Abstract
Several OPRD1 intronic variants were associated with opioid addiction (OD) in a population-specific manner. This follow-up study aims to further characterize the OPRD1 haplotype pattern of the risk variants in different populations and apply in silico analysis to identify potential causal variants. A population-specific haplotype pattern was revealed based on six OPRD1 eQTL SNPs and five common haplotypes were identified in a sample of European ancestry (CEU). A European-specific haplotype (‘Hap 3’) that includes SNPs previously associated with OD and is tagged by SNP rs2236861 is more common in subjects with OD. It is quite common (10%) in CEU but is absent in the African sample (YRI) and extends upstream of OPRD1. SNP rs2236857 is most probably a non-causal variant in LD with the causal SNP/s in a population-specific manner. The study provides an explanation for the lack of association in African Americans, despite its high frequency in this population. OD samples homozygous for ‘Hap 3’ were reanalyzed using a denser coverage of the region and revealed at least 25 potentially regulatory SNPs in high LD. Notably, GTEx data indicate that some of the SNPs are eQTLs for the upstream phosphatase and actin regulator 4 (PHACTR4), in the cortex, and others are eQTLs for OPRD1 and the upstream lncRNA ENSG00000270605, in the cerebellum. The study highlights the limitation of single SNP analysis and the sensitivity of association studies of OPRD1 to a genetic background. It proposes a long-range functional connection between OPRD1 and PHACTR4. PHACTR4, a mediator of cytoskeletal dynamics, may contribute to drug addiction by modulating synaptic plasticity.
Similar content being viewed by others
Introduction
Opioids act via the opioid receptors that have a major role in reward mechanisms, pain transmission, and drug addiction1,2. Opioid abuse is a major public health problem. The G-protein-coupled delta-opioid receptor (DOPr, DOR) is targeted by enkephalins and is widely distributed in the brain. It is involved in several brain processes, including learning and memory, anxiety, depression, and impulsivity3. It is also involved in regulating hyperalgesia as well as chronic inflammatory pain4. The opioid system has been well conserved throughout the evolution of vertebrates5. DOR is a target for the development of new pain therapies6. DOR is encoded by OPRD1 (NM_000911.4) on chromosome 1 and translates a 372-amino-acid protein that is highly conserved in vertebrates.
Susceptibility to opioid dependence (OD) includes genetic and non-genetic components. Several OPRD1 polymorphisms were associated with OD but many of the findings have not been replicated (for a recent review see ref. 7). Diverse genetic backgrounds and variation in linkage disequilibrium (LD) between populations may contribute to variable results in association studies. The OPRD1 intronic SNP rs2236857 was associated with OD in our original association study of subjects of European descent (EA)8. This result was replicated in an independent Australian cohort9. SNP rs2236857 was also associated with response to life stress in subjects with OD in Han Chinese10. A second intronic SNP, rs2236861, was associated with OD in EA in our original study and this result was replicated in a study from Austria11. Association study of a cohort of African descent (AA) did not detect any association of OPRD1 SNPs with OD12. One of the problems of association studies is that it is difficult to identify the causal variants since it is impossible to distinguish the signal of a marker from that of the other SNPs that are in LD with it. This challenge is greater when the LD structure differs between the populations. Non-causal markers may have different effects in different populations. Different approaches for fine mapping exist, including overlapping variants with functional elements13. Variants can disrupt the binding of transcription factors, resulting in a change in gene expression. Another approach is the use of quantitative trait loci (eQTL) information.
Our hypothesis was that the OD risk SNPs indicated in the original study are LD proxies for functional variant/s that are population-specific. To explore this hypothesis, we utilized publicly available functional and genetic data as well as data from our OD and control samples to obtain a higher resolution of the genetic structure of OPRD1 and to provide an explanation for the results of the association studies.
Materials and methods
Sample
The sample includes subjects with opioid addiction (OD) and controls and was described in detail elsewhere14. Briefly, OD subjects (cases) were recruited at several opiate treatment programs in the US (e.g., Manhattan Campus of VA NY Harbor Health Care System, Weill Medical College of Cornell University, and Dr. Miriam and Sheldon G. Adelson Clinic for Drug Abuse Treatment and Research, in Las Vegas) or at the Rockefeller University. All cases had a history of at least 1 year of multiple daily uses of heroin and were on methadone maintenance treatment at the time of recruitment. The European American (EA) sample included subjects with >50% European/Middle Eastern ancestry contributions, based on structure or PC analysis as described14,15 (OD, n = 545; Controls, n = 196). The African American (AA) sample included subjects with >50% African ancestry contribution (OD, n = 308; Controls, n = 190); self-identified Hispanics were excluded.
The study was approved by the institutional review boards of the VA New York Harbor Health Care System and the Rockefeller University (for Rockefeller University and the Las Vegas clinic). All subjects signed informed consent for genetic studies.
Genotyping
All samples were genotyped with the Illumina® 1536-plex GoldenGate custom panel that included a limited number of OPRD1 tag SNPs, as described8. A subsample was genotyped with the genome-wide custom Smokescreen® array, as described15,16.
Haplotype analysis and in silico functional analysis
Six SNPs were selected for haplotype analysis based on frequency, location, LD, and data from previous studies. Phased genotypes were obtained from the Ensembl genome browser. The main analysis was performed with the CEU (Utah residents with Northern and Western European ancestry) sample. The YRI (Yoruba in Ibadan, Nigeria), and CHB (Han Chinese in Beijing, China) samples were used for comparison.
The following databases were used for the analysis:
The Ensembl genome browser release 103 (www.ensembl.org)
IGSR: The International Genome Sample Resource (The 1000 Genomes Project) (https://www.internationalgenome.org/)
The Genome Aggregation Database (GnomeAD) V2.1.1 (https://gnomad.broadinstitute.org/)
The University of California, Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu/)
The Allele Frequency Database (Alfred) (alfred.med.yale.edu)
LDlink (https://ldlink.nci.nih.gov/)
RegulomeDB (https://www.regulomedb.org/)
ENCODE (https://screen.encodeproject.org)
GeneCards (https://www.genecards.org/)
The Human Protein Atlas (https://www.proteinatlas.org/)
TarBase v.8, DIANA Tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/)
The Genotype-Tissue Expression (GTEx)(https://gtexportal.org)
EMBL-EBI (https://www.ebi.ac.uk/).
Results
Selected SNPs for haplotype analysis
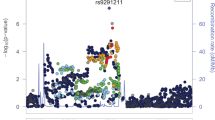
Six SNPs were selected for haplotype analysis based on previous association studies, LD pattern, frequency, location, and potential functionality (Table 1). Based on GTEx, SNPs rs2236857 and rs2236861, indicated in our original association study of OD8, are the expression quantitative trait loci (eQTLs) for the phosphatase and actin regulator 4 gene, PHACTR4, located ~300 kb upstream of OPRD1 (Fig. 1). The eQTL SNP rs10753331 is a proxy of SNP rs590013 that was associated with educational attainment17, as well as SNP rs419335 that was associated with OD9, and with decreased oxycodone analgesic response18. SNP rs67244013 is eQTL for OPRD1 in the cerebellum and testis. The synonymous SNP rs2234918 (p.Gly307=) on exon 3 is one of the only two common coding OPRD1 SNPs. It was indicated in several studies in association with OD, pain, and treatment outcome10,19,20,21, and is eQTL for OPRD1 in the testis, and the erythrocyte membrane protein band 4.1 gene, EPB41, in the cerebellum. SNP rs2234918 is in high LD with SNP rs4654327, indicated in association with OD in Han Chinese22. The 3′ UTR SNP rs204076 is eQTL for OPRD1 in the cortex.
LD blocks and cis-eQTLs
Several LD blocks, tagged by the selected SNPs, were identified in CEU (Fig. 1). They extend upstream and downstream of OPRD1. LD Block 1 extends from a region ~40 kb upstream to intron 1 of OPRD1, is tagged by SNP rs2236861, and includes several regulatory variants, including the regulatory SNP rs12133192. LD Block 2, on intron 1, is tagged by the regulatory SNP rs10753331 and extends ~21 kb. LD Block 3 is tagged by the 3′ UTR SNP rs204076 and extends ~30 kb from intron 2 of OPRD1 downstream to EPB41 (Fig. 1). The different LD pattern between populations affects the range of these LD blocks.
OPRD1 is expressed primarily in the brain with high expression in the cortex, caudate, hippocampus, nucleus accumbens, and putamen. Moderate to low expression was shown in the cerebellum, hypothalamus, substantia nigra, and thalamus. There is a complex relationship between eQTLs and their target genes in the extended OPRD1 region. While some OPRD1 SNPs are cis-eQTLs for other genes, several SNPs in the OPRD1 vicinity are cis-eQTLs for OPRD1. GTEx data indicate two distinct groups of eQTLs for OPRD1. The first group includes eQTLs for OPRD1 in the cortex. This group is tagged by the selected 3′ UTR SNP rs204076 and is related to LD Block 3 (Fig. 1). These SNPs are also eQTLs for EPB41 in the cerebellum, where it is highly expressed.
The second group represents eQTLs for OPRD1 in the cerebellum. This group includes SNPs in OPRD1 intron 1 and the upstream region. It includes SNP rs67244013 and its proxy SNP rs12749204 that are also eQTLs for an upstream lncRNA ENSG00000270605 (RP5-1092A3.4), as well as the regulatory SNP rs204048 that is eQTL for PHACTR4 in the cortex. It also includes the OPRD1 eQTL rs12089573. The significance of this group is undetermined since the function of OPRD1 in the cerebellum is still unknown. Interestingly, lncRNA ENSG00000270605 is mainly expressed in the cerebellum, but its function is not known.
OPRD1 is present in most vertebrates and its three exons are highly conserved. Comparative genomics data of the extended OPRD1 region indicate a strongly conserved synteny of the OPRD1 region on human chromosome 1, mouse chromosome 4, and rat chromosome 5. Notably, PHACTR4 and OPRD1 are adjacent in zebrafish whose common ancestors diverged ∼450 million years ago23. Zebrafish has two copies of oprd1 on chromosome 19:14,921,000-14,951,756 (oprd1a) and on chromosome 16:34,160,835-34,174,260 (oprd1b), as a result of a known whole-genome duplication5,24. There are also two phactr4 genes in these regions: phactr4a on chromosome 19, 191 kb from oprd1a, and phactr4b on chromosome 16, 25 kb from oprd1b.
Major haplotypes
Phased genotype data from the CEU sample were used to determine the major haplotypes from the selected SNPs and compared to representative samples from Asia (CHB) and Africa (YRI). Five major haplotypes (Haps 1−5) with a frequency of >0.05 were identified in CEU (Table 2 and Fig. 2). Hap 3 includes the variant C allele of SNP rs2236857, as well as the variant alleles of SNPs rs10753331, rs2234918, rs2236861, and rs67244013. It is uniquely tagged by rs2236861 among the common five haplotypes in CEU. SNPs rs67244013 and rs2236861 are redundant for defining the common haplotypes in this sample. Hap 3 is absent in YRI and is more common (10%) in CEU than in CHB (5%) (Table 2 and Fig. 2). It extends far upstream of OPRD1 and includes numerous eQTLs for other genes in the region (e.g., PHACTR4). The variant C allele of SNP rs2236857 appears on two common haplotypes (Hap 3 and Hap 4) and several haplotypes that are rare in CEU and more common in YRI.
There is a significant difference in the haplotype pattern between CEU and YRI and some similarities between CEU and CHB. The only common haplotype among the three samples is Hap 1. Haps 2 and 3 are rare in the African sample. Haps 7 and 8 are very frequent in YRI and rare in CEU and CHB.
This data highlights the higher resolution obtained by haplotype analysis compared to a single SNP analysis. The different pattern of haplotypes carrying the SNP rs2236857 variant allele in diverse populations and its appearance on more than one haplotype may explain in part the different association results in studies that were based on single SNPs. The intriguing finding that SNP rs2236857 was not associated with OD in African Americans despite its high frequency in Africa may be partially explained by the different haplotype patterns in different populations. It also suggests that SNP rs2236857 is not the causal SNP and may be a marker for a causal proxy SNP in a population-specific manner. Based on this information, association studies of OPRD1 are sensitive to ancestry contribution. The LD between rs2236861 and rs2236857 differs between populations (r2 = 0.3 in CEU, r2 = 0.8 in CHB), as reflected in the haplotype architecture.
Analysis of subjects with OD and controls
Subjects with OD and controls from two main ancestries (European and African Americans) were analyzed for OPRD1 haplotypes. There was a higher frequency (6%) of homozygotes for the variant alleles of SNPs rs2236861 and rs2236857 (AA−CC) in the EA OD sample compared to controls (2%). There were fewer homozygotes for the reference alleles (GG−TT) in the EA OD (41%) compared to controls (48%), corroborating the original results. This SNP combination is rare in AA, as expected based on allele frequencies. The haplotype pattern of the EA and AA samples was similar to that of CEU and YRI, respectively (Table 2), except for Hap 3 that was at higher frequency in the OD EA group compared to controls and the CEU sample.
A selected region overlapping OPRD1 was analyzed in-depth in samples that were homozygous for the main haplotypes and were genotyped by the genome-wide SmokeScreen® array15,16. Additional information was obtained from the two CEU samples that were homozygous for Hap 3. Numerous informative high-quality SNPs were found to be carried specifically by the samples homozygous to Hap 3. Excluding SNPs in which the Hap 3 homozygous samples include the ancestor allele (e.g., SNP rs3766951), the emerging picture is of a long haplotype with at least 25 potentially regulatory SNPs (Table 3). The majority of the SNPs are in OPRD1 intron 1 and some of them are shared by other less frequent haplotypes.
Notably, based on GTEx data, the majority of the SNPs are eQTLs for reduced expression of PHACTR4 in several brain tissues. Several SNPs are eQTLs for OPRD1 in the cerebellum. Two regulatory SNPs (rs509577 and rs72665504) on intron 1 that have no proxies in CEU were identified as unique to Hap 3 based on these samples. SNP rs509577 is located in a regulatory region ENSR00000352114 and is an eQTL for several genes in non-brain tissues. SNP rs72665504 is an eQTL for OPRD1 in testis.
Although no association of PHACTR4 SNPs was reported with OD and the LD blocks that define the major OPRD1 haplotypes do not extend to PHACTR4, we explored the relationship between two PHACTR4 eQTLs (rs7518249 and rs4654365) and OPRD1 haplotypes. There is low LD between SNPs rs7518249 and rs4654365 in CEU. The variant T allele of SNP rs7518249 is associated with reduced PHACTR4 expression in several brain regions (e.g., P = 7.8e−13, caudate, GTEx). It is located in a functional CTCF binding site and promoter flanking region. SNP rs7518249 is in moderate LD (r2 = 0.33) with the OPRD1 SNP rs2236861 in CEU. The variant G allele of the GMEB1 SNP rs4654365 is also associated with the reduced PHACTR4 brain expression (e.g., P = 1.0e−8, caudate, GTEx). SNP rs4654365 is in low LD (r2 = 0.13) with SNP rs2236861 in CEU. Haplotype analysis in our sample as well as in CEU revealed that the variant alleles of these PHACTR4 eQTLs are part of Hap 3, although they are not unique to it.
Several SNPs that were reported in the literature were not included in this study for simplicity and technical reasons. SNP rs1042114 is a functional missense variant in exon 1 (c.80T>Gp.Phe27Cys). The minor frequency of the G allele is 13% in CEU and is very low in Africa and East Asia indicating that it has probably arisen after the split between European and Asian populations. In CEU, it appears mostly on Hap 2, sometimes on Hap 1, and on rare haplotypes that are independent of Hap 3. It is in complete LD with the upstream SNP rs61787581 located in a TF binding site and is also in high LD with several SNPs in intron 1, including the regulatory SNPs rs569356 and rs204051, as well as SNP rs533123 that was associated in a large-scale GWAS with schizophrenia25,26 and educational attainment17. SNP rs590013 that was associated with educational attainment17 is a proxy of SNP rs10753331 included in this study. SNP rs678849 was shown to predict OD treatment response in African Americans, but not in European Americans21,27. Hap 3 includes the ancestral C allele of SNP rs678849.
Discussion
Association studies indicated OPRD1 risk variants for drug addiction in a population-specific manner. Specifically, two intronic SNPs indicated in our original study in a cohort of European descent8 were replicated in independent samples of European ancestry9,11 but not in an African cohort12. These findings led to the hypothesis that these SNPs are LD proxies for functional variants in a population-specific manner. To explore this hypothesis, we have used publicly available data as well as data from our samples to define the haplotype structure of OPRD1 and to perform in silico functional analysis.
Haplotype analysis suggested a large shift in the OPRD1 genetic architecture between populations and showed the limitation of the original single SNP analyses. One of the target SNPs, rs2236857, was found to be a part of two common haplotypes in Europeans and two additional African-specific haplotypes. This population-specific pattern can explain the negative result obtained in the African American sample and highlights the sensitivity of association studies of OPRD1 to population stratification. Notably, one of these haplotypes that is tagged by one of the original identified SNPs, rs2236861, in addition to SNP rs2236857, is quite common in European samples but is absent in African samples. The frequency of this haplotype was higher in the OD sample compared to controls, suggesting that it contributed to the association signal. In silico functional analysis revealed that SNPs within this haplotype are associated with a reduced expression for the upstream PHACTR4, encoding phosphatase and actin regulator 4, in the cortex. Additionally, some of the SNPs within this haplotype are eQTLs for OPRD1 and an upstream lncRNA, in the cerebellum.
The study proposes a long-range functional connection between noncoding regions of OPRD1 and PHACTR4 expression that may provide an alternative explanation for the effect of OD risk variants. This connection was previously suggested, but not further explored9. This kind of long-range connection was shown in studies of other genes. For example, obesity-associated SNPs within the FTO gene that was known to be related to obesity were shown to form long-range functional connections with IRX328. These findings have implications beyond these specific genes. It points to the need for careful interpretations of association studies, especially for noncoding risk variants. Long-range functional connections can be formed by long-range chromatin interactions that may skip over genes29.
Opioid addiction, cytoskeletal dynamics, and synaptic plasticity
Neurons rely on cytoskeletal dynamics for synaptic plasticity. Synaptic plasticity is regulated by the polymerization of a cytoskeletal protein, actin. Actin cycling is controlled by transcription and epigenetic regulation of cytoskeletal proteins encoding genes30. Brain plasticity allows an individual to adapt to changes in the environment. Dendritic spines are dynamic post-synaptic structures that are thought to store memory. Drugs of abuse can alter actin dynamics and cause structural changes to dendrites, as was recently shown for the actin-binding protein, drebrin31,32. Drug relapse was associated with the involuntary retrieval of associative memories33.
The proposed association between PHACTR4 and opioid addiction is intriguing. PHACTR4 is part of a family of four structurally related members (PHACTR1-4)34. This phosphatase and actin regulator inhibits protein phosphatase-1 (PP-1), which is one of the main serine/threonine dephosphorylating enzymes in the cell35. Phactr4 was shown to have a role in a feedback loop that maintains actin monomers level36. Phactr4 was shown to regulate cytoskeletal remodeling during enteric neural crest cells37, and mice with a missense mutation in Phactr4 had defective neural tubes and optic fissure closures38. Phactr4 may have a different role in adult plasticity in addition to its developmental role.
In summary, the study suggested a large shift in the OPRD1 genetic architecture between populations, identified a European-specific haplotype that is associated with OD, and showed the limitations of single SNP analyses. It proposes a long-range functional connection between noncoding regions of OPRD1 and PHACTR4, and suggests that PHACTR4, as a mediator of cytoskeletal dynamics, contributes to drug addiction. If proven experimentally, this study can advance our understanding of the molecular mechanisms of synaptic plasticity in drug addiction and may provide an avenue for treatment.
References
Darcq, E. & Kieffer, B. L. Opioid receptors: drivers to addiction? Nat. Rev. Neurosci. 19, 499–514 (2018).
Kreek, M. J. et al. Opiate addiction and cocaine addiction: underlying molecular neurobiology and genetics. J. Clin. Invest. 122, 3387–3393 (2012).
Chu Sin Chung, P. & Kieffer, B. L. Delta opioid receptors in brain function and diseases. Pharmacol. Ther. 140, 112–120 (2013).
Gendron, L., Cahill, C. M., von Zastrow, M., Schiller, P. W. & Pineyro, G. Molecular pharmacology of delta-opioid receptors. Pharmacol. Rev. 68, 631–700 (2016).
Dreborg, S., Sundstrom, G., Larsson, T. A. & Larhammar, D. Evolution of vertebrate opioid receptors. Proc. Natl Acad. Sci. USA 105, 15487–15492 (2008).
Quirion, B., Bergeron, F., Blais, V. & Gendron, L. The delta-opioid receptor; a target for the treatment of pain. Front. Mol. Neurosci. 13, 52 (2020).
Burns, J. A. et al. Molecular imaging of opioid and dopamine systems: insights into the pharmacogenetics of opioid use disorders. Front. Psychiatry 10, 626 (2019).
Levran, O. et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 7, 720–729 (2008).
Nelson, E. C. et al. Association of OPRD1 polymorphisms with heroin dependence in a large case-control series. Addict. Biol. 19, 111–121 (2014).
Huang, C. C. et al. OPRD1 gene affects disease vulnerability and environmental stress in patients with heroin dependence in Han Chinese. Prog. Neuropsychopharmacol. Biol. Psychiatry 89, 109–116 (2019).
Beer, B. et al. Association of polymorphisms in pharmacogenetic candidate genes (OPRD1, GAL, ABCB1, OPRM1) with opioid dependence in European population: a case-control study. PLoS ONE 8, e75359 (2013).
Levran, O. et al. Heroin addiction in African Americans: a hypothesis-driven association study. Genes Brain Behav. 8, 531–540 (2009).
Broekema, R. V., Bakker, O. B. & Jonkers, I. H. A practical view of fine-mapping and gene prioritization in the post-genome-wide association era. Open Biol. 10, 190221 (2020).
Levran, O. et al. African-specific variability in the acetylcholine muscarinic receptor M4: association with cocaine and heroin addiction. Pharmacogenomics 17, 995–1003 (2016).
Levran, O. et al. A non-coding CRHR2 SNP rs255105, a cis-eQTL for a downstream lincRNA AC005154.6, is associated with heroin addiction. PLoS ONE 13, e0199951 (2018).
Baurley, J. W., Edlund, C. K., Pardamean, C. I., Conti, D. V. & Bergen, A. W. Smokescreen: a targeted genotyping array for addiction research. BMC Genomics 17, 145 (2016).
Lee, J. J. et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 50, 1112–1121 (2018).
Olesen, A. E. et al. The genetic influences on oxycodone response characteristics in human experimental pain. Fundam. Clin. Pharmacol. 29, 417–425 (2015).
Olesen, A. E. et al. Association between genetic polymorphisms and pain sensitivity in patients with hip osteoarthritis. Pain Pract. 18, 587–596 (2018).
Alongkronrusmee, D., Chiang, T. & van Rijn, R. M. Delta opioid pharmacology in relation to alcohol behaviors. Handb. Exp. Pharmacol. 247, 199–225 (2018).
Crist, R. C. et al. An intronic variant in OPRD1 predicts treatment outcome for opioid dependence in African-Americans. Neuropsychopharmacology 38, 2003–2010 (2013).
Gao, X. et al. Contribution of genetic polymorphisms and haplotypes in DRD2, BDNF, and opioid receptors to heroin dependence and endophenotypes among the Han Chinese. OMICS 21, 404–412 (2017).
Fishman, M. C. Genomics. Zebrafish—the canonical vertebrate. Science 294, 1290–1291 (2001).
Pinal-Seoane, N. et al. Characterization of a new duplicate delta-opioid receptor from zebrafish. J. Mol. Endocrinol. 37, 391–403 (2006).
Lam, M. et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 51, 1670–1678 (2019).
Goes, F. S. et al. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am. J. Med. Genet. B Neuropsychiatr. Genet. 168, 649–659 (2015).
Crist, R. C. et al. Replication of the pharmacogenetic effect of rs678849 on buprenorphine efficacy in African-Americans with opioid use disorder. Pharmacogenomics J. 19, 260–268 (2019).
Smemo, S. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375 (2014).
Chandra, V. et al. Promoter-interacting expression quantitative trait loci are enriched for functional genetic variants. Nat. Genet. 53, 110–119 (2021).
Carlisle, H. J. & Kennedy, M. B. Spine architecture and synaptic plasticity. Trends Neurosci. 28, 182–187 (2005).
Martin, J. A. et al. A novel role for the actin-binding protein drebrin in regulating opiate addiction. Nat. Commun. 10, 4140 (2019).
Rothenfluh, A. & Cowan, C. W. Emerging roles of actin cytoskeleton regulating enzymes in drug addiction: actin or reactin’? Curr. Opin. Neurobiol. 23, 507–512 (2013).
Milton, A. L. & Everitt, B. J. The persistence of maladaptive memory: addiction, drug memories and anti-relapse treatments. Neurosci. Biobehav. Rev. 36, 1119–1139 (2012).
Allen, P. B., Greenfield, A. T., Svenningsson, P., Haspeslagh, D. C. & Greengard, P. Phactrs 1-4: a family of protein phosphatase 1 and actin regulatory proteins. Proc. Natl Acad. Sci. USA 101, 7187–7192 (2004).
Ceulemans, H. & Bollen, M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 84, 1–39 (2004).
Huet, G. et al. Actin-regulated feedback loop based on Phactr4, PP1 and cofilin maintains the actin monomer pool. J. Cell. Sci. 126, 497–507 (2013).
Zhang, Y., Kim, T. H. & Niswander, L. Phactr4 regulates directional migration of enteric neural crest through PP1, integrin signaling, and cofilin activity. Genes Dev. 26, 69–81 (2012).
Kim, T. H., Goodman, J., Anderson, K. V. & Niswander, L. Phactr4 regulates neural tube and optic fissure closure by controlling PP1-, Rb-, and E2F1-regulated cell-cycle progression. Dev. Cell 13, 87–102 (2007).
Dong, S. & Boyle, A. P. Predicting functional variants in enhancer and promoter elements using RegulomeDB. Hum. Mutat. 40, 1292–1298 (2019).
Acknowledgements
This paper is dedicated to the memory of our mentor Prof. Mary Jeanne Kreek, in recognition of her incredible contributions and dedication to the research and treatment of drug addiction. This work was supported by Dr. Miriam and Sheldon G. Adelson Medical Research Foundation and the Clinical and Translational Science Award UL1RR024143 from the National Center for Advancing Translational Sciences of the NIH (B. Coller).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Levran, O., Randesi, M., Adelson, M. et al. OPRD1 SNPs associated with opioid addiction are cis-eQTLs for the phosphatase and actin regulator 4 gene, PHACTR4, a mediator of cytoskeletal dynamics. Transl Psychiatry 11, 316 (2021). https://doi.org/10.1038/s41398-021-01439-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-021-01439-y