Abstract
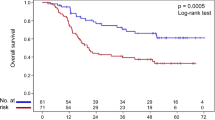
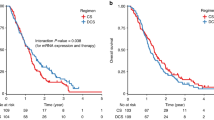
Genetic variations in DNA base excision repair (BER) genes may affect tumor sensitivity to chemotherapy and radiotherapy. Thus, we investigated the effects of single-nucleotide polymorphisms (SNPs) in key BER pathway genes on clinical outcomes in male patients who received concurrent chemoradiotherapy (CCRT). Seven SNPs from XRCC1, OGG1, APEX1, and MUTYH were genotyped using the Sequenom iPLEX MassARRAY system in samples from 319 men with advanced oral squamous cell carcinoma. The disease-free survival (DFS) rates of the MUTYH rs3219489 genotypes and those of the other genotypes differed significantly (log-rank test p = 0.027). Multivariate Cox proportional hazard analysis showed that the MUTYH rs3219489 GG genotype was associated with poor DFS (recessive model: hazard ratio [HR] = 2.01, 95% confidence interval [CI] = 1.31–3.10; p = 0.002). The CT + TT genotypes of XRCC1 rs1799782 (dominant model: HR = 0.65, 95% CI = 0.43–0.99; p = 0.044) and GG genotype of APEX1 rs1760944 (recessive model: HR = 1.64, 95% CI = 1.00–2.70; p = 0.050) were associated with overall survival (OS). Carrying the two risk genotypes, CC and GG of XRCC1 rs1799782 and APEX1 rs1760944, respectively, (HR = 2.95, 95% CI = 1.47–5.88; p = 0.002) increased mortality risk. Our findings showed that carrying the two risk genotypes of XRCC1 rs1799782 and APEX1 rs1760944 was associated with poor OS, while the GG genotype of MUTYH rs3219489 was associated with poor DFS. Patients carrying the risk genotypes may not benefit from CCRT; therefore, they will need alternative treatments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Chiang C-J, Lo W-C, Yang Y-W, You S-L, Chen C-J, Lai M-S. Incidence and survival of adult cancer patients in Taiwan, 2002–2012. J Formos Med Assoc. 2016;115:13.
Huang C-C, Ou C-Y, Lee W-T, Hsiao J-R, Tsai S-T, Wang J-D. Life expectancy and expected years of life lost to oral cancer in Taiwan: a nation-wide analysis of 22,024 cases followed for 10 years. Oral Oncol. 2015;51:349–54.
Chuang S-L, Su WW-Y, Chen SL-S, Yen AM-F, Wang C-P, Fann JC-Y, et al. Population-based screening program for reducing oral cancer mortality in 2,334,299 Taiwanese cigarette smokers and/or betel quid chewers. Cancer. 2017;123:1597–609.
Tsai K-Y, Su C-C, Lin Y-Y, Chung J-A, Lian I-B. Quantification of betel quid chewing and cigarette smoking in oral cancer patients. Community Dent Oral Epidemiol. 2009;37:555–61.
Güneri P, Epstein JB. Late stage diagnosis of oral cancer: components and possible solutions. Oral Oncol. 2014;50:1131–6.
Ferris RL, Geiger JL, Trivedi S, Schmitt NC, Heron DE, Johnson JT, et al. Phase II trial of post-operative radiotherapy with concurrent cisplatin plus panitumumab in patients with high-risk, resected head and neck cancer. Ann Oncol. 2016;27:2257–62.
Adelstein D, Gillison ML, Pfister DG, Spencer S, Adkins D, Brizel DM, et al. NCCN guidelines insights: head and neck cancers, version 2.2017. J Natl Compr Canc Netw. 2017;15:761–70.
Chitapanarux I, Traisathit P, Komolmalai N, Chuachamsai S, Sittitrai P, Pattarasakulchai T, et al. Ten-year outcome of different treatment modalities for squamous cell carcinoma of oral cavity. Asian Pac J Cancer Prev. 2017;18:1919–24.
Zhang H, Dziegielewski PT, Biron VL, Szudek J, Al-Qahatani KH, O’Connell DA, et al. Survival outcomes of patients with advanced oral cavity squamous cell carcinoma treated with multimodal therapy: a multi-institutional analysis. J Otolaryngol Head Neck Surg. 2013;42:30.
Santosh A, Jones T, Harvey J. A review on oral cancer biomarkers: understanding the past and learning from the present. J Cancer Res Ther. 2016;12:486–92.
Taghavi N, Yazdi I. Prognostic factors of survival rate in oral squamous cell carcinoma: clinical, histologic, genetic and molecular concepts. Arch Iran Med. 2015;18:314–9.
Quintela-Fandino M, Hitt R, Medina PP, Gamarra S, Manso L, Cortes-Funes H, et al. DNA-repair gene polymorphisms predict favorable clinical outcome among patients with advanced squamous cell carcinoma of the head and neck treated with cisplatin-based induction chemotherapy. J Clin Oncol. 2006;24:4333–9.
Jadhav KB, Gupta N. Clinicopathological prognostic implicators of oral squamous cell carcinoma: need to understand and revise. N Am J Med Sci. 2013;5:671–9.
Hornhardt S, Rossler U, Sauter W, Rosenberger A, Illig T, Bickeboller H, et al. Genetic factors in individual radiation sensitivity. DNA Repair. 2014;16:54–65.
Wallace SS. Base excision repair: a critical player in many games. DNA Repair. 2014;19:14–26.
Gavande NS, VanderVere-Carozza PS, Hinshaw HD, Jalal SI, Sears CR, Pawelczak KS, et al. DNA repair targeted therapy: the past or future of cancer treatment? Pharm Ther. 2016;160:65–83.
Visnes T, Grube M, Hanna BMF, Benitez-Buelga C, Cazares-Korner A, Helleday T. Targeting BER enzymes in cancer therapy. DNA Repair. 2018;71:118–26.
López-Verdín S, Lavalle-Carrasco J, Carreón-Burciaga RG, Serafín-Higuera N, Molina-Frechero N, González-González R, et al. Molecular markers of anticancer drug resistance in head and neck squamous cell carcinoma: a literature review. Cancers. 2018;10:376.
Hu JJ, Smith TR, Miller MS, Mohrenweiser HW, Golden A, Case LD. Amino acid substitution variants of APE1 and XRCC1 genes associated with ionizing radiation sensitivity. Carcinogenesis. 2001;22:917–22.
Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–8.
Li DJ, Xiao D. Association between the XRCC1 polymorphisms and clinical outcomes of advanced NSCLC treated with platinum-based chemotherapy: a meta-analysis based on the PRISMA statement. BMC Cancer. 2017;17:501.
Mahimkar MB, Samant TA, Kannan S, Tulsulkar J, Pai PS, Anantharaman D. Polymorphisms in GSTM1 and XPD genes predict clinical outcome in advanced oral cancer patients treated with postoperative radiotherapy. Mol Carcinog. 2012;51:E94–103.
Senghore T, Wang WC, Chien HT, Chen YX, Young CK, Huang SF, et al. Polymorphism of Mismatch Repair Pathway Genes Predict Clinical Outcomes in Oral Squamous Cell Carcenoma Patients receiving Ajuvant Concurrent Chemradiotherapy. Cancers. 2019;11:598.
Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–4.
Wu Y-G, Li H-F, Ren Y-J, Zou D-B, Zhang K-N, Xiao X. The association of XRCC1 polymorphism with osteosarcoma risk, clinicopathologic features, and prognosis in a Chinese Han population. Cancer Manag Res. 2018;10:4959–67.
Thakur S, Sarkar B, Cholia RP, Gautam N, Dhiman M, Mantha AK. APE1/Ref-1 as an emerging therapeutic target for various human diseases: phytochemical modulation of its functions. Exp Mol Med. 2014;46:e106.
Wang J, Guo C, Gong X, Ao F, Huang Y, Huang L, et al. The impacts of genetic polymorphisms in genes of base excision repair pathway on the efficacy and acute toxicities of (chemo)radiotherapy in patients with nasopharyngeal carcinoma. Oncotarget. 2017;8:78633–41.
Costa EF, Santos ES, Liutti VT, Leal F, Santos VC, Rinck-Junior JA, et al. Association between polymorphisms in genes related to DNA base-excision repair with risk and prognosis of oropharyngeal squamous cell carcinoma. J Cancer Res Clin Oncol. 2016;142:1917–26.
Yen CY, Liu SY, Chen CH, Tseng HF, Chuang LY, Yang CH, et al. Combinational polymorphisms of four DNA repair genes XRCC1, XRCC2, XRCC3, and XRCC4 and their association with oral cancer in Taiwan. J Oral Pathol Med. 2008;37:271–7.
Senghore T, Chien HT, Wang WC, Chen YX, Young CK, Huang SF, et al. Polymorphisms in ERCC5 rs17655 and ERCC1 rs735482 Genes Associated with the Survival of Male Patients with Postoperative Oral Squamous Cell Carcenoma Treated with Adjuvant Concurrent Chemoradiotherapy. J Clin Med. 2019;8:33.
Cui Z, Yin Z, Li X, Wu W, Guan P, Zhou B. Association between polymorphisms in XRCC1 gene and clinical outcomes of patients with lung cancer: a meta-analysis. BMC Cancer. 2012;12:71.
Gu AQ, Wang WM, Chen WY, Shi CL, Lu JH, Han JQ. XRCC1 genetic polymorphisms and sensitivity to platinum-based drugs in non-small cell lung cancer: an update meta-analysis based on 4708 subjects. Int J Clin Exp Med. 2015;8:145–54.
Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583.
Hsia KT, Liu CJ, Mar K, Lin LH, Lin CS, Cheng MF, et al. Impact of apurinic/apyrimidinic endonuclease 1/redox factor-1 on treatment response and survival in oral squamous cell carcinoma. Head Neck. 2016;38:550–9.
Koukourakis MI, Giatromanolaki A, Kakolyris S, Sivridis E, Georgoulias V, Funtzilas G, et al. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/Ref-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. Int J Radiat Oncol Biol Phys. 2001;50:27–36.
Xiao X, Yang Y, Ren Y, Zou D, Zhang K, Wu Y. rs1760944 polymorphism in the APE1 region is associated with risk and prognosis of osteosarcoma in the Chinese Han population. Scic Rep. 2017;7:9331.
Mazzei F, Viel A, Bignami M. Role of MUTYH in human cancer. Mutat Res. 2013;743-744:33–43.
Turco E, Ventura I, Minoprio A, Russo MT, Torreri P, Degan P, et al. Understanding the role of the Q338H MUTYH variant in oxidative damage repair. Nucleic Acids Res. 2013;41:4093–103.
Shinmura K, Goto M, Suzuki M, Tao H, Yamada H, Igarashi H, et al. Reduced expression of MUTYH with suppressive activity against mutations caused by 8-hydroxyguanine is a novel predictor of a poor prognosis in human gastric cancer. J Pathol. 2011;225:414–23.
Singh A, Singh N, Behera D, Sharma S. Genetic Investigation of Polymorphic OGG1 and MUTYH Genes Towards Increased Susceptibility in Lung Adenocarcinoma and its Impact on Overall Survival of Lung Cancer Patients Treated with Platinum Based Chemotherapy. Pathol Oncol Res. 2017;25:1327–40.
Acknowledgements
The authors are grateful to the study participants.
Funding
Support for this study was obtained from Ministry of Science and Technology, Taiwan (MOST107-2314-B-038-071, MOST108-2314-B-038-088, and MOST106-2314-B-182-025-MY3), Health and Welfare Surcharge on Tobacco Products (MOHW109-TDU-B-212-134016), and Chang Gung Memorial Hospital (CMRPG3H0793, CMRPG3J0591, CMRPG3J0592 and CMRPB53).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Senghore, T., Chien, HT., Wang, WC. et al. Predictive value of genetic variants XRCC1 rs1799782, APEX1 rs1760944, and MUTYH rs3219489 for adjuvant concurrent chemoradiotherapy outcomes in oral squamous cell carcinoma patients. Pharmacogenomics J 20, 813–822 (2020). https://doi.org/10.1038/s41397-020-0170-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-020-0170-5