Abstract
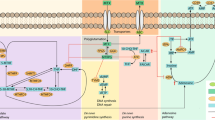
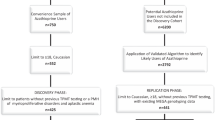
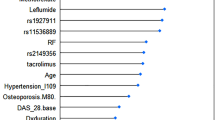
The objective of the study is to develop genetic and clinical prediction models for the efficacy and hepatotoxicity of methotrexate (MTX) in patients with rheumatoid arthritis (RA). Among RA patients treated with MTX, 1966 polymorphisms of 246 enzymes/transporters relevant to pharmacokinetics and pharmacodynamics were measured by the Drug Metabolism Enzymes and Transporters (DMET) microarray and direct sequencing, and clinical variables at baseline were collected. For efficacy, response criteria of the European League Against Rheumatism were used to classify patients as responders or non-responders. Hepatotoxicity was defined as elevations of aspartate aminotransferase or alanine aminotransferase ≥1.5 times the reference range upper limit. Among 166 patients, a genetic prediction model for efficacy using seven polymorphisms showed the area under the receiver operating characteristic curve (AUC) was 0.822, with 74.3% sensitivity and 76.8% specificity. A combined genetic and clinical model indicated the AUC was 0.844, with 81.5% sensitivity and 76.9% specificity. By incorporating clinical variables into the genetic model, the overall category-free net reclassification improvement (NRI) was 0.663 (P < 0.0001) and the overall integrated discrimination improvement (IDI) was 0.083 (P = 0.0009). For hepatotoxicity, a genetic prediction model using seven polymorphisms showed the AUC was 0.783 with 70.0% sensitivity and 80.0% specificity, while the combined model indicated the AUC was 0.906 with 85.1% sensitivity and 87.8% specificity (overall category-free NRI: 1.002, P < 0.0001; overall IDI: 0.254, P < 0.0001). Our genetic and clinical models demonstrated moderate diagnostic accuracy for MTX efficacy and high accuracy for hepatotoxicity. These findings should, however, be validated and interpreted with a caution until external validation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availablility
Allele frequencies are available in https://humandbs.biosciencedbc.jp/.
References
Singh JA, Saag KG, Bridges SL Jr, Akl EA, Bannuru RR, et al. 2015 American College of Rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Rheumatol. 2016;68:1–26.
Smolen JS, Landewe R, Bijlsma J, Burmester G, Chatzidionysiou K, Dougados M, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2016 update. Ann Rheum Dis. 2017;76:960–77.
Lopez-Olivo MA, Siddhanamatha HR, Shea B, Tugwell P, Wells GA, Suarez-Almazor ME. Methotrexate for treating rheumatoid arthritis. Cochrane Database Syst Rev. 2014;10:Cd000957.
Curtis JR, Beukelman T, Onofrei A, Cassell S, Greenberg JD, Kavanaugh A, et al. Elevated liver enzyme tests among patients with rheumatoid arthritis or psoriatic arthritis treated with methotrexate and/or leflunomide. Ann Rheum Dis. 2010;69:43–7.
Brown PM, Pratt AG, Isaacs JD. Mechanism of action of methotrexate in rheumatoid arthritis, and the search for biomarkers. Nat Rev Rheumatol. 2016;12:731–42.
Qiu Q, Huang J, Shu X, Fan H, Zhou Y, Xiao C. Polymorphisms and pharmacogenomics for the clinical efficacy of methotrexate in patients with rheumatoid arthritis: a systematic review and meta-analysis. Sci Rep. 2017;7:44015.
Qiu Q, Huang J, Lin Y, Shu X, Fan H, Tu Z, et al. Polymorphisms and pharmacogenomics for the toxicity of methotrexate monotherapy in patients with rheumatoid arthritis: a systematic review and meta-analysis. Medicine. 2017;96:e6337.
Umicevic Mirkov M, Janss L, Vermeulen SH, van de Laar MA, van Riel PL, Guchelaar HJ, et al. Estimation of heritability of different outcomes for genetic studies of TNFi response in patients with rheumatoid arthritis. Ann Rheum Dis. 2015;74:2183–7.
Plant D, Wilson AG, Barton A. Genetic and epigenetic predictors of responsiveness to treatment in RA. Nat Rev Rheumatol. 2014;10:329–37.
Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO 3rd, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism Collaborative Initiative. Ann Rheum Dis. 2010;69:1580–8.
Kameda H, Fujii T, Nakajima A, Koike R, Sagawa A, Kanbe K. et al. Japan College of Rheumatology guideline for the use of methotrexate in patients with rheumatoid arthritis. Mod Rheumatol. 2019;29:31–40.
Inoue E, Yamanaka H, Hara M, Tomatsu T, Kamatani N. Comparison of Disease Activity Score (DAS)28- erythrocyte sedimentation rate and DAS28- C-reactive protein threshold values. Ann Rheum Dis. 2007;66:407–9.
Fransen J, van Riel PL. The Disease Activity Score and the EULAR response criteria. Clin Exp Rheumatol. 2005;23:S93–9.
Guzzi PH, Agapito G, Di Martino MT, Arbitrio M, Tassone P, Tagliaferri P, et al. DMET-analyzer: automatic analysis of Affymetrix DMET data. BMC Bioinform. 2012;13:258.
den Boer E, de Rotte MC, Pluijm SM, Heil SG, Hazes JM, de Jonge R. Determinants of erythrocyte methotrexate polyglutamate levels in rheumatoid arthritis. J Rheumatol. 2014;41:2167–78.
Kumagai S, Nishida M, Uemura Y, Izumi M, Abe K, Yoneda K, et al. Methotrexate polyglutamates levels in erythrocytes were genetically affected in RA patients with low disease activity for long period. Ann Rheum Dis. 2017;76:282.
Cook NR, Paynter NP. Performance of reclassification statistics in comparing risk prediction models. Biom J. 2011;53:237–58.
Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Mak. 2006;26:565–74.
Agapito G, Guzzi PH, Cannataro M. DMET-Miner: efficient discovery of association rules from pharmacogenomic data. J Biomed Inform. 2015;56:273–83.
Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–93.
Hazlewood GS, Barnabe C, Tomlinson G, Marshall D, Devoe D, Bombardier C. Methotrexate monotherapy and methotrexate combination therapy with traditional and biologic disease modifying antirheumatic drugs for rheumatoid arthritis: abridged Cochrane systematic review and network meta-analysis. Br Med J. 2016;353:i1777.
Visser K, van der Heijde D. Optimal dosage and route of administration of methotrexate in rheumatoid arthritis: a systematic review of the literature. Ann Rheum Dis. 2009;68:1094–9.
van der Straaten RJ, Wessels JA, de Vries-Bouwstra JK, Goekoop-Ruiterman YP, Allaart CF, Bogaartz J, et al. Exploratory analysis of four polymorphisms in human GGH and FPGS genes and their effect in methotrexate-treated rheumatoid arthritis patients. Pharmacogenomics. 2007;8:141–50.
Hakamata J, Kaneko Y, Shimizu M, Yamaoka K, Maruyama J, Takeuchi T, et al. Factors predicting the therapeutic response to methotrexate in Japanese patients with rheumatoid arthritis: a hospital-based cohort study. Biol Pharm Bull. 2018;41:1414–22.
Wei SC, Tan YY, Weng MT, Lai LC, Hsiao JH, Chuang EY, et al. SLCO3A1, A novel crohn’s disease-associated gene, regulates nf-kappaB activity and associates with intestinal perforation. PLoS One. 2014;9:e100515.
Pan Q, Zhang X, Zhang L, Cheng Y, Zhao N, Li F, et al. Solute carrier organic anion transporter family member 3A1 is a bile acid efflux transporter in cholestasis. Gastroenterology. 2018;155:1578–92.e16.
Dervieux T, Orentas Lein D, Marcelletti J, Pischel K, Smith K, Walsh M, et al. HPLC determination of erythrocyte methotrexate polyglutamates after low-dose methotrexate therapy in patients with rheumatoid arthritis. Clin Chem. 2003;49:1632–41.
Takahashi C, Kaneko Y, Okano Y, Taguchi H, Oshima H, Izumi K, et al. Association of erythrocyte methotrexate-polyglutamate levels with the efficacy and hepatotoxicity of methotrexate in patients with rheumatoid arthritis: a 76-week prospective study. RMD Open. 2017;3:e000363.
Yamamoto T, Shikano K, Nanki T, Kawai S. Folylpolyglutamate synthase is a major determinant of intracellular methotrexate polyglutamates in patients with rheumatoid arthritis. Sci Rep. 2016;6:35615.
Li R, Zhao JX, Su Y, He J, Chen LN, Gu F, et al. High remission and low relapse with prolonged intensive DMARD therapy in rheumatoid arthritis (PRINT): a multicenter randomized clinical trial. Medicines. 2016;95:e3968.
Acknowledgements
The DMET plus microarray measurement, direct sequencing, and statistical analyses were partly supported by Sysmex Corporation under the Collaborative Research Agreement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Onishi, A., Kamitsuji, S., Nishida, M. et al. Genetic and clinical prediction models for the efficacy and hepatotoxicity of methotrexate in patients with rheumatoid arthritis: a multicenter cohort study. Pharmacogenomics J 20, 433–442 (2020). https://doi.org/10.1038/s41397-019-0134-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-019-0134-9
This article is cited by
-
Clinical prediction models of rheumatoid arthritis and its complications: focus on cardiovascular disease and interstitial lung disease
Arthritis Research & Therapy (2023)