) version 16 as the reference library and ECFP4 as the fingerprint [19, 30]. We predicted targets for each of the 197 herbs and recorded those with E-values of less than 10−10 for herb-target analysis. The associated targets were further linked to diseases on the DisGeNET platform (http://www.disgenet.org), one of the largest and most comprehensive repositories of human gene-disease associations (DGAs) [31, 32]. Each association is measured by a DGA score from 0 to 1. With the cutoff set to 0.08, we associated the herbs with diseases via the corresponding targets. Finally, we can plot the CHM target profile for each disease and design new herbal formulas in a target-oriented manner.
Results
Comparison of herbal compounds and the reference library
Since natural compounds are diverse in structure, we first compared the chemical properties of herbal compounds and the annotated ligand sets from the ChEMBL database. We calculated six properties for these compounds, including the molecular weight, LogP, number of hydrogen bond acceptors, number of hydrogen bond donors, number of rotatable bonds, and number of rings. The distributions were plotted and compared (Supplementary Fig. S1). For all six properties, the distributions from the herbal compounds and ChEMBL compounds largely overlapped, and the corresponding average values were similar in both sets. Our analysis agreed well with a recent study that natural products populate regions of chemical space that are of high relevance to drug discovery [33].
We further analyzed the chemical scaffolds covered by the herbal and ChEMBL compounds. For the 4528 unique herbal compounds from the 197 herbs, 1674 Bemis-Murcko scaffolds were obtained [34], among which 988 scaffolds (~59%) were shared by compounds in the annotated ligand sets. In our prediction for each of the 4528 herbal compounds, only 583 compounds (~13%) could not be associated with any other scaffold in the annotated ligand sets. Therefore, one can use SEA to predict the target for the majority of the herbal compounds. Only a small fraction of the compounds were not able to be applied to SEA due to their unique topology. The maximum Tanimoto coefficient (maxTC) for each pairwise association was also calculated, with ~89% of the compounds having values of at least 0.4. In other words, similar compounds occur in both the herbs and the reference library.
Analysis of herb-target associations
Herb-target associations with E-values of less than 10−10 were documented (Supplementary Table S2). At this E-value level, the associations are significant from a statistical point of view. In total, we obtained 3172 associations among 197 herbs and 406 drug targets. The top 10 herb-associated targets were adhesin protein fimH, cytochrome P450 1B1, 3-oxoacyl-[acyl-carrier protein] reductase, arachidonate 5-lipoxygenase, fatty acid synthase, aldose reductase, arachidonate 12-lipoxygenase, sodium/glucose cotransporter 1, xanthine dehydrogenase and cytochrome P450 17A1. Except for the first target from E. coli, all the others exist in the human body. Many targets bind to various glycosides and flavonoid derivatives, which are quite popular as natural compounds. As a result, this may account for the polypharmacology in herbal medicine.
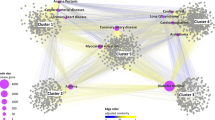
Herb-target associations with a smaller E-value threshold of 10−60 are depicted in Fig. 2. At this cutoff, only 195 associations remained among 79 herbs and 54 targets, with cytochrome P450 1B1 being the most associated target with 27 herbs. This protein belongs to the cytochrome P450 superfamily of enzymes, which catalyzes many reactions involved in drug metabolism [35]. Therefore, herbs linked to such targets are expected to either be well metabolized or inhibit cytochrome P450 in the human body.
Herb-target associations predicted by SEA with an E-value less than 10−60. This figure displays a subset of the herb-target associations from Supplementary Table S2. Targets of adhesin protein fimH in E. coli and CG8425-PA in Drosophila are not shown for clarity. The red nodes represent the herbs while the blue nodes represent the targets. The node size of the target is scaled by the number of associated herbs and the thickness of the edge is scaled by the E-value (the smaller the E-value, the thicker the edge)
Verification of the predicted targets for three representative herbs
To examine the putative targets revealed by SEA, we manually checked three well-known herbs: Radix Glycyrrhizae, Flos Lonicerae, and Rhizoma Coptidis (Supplementary Table S3). Radix Glycyrrhizae is the most frequent ingredient prescribed in diverse herbal formulas for a spectrum of diseases [36, 37]. SEA revealed that Radix Glycyrrhizae may associate with 27 targets in different pathways. Among the 27 identified targets, 19 were reported to interact with Radix Glycyrrhizae, and the remaining targets might warrant further exploration. Flos Lonicerae is often used as an anti-inflammatory, antibacterial, and antidiabetic herb [38, 39], which agrees well with the predicted targets, including arachidonate 5-lipoxygenase, adhesin protein fimH, protein-tyrosine phosphatase 1B, and aldose reductase. Rhizoma Coptidis is usually prescribed for neurological disorders (e.g., Alzheimer’s disease), inflammation, and skin disorders [40, 41]. Consistently, this herb was associated with acetylcholinesterase, cholinesterase, butyrylcholinesterase, arachidonate 5-lipoxygenase, and tyrosinase. Recently, Rhizoma Coptidis has attracted much attention for the treatment of obesity and diabetes due to the effective compound berberine [42]. Its possible targets are not clarified but are suggested to be AMP-activated protein kinase, gut microbiota, etc. [43, 44]. Our SEA analysis showed that berberine might be an inhibitor of butyrylcholinesterase, which has been linked to obesity as reported in some studies [45,46,47].
Generally, the precision of SEA is satisfactory since the majority of targets can be confirmed in the scientific literature, while the remaining targets might also be true in future studies. In terms of recall, it is largely dependent on the currently known ligands from both the targets and herbs. With the databases becoming more comprehensive or with new methods for ligand similarity calculations, SEA will consequently have a higher recall. As a proof of concept, our analysis demonstrated that SEA can be reliably applied to predict drug targets for a given set of ligands from herbal medicine. Moreover, the E-value threshold of 10−10 is a reasonable cutoff in our analysis.
Analysis of herb-disease associations
Based on the targets revealed by SEA, we further linked the targets to diseases on the DisGeNET platform. DisGeNET has a comprehensive collection of human gene-disease associations, integrating resources from expert-curated databases (UniProt, CTD, PSYGENET, ORPHANET, HPO), animal models (RGD, MGD, CTD) and text-mining results (GAD, LHGDN, BEFREE) [31, 32]. Data from different resources are scored at different scales. For instance, one record from the curated database has a partial score of 0.2, while one from the animal model has a partial score of 0.08. The DGA score is computed by summing all the partial scores. Herein, we recorded all the associations at a cutoff of 0.08. With this criterion, 41628 associations (Supplementary Table S4) were made among 192 herbs and 2439 diseases, with 16% being orphan diseases [48]. Orthodox medicine lacks drugs for orphan diseases due to the various challenges in research and development [49]. However, herbal medicine may provide complementary solutions to the current situation and future drug discovery.
We have shown several well-known diseases and their associated herbs with E-values less than 10−30 and DGA scores of at least 0.3 (Fig. 3). One common disease was alcoholic intoxication (chronic) with nine associated herbs. Alcohol-related harms, either chronic or acute, are a huge public health problem in China [50]. Alcoholic intoxication-related targets include aldehyde dehydrogenase, alcohol dehydrogenase beta chain, serotonin transporter, alcohol dehydrogenase gamma chain, GABA receptor alpha-2 subunit, mu opioid receptor, etc. Here, we would like to suggest the herbs revealed from our computational study as alternative medicines. On the other hand, the herbs were linked to various diseases via different targets. For example, Sophora Japonica and Radix Puerariae were associated with alcoholic intoxication (chronic) by the target aldehyde dehydrogenase, with asthma by interleukin-5, and with melanoma by tyrosinase. Not surprisingly, herbal medicine, as a collection of many natural compounds, is more likely to interact with diverse diseases in a polypharmacological manner.
Target-oriented herbal formula design
With the herb-target and herb-disease associations in hand, we can formalize the CHM target profile for the diseases of interest. We illustrated the profiles for four common chronic diseases with E-values of less than 10−30 and DGA scores of at least 0.2 (Fig. 4). At different levels of the DGA score, the number of associated targets varied. While many targets are associated with the disease of interest, the DGA score informs us as to how relevant the targets are. Here, we chose a cutoff of 0.2 because associations from a manually curated source are scored at the level of at least 0.2.
CHM target profiles for four common chronic diseases. Herb-target associations with an E-value less than 10−30 and a DGA score of at least 0.2 are displayed for (a) Alzheimer’s disease, (b) depressive disorder, (c) hypertensive disease, and (d) non-insulin-dependent diabetes mellitus. The nodes with light colors represent the herbs while the dark nodes represent the targets. The node size of the target is scaled by the number of associated herbs
Again, by checking the research literature, we verified the predicted herbs in the CHM target profiles (Supplementary Table S5). To the best of our knowledge, some herbs have been prescribed or studied for the corresponding diseases, while other associations are new. For instance, four herbs (not including Fructus Hordei Germinatus) have been used in treating Alzheimer’s disease. Among the 15 herbs associated with depressive disorder, 10 have demonstrated antidepressive functions in previous usage or experiments. For hypertensive disease, 20 out of 33 herbs found to be associated were mentioned in the literature for this disease. In the case of non-insulin-dependent diabetes mellitus, 19 out of 24 herbs occurred previously in different herbal treatments or studies. Therefore, our prediction agreed well with the experimental studies and simultaneously provided novel findings.
From the CHM target profile, we can propose a method of target-oriented herbal formula design. In contrast to the traditional design approach, which focuses on the syndrome of the patient while balancing the nature and flavor of the herbal ingredients, our method is based on the drug targets that are associated with the disease of interest. The first example is Alzheimer’s disease (Fig. 4a), which occurs in a large part of the population above 70 years of age, yet lacks efficient drugs that are able to cure or prevent it [51, 52]. From computational predictions, five herbs were associated with six targets. Therefore, our designed formula would include the combination of Radix Aconiti Lateralis Preparata, Fructus Hordei Germinatus, Rhizoma Curcumae Longae, and Rhizoma Coptidis or Rhizoma Corydalis because the last two are close in pharmacology.
The second example is depressive disorder (Fig. 4b), which is associated with as many as 15 targets. In this case, we have more flexibility in the formula design depending on the understanding of the mechanism, which also echoes personalized medicine in depressive disorder treatment [53]. Nevertheless, the same principles still apply. For example, we can choose Semen Nelumbinis, Fructus Lycii, Fructus Quisqualis, and Fructus Hordei Germinatus since they could interact with more than one target. For the five herbs linked to P-glycoprotein 1, only one herb is suggested. Similar strategies can be adopted for hypertensive disease and non-insulin-dependent diabetes mellitus, as well. The philosophy of target-oriented herbal formula design is to cover as many targets as possible with herbs that can associate with multiple targets, while the E-value and DGA score serve as quantitative indexes.
Discussion
The modernization of CHM requires a postmodern understanding of the ancient narratives of the healing herbs. Although tremendous efforts have been made to reveal the rationale behind CHM treatment, a systematic pharmacological study on individual herbs is still a huge challenge. Herein, we employed the SEA to reveal 406 potential targets for 197 frequently prescribed herbs. To verify the results, we searched the predictions in the scientific literature for three well-known herbs and found that approximately 70% of the putative targets have been reported.
We further linked the drug targets on the DisGeNET platform so that the various diseases were related to herbs via the corresponding targets. At different DGA score cutoffs, herbs were suggested to be the alternative solutions to 2439 diseases, with 16% being orphan diseases. Consequently, the herb-target and herb-disease analyses laid the foundation for the disease-oriented herbal formula design, a modern design strategy leveraging and driven by the pharmacological data. This strategy enables complex diseases to be approached from multiple drug targets associated with different herbs. The method is also quantitative with E-value and DGA score describing how strong the herb-target-disease associations are.
In conclusion, our study provides a novel approach for rational herbal formula design based on the pharmacological predictions of herbs. This method holds great potential for applications to understand and reconstruct herbal medicine from a molecular level. It may serve as the initial step in the pipeline of natural compound-inspired drug discovery [54]. Follow-up in vitro and in vivo tests can further confirm and improve these predictions. Therefore, the ancient knowledge of CHM can be inherited and appreciated in line with modern biomedical research.
References
Normile D. Asian medicine: the new face of traditional Chinese medicine. Science. 2003;299:188–90.
Li Z, Xu C. The fundamental theory of traditional Chinese medicine and the consideration in its research strategy. Front Med. 2011;5:208–11.
Gu S, Pei J. Innovating Chinese herbal medicine: from traditional health practice to scientific drug discovery. Front Pharmacol. 2017;8:381.
Neill US. From branch to bedside: Youyou Tu is awarded the 2011 Lasker~DeBakey Clinical Medical Research Award for discovering artemisinin as a treatment for malaria. J Clin Investig. 2011;121:3768–73.
Su XZ, Miller LH. The discovery of artemisinin and the Nobel Prize in Physiology or Medicine. Sci China Life Sci. 2015;58:1175–9.
Gu S, Pei J. Chinese herbal medicine meets biological networks of complex diseases: a computational perspective. Evid Based Complement Altern Med. 2017;2017:7198645.
Xie T, Song S, Li S, Ouyang L, Xia L, Huang J. Review of natural product databases. Cell Prolif. 2015;48:398–404.
Feng Y, Wu ZH, Zhou XZ, Zhou ZM, Fan WY. Knowledge discovery in traditional Chinese medicine: State of the art and perspectives. Artif Intell Med. 2006;38:219–36.
Gu S, Yin N, Pei J, Lai L. Understanding molecular mechanisms of traditional Chinese medicine for the treatment of influenza viruses infection by computational approaches. Mol Biosyst. 2013;9:2696–700.
Chang KW, Tsai TY, Chen KC, Yang SC, Huang HJ, Chang TT, et al. iSMART: an integrated cloud computing web server for traditional chinese medicine for online virtual screening, de novo evolution and drug design. J Biomol Struct Dyn. 2011;29:243–50.
Liang H, Ruan H, Ouyang Q, Lai L. Herb-target interaction network analysis helps to disclose molecular mechanism of traditional Chinese medicine. Sci Rep. 2016;6:36767.
Gu S, Yin N, Pei J, Lai L. Understanding traditional Chinese medicine anti-inflammatory herbal formulae by simulating their regulatory functions in the human arachidonic acid metabolic network. Mol Biosyst. 2013;9:1931–8.
Li J, Lu C, Jiang M, Niu XY, Guo HT, Li L, et al. Traditional Chinese medicine-based network pharmacology could lead to new multicompound drug discovery. Evid-Based Complement Alternat Med. 2012;2012:149762.
Zobir SZM, Fauzi FM, Liggi S, Drakakis G, Fu XJ, Fan TP, et al. Global mapping of traditional Chinese medicine into bioactivity space and pathways annotation improves mechanistic understanding and discovers relationships between therapeutic action (sub)classes. Evid-Based Complement Alternat Med. 2016;2016:2106465.
Huang T, Mi H, Lin CY, Zhao L, Zhong LL, Liu FB, et al. MOST: most-similar ligand based approach to target prediction. BMC Bioinforma. 2017;18:165.
Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206.
Gong J, Cai C, Liu X, Ku X, Jiang H, Gao D, et al. ChemMapper: a versatile web server for exploring pharmacology and chemical structure association based on molecular 3D similarity method. Bioinformatics. 2013;29:1827–9.
Lo YC, Senese S, Li CM, Hu Q, Huang Y, Damoiseaux R, et al. Large-scale chemical similarity networks for target profiling of compounds identified in cell-based chemical screens. PLoS Comput Biol. 2015;11:e1004153.
Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50:742–54.
Willett P, Barnard JM, Downs GM. Chemical similarity searching. J Chem Inf Comp Sci. 1998;38:983–96.
Pearson WR. Empirical statistical estimates for sequence similarity searches. J Mol Biol. 1998;276:71–84.
Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–81.
Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361.
Zhou J, Xie G, Yan X. Encyclopedia of traditional Chinese medicines. Isol Compd AB. 2011;1:455.
He M, Yan X, Zhou J, Xie G. Traditional Chinese medicine database and application on the Web. J Chem Inf Comput Sci. 2001;41:273–7.
Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13.
Chen CY. TCM Database@Taiwan: the world’s largest traditional Chinese medicine database for drug screening in silico. PLoS One. 2011;6:e15939.
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33.
Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Comp Sci. 1988;28:31–6.
Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2011;40:D1100–7.
Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2016;45: gkw943.
Piñero J, Queralt-Rosinach N, Bravo À, Deu-Pons J, Bauer-Mehren A, Baron M, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database. 2015;2015:bav028.
Chen Y, Garcia de Lomana M, Friedrich N-O, Kirchmair J. Characterization of the chemical space of known and readily obtainable natural products. J Chem Inf Model. 2018;58:1518–32.
Bemis GW, Murcko MA. The properties of known drugs. 1. Mol Framew J Med Chem. 1996;39:2887–93.
Rendic S, Guengerich FP. Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem Res Toxicol. 2014;28:38–42.
Ji S, Li Z, Song W, Wang Y, Liang W, Li K, et al. Bioactive constituents of Glycyrrhiza uralensis (Licorice): Discovery of the effective components of a traditional herbal medicine. J Nat Products. 2016;79:281–92.
Asl MN, Hosseinzadeh H. Review of pharmacological effects of Glycyrrhiza sp. and its bioactive compounds. Phytother Res. 2008;22:709–24.
Muluye RA, Bian Y, Alemu PN. Anti-inflammatory and antimicrobial effects of heat-clearing chinese herbs: a current review. J Tradit Complement Med. 2014;4:93–8.
Shang X, Pan H, Li M, Miao X, Ding H. Lonicera japonica Thunb.: ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J Ethnopharmacol. 2011;138:1–21.
Howes M-JR, Houghton PJ. Plants used in Chinese and Indian traditional medicine for improvement of memory and cognitive function. Pharmacol Biochem Behav. 2003;75:513–27.
Parsaeimehr A, Martinez-Chapa S, Parra-Saldívar R. Medicinal plants versus skin disorders: a survey from ancient to modern herbalism. In: Kateryna K, Mahendra R, editors. The microbiology of skin, soft tissue, bone and joint infections. Amsterdam: Elsevier; 2017. p. 205–221.
Tang LQ, Wei W, Chen LM, Liu S. Effects of berberine on diabetes induced by alloxan and a high-fat/high-cholesterol diet in rats. J Ethnopharmacol. 2006;108:109–15.
Hwang JT, Kwon DY, Yoon SH. AMP-activated protein kinase: a potential target for the diseases prevention by natural occurring polyphenols. N Biotechnol. 2009;26:17–22.
Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One. 2012;7:e42529.
Brunhofer G, Fallarero A, Karlsson D, Batista-Gonzalez A, Shinde P, Mohan CG, et al. Exploration of natural compounds as sources of new bifunctional scaffolds targeting cholinesterases and beta amyloid aggregation: the case of chelerythrine. Bioorg Med Chem. 2012;20:6669–79.
Furtado-Alle L, Andrade FA, Nunes K, Mikami LR, Souza RL, Chautard-Freire-Maia EA. Association of variants of the -116 site of the butyrylcholinesterase BCHE gene to enzyme activity and body mass index. Chem Biol Interact. 2008;175:115–8.
Benyamin B, Middelberg RP, Lind PA, Valle AM, Gordon S, Nyholt DR, et al. GWAS of butyrylcholinesterase activity identifies four novel loci, independent effects within BCHE and secondary associations with metabolic risk factors. Hum Mol Genet. 2011;20:4504–14.
Aymé S. Orphanet, an information site on rare diseases. Soins. 2003;169:46–7.
Nwaka S, Ridley RG. Science & society: Virtual drug discovery and development for neglected diseases through public–private partnerships. Nat Rev Drug Discov. 2003;2:919.
Cochrane J, Chen H, Conigrave KM, Hao W. Alcohol use in China. Alcohol Alcohol. 2003;38:537–42.
Alzheimer’s A. 2015 Alzheimer’s disease facts and figures. Alzheimer’s Dementia: J Alzheimer’s Assoc. 2015;11:332.
Zhou X, Chen Y, Mok KY, Zhao Q, Chen K, Chen Y, et al. Identification of genetic risk factors in the Chinese population implicates a role of immune system in Alzheimer’s disease pathogenesis. Proc Natl Acad Sci U S A. 2018;115:1697–706.
Saltiel PF, Silvershein DI. Major depressive disorder: mechanism-based prescribing for personalized medicine. Neuropsychiatr Dis Treat. 2015;11:875.
Zhang J, He Y, Jiang X, Jiang H, Shen J. Nature brings new avenues to the therapy of central nervous system diseases—An overview of possible treatments derived from natural products. Sci China Life Sci. 2019;62:1332–67.
Acknowledgements
The authors would like to thank Dr. Hao Liang and other members of the Lai group for helpful discussions. This work was supported, in part, by the Ministry of Science and Technology (2016YFA05023032) and the National Natural Science Foundation of China (21633001).
Author information
Authors and Affiliations
Contributions
SG and LHL conceptualized the project. SG performed the experiments. SG and LHL wrote the manuscript together.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Gu, S., Lai, Lh. Associating 197 Chinese herbal medicine with drug targets and diseases using the similarity ensemble approach. Acta Pharmacol Sin 41, 432–438 (2020). https://doi.org/10.1038/s41401-019-0306-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-019-0306-9