Abstract
Staphylococcus aureus colonizes the same ecological niche as many commensals. However, little is known about how such commensals modulate staphylococcal fitness and persistence. Here we report a new mechanism that mediates dynamic interactions between a commensal streptococcus and S. aureus. Commensal Streptococcus parasanguinis significantly increased the staphylococcal biofilm formation in vitro and enhanced its colonization in vivo. A streptococcal biofilm-associated protein BapA1, not fimbriae-associated protein Fap1, is essential for dual-species biofilm formation. On the other side, three staphylococcal virulence determinants responsible for the BapA1-dependent dual-species biofilm formation were identified by screening a staphylococcal transposon mutant library. The corresponding staphylococcal mutants lacked binding to recombinant BapA1 (rBapA1) due to lower amounts of eDNA in their culture supernatants and were defective in biofilm formation with streptococcus. The rBapA1 selectively colocalized with eDNA within the dual-species biofilm and bound to eDNA in vitro, highlighting the contributions of the biofilm matrix formed between streptococcal BapA1 and staphylococcal eDNA to dual-species biofilm formation. These findings have revealed an additional new mechanism through which an interspecies biofilm matrix network mediates polymicrobial interactions.
Similar content being viewed by others
Introduction
Most host-associated microorganisms in their habitats exist in multispecies communities known as biofilms; for instance, the human oral cavity is colonized by over 700 prokaryotic species known as the indigenous microbiota [1]. This flora ordinarily maintains ecological homeostasis through intra- and interspecies interactions [2]. However, such balance can be disrupted under pathogenic conditions to create a dysbiotic community [3]. Due to diverse and complex microbial dynamics, the study of precise interspecies interactions in the local microbiota niche and the role of biofilm matrix components would improve our mechanistic understanding of the development of a dysbiotic community in polymicrobial diseases.
Staphylococcus aureus is a prevalent human pathogen related to hospital-acquired infections [4, 5]. Although infections tend to repeat and undergo a persistent recurring cycle [6], the underlying mechanisms are not well-understood. However, studies have revealed a link between S. aureus biofilm and its persistence [7]. Significant advances have been made in understanding biofilm formation mechanisms for single S. aureus species, and emerging findings [8, 9] have shown that S. aureus often coexists with other microorganisms in a complex polymicrobial environment where dynamic synergistic and antagonistic interactions modulate the community homeostasis. Co-infection of S. aureus with Pseudomonas aeruginosa enhances the community resistance and provides a fitness advantage for P. aeruginosa [8]. The coinfection also alters S. aureus antibiotic susceptibility [10,11,12,13]. S. aureus peptidoglycans stimulate the production of P. aeruginosa antibiotics and toxins that not only increase P. aeruginosa-induced killing, but also alter the composition of the polymicrobial community [14]. Both S. aureus and P. aeruginosa interact cooperatively and competitively in P. aeruginosa strain- and signaling-dependent manners. In addition, S. aureus exhibits an inverse correlation with S. pneumoniae in infection of children [9, 15]. This carriage pattern represents the outcome of active interspecies microbial competition [16]. S. pneumoniae forms stable biofilms with S. aureus in vitro and in vivo and inhibits S. aureus dispersal and transition to disease [17]. Probiotic bacilli inhibit S. aureus colonization through interference with quorum-sensing signals [18]. In contrast, human skin commensals significantly enhance S. aureus virulence [19]. Despite reports of those dynamic polymicrobial interactions, little is understood about the underlying molecular mechanisms of polymicrobial biofilm formation.
S. aureus is versatile and often colonizes different human body sites, including the oral cavity, varying from 24 to 36% [20]. Especially, S. aureus is more likely to colonize the oral cavity during mechanical ventilation [21]. Thus, the human upper respiratory tract appears to be the main reservoir for S. aureus [22, 23]. It is well-known that the respiratory tract harbors complex polymicrobial consortia, of which the majority of bacteria belong to streptococcal genera. Streptococcus parasanguinis represents the predominant streptococcal species in human oral cavity [24]. Our previous studies have determined that S. parasanguinis exhibits probiotic activity against various pathogens. It inhibits the opportunistic pathogens P. aeruginosa [25]. In addition, S. parasanguinis inhibits Aggregatibacter actinomycetemcomitans, an oral periodontal pathogen that associates with S. parasanguinis in vivo [26], but enhances its own biofilm formation through a metabolite-dependent signaling pathway [27]. Thus S. parasanguinis appears to be a versatile interacting partner with a wide variety of pathogens. Given that S. parasanguinis and S. aureus shared colonization niches and their distinct features as opportunistic commensal and pathogen, it is not surprising that the nasal carriage of S. aureus is positively correlated with S. parasanguinis [28]. However, it is unknown how they interact with each other, whether their interactions impact biofilm formation.
In this study, we report that S. parasanguinis promotes S. aureus biofilm formation. A key streptococcal surface protein, BapA1, mediated the enhanced S. aureus biofilm formation. Three staphylococcal virulence factors involved in the dual-species biofilm formation were identified by screening an S. aureus transposon mutant library. Mechanistically, the binding of streptococcal BapA1 to a critical staphylococcal biofilm matrix, eDNA, shapes the formation of dual-species biofilms between S. parasanguinis and S. aureus. These results highlight a dynamic polymicrobial interaction that is crucial for understanding S. aureus colonization and persistence. The elucidation of molecular mechanisms responsible for the microbial interactions should facilitate the development of novel therapeutics preventing or treating S. aureus persistence.
Materials and methods
Bacterial strains and culture conditions
Bacterial strains used in this study are listed in Supplementary Table S1. S. aureus and S. parasanguinis strains were grown in Miller (Luria-Bertani) broth (BD Difco™, Catalog No. DF0446-17-3) and Todd-Hewitt broth (THB) (BD), respectively, with 5% CO2 at 37 °C, unless otherwise mentioned. Appropriate antibiotics, kanamycin (125 μg/mL) and erythromycin (5 μg/mL), were used to select corresponding S. parasanguinis [29] and S. aureus mutant strains [30].
Biofilm assay
Biofilm formation was assessed by a microtiter plate assay. Briefly, overnight cultures of S. aureus and S. parasanguinis were sub-cultured separately and grown to the middle log phase (OD600 of 0.6). Staphylococcal and streptococcal cells were 1000-fold diluted in tryptic soy broth containing 0.5% yeast extract and 0.5% glucose (TSBYG, BD) either separately for single-species or mixed for dual-species biofilms. Aliquots (200 µl) of diluted culture (approximately 106 CFU/ml) were added into sterile 96-well flat-bottom plates (Nunc) and incubated without shaking for 24 h to form biofilms. The biofilms were stained with 0.1% crystal violet and then measured at 562 nm (BioTek, Synergy 2 Multi-Detection Microplate Reader). The results were presented as OD562 unless otherwise mentioned. Each assay was repeated at least three times, and each data was averaged from the results of three triplicate wells performed.
Viability of S. aureus and S. parasanguinis in biofilms
S. aureus and S. parasanguinis cells from single or dual-species biofilms were serially diluted and plated on agar to enumerate colony-forming units (CFUs) as reported [25]. In brief, 24-h biofilms were washed three times using sterile phosphate-buffered saline (PBS), and biofilm cells were scraped off the wells and resuspended in 1 mL of PBS, followed by water bath sonication for 20 s to disturb the cell clusters. Finally, collected bacterial cells were serially diluted and plated on Staphylococcus Medium 110 (BD) and Mitis salivarius (BD) agars to enumerate staphylococcal and streptococcal CFUs, respectively.
Confocal laser scanning microscopy (CLSM) analysis
Biofilms were grown in a µ-Slide 8 well IbiTreat (Ibidi, Germany) as described in the above biofilm assays. S. aureus was stained by its specific polyclonal antibody PA1-7246 (Invitrogen), and S. parasanguinis was detected with a monoclonal antibody specific for Fap1 [31]. Both primary antibodies were added into biofilms in 1% BSA PBS and incubated for 30 min. Subsequently, the biofilms were incubated with fluorescent-conjugated secondary antibodies (Thermo Fisher Scientific) for 20 min after washing with PBS. Alexa Fluor 594-conjugated goat anti-mouse IgG and Alexa Fluor 488-conjugated goat anti-rabbit IgG were used to stain S. parasanguinis and S. aureus, respectively. Stained biofilms were examined with CLSM (Nikon Eclipse TE2000 inverted microscope) with a 60 × objective magnification. Representative image stacks were selected from three independent experiments and used for the imaging analysis. Biofilm thickness and biovolume were quantified using NIS Elements imaging software and BiofilmQ software [32].
Effects of DNase I on dual-species biofilm formation
To evaluate the involvement of eDNA in dual-species biofilms, we first added DNase I (25 or 50 U/mL) into biofilm cultures at the beginning of incubation to determine its effects on biofilm development. In addition, we detected the effect of DNase I on mature biofilms. Briefly, the pre-formed 24 h biofilms continued to incubate for 6 h after adding DNase I (25 or 50 U/mL), followed by the crystal violet staining for biofilm measurement.
Quantification of planktonic extracellular DNA (eDNA)
Culture supernatants harvested from bacteria grown to stationary phase (OD600 of 1.0) in TSBYG were filtered with a 0.22 µm filter and used to measure eDNA. Quantitative PCR was performed using SYBR green master mix on an iCycler iQ5 machine (Bio-Rad). The S. aureus 16 S rRNA gene amplified with a species-specific primer set (Supplementary Table S2) was used to quantify DNA amounts.
Biofilm DNA and polysaccharide matrix staining
As described above, the S. aureus biofilm was grown onto an 8-well plate in TSBYG medium. Biofilm DNA was stained with 1 µM of 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) and 1 µM of propidium iodide (PI) (Invitrogen) for 10 min, whereas the polysaccharide matrix was probed with 1 µM of CF®594 conjugated wheat germ agglutinin (WGA) (Biotium, CA, USA) for 10 min. The stained biofilms were then examined under a fluorescence microscope (BioTek Cytation 5) with a 40× objective magnification. Furthermore, colocalization analysis was performed with an ImageJ and presented as a Pearson correlation coefficient [33].
Colonization of Drosophila. melanogaster by S. aureus and S. parasanguinis
Bacterial infection of D. melanogaster flies was studied as previously described [34]. Briefly, S. aureus and S. parasanguinis cultures were grown to an OD600 of 1.0, and 1 mL of the culture was harvested to infect flies. The harvested cell pellets were resuspended, separately, or mixed in 100 µl of sterile 5% sucrose, then spotted on a sterile 21-mm filter paper disc (Whatman) placed on the surface of 5 mL of solidified 5% sucrose agar in a plastic vial (Fisher scientific). Male flies (3–5 days old) were fed with antibiotics (erythromycin, vancomycin, and ampicillin at 50 µg/mL) for 3 days and then transferred to fresh food for 3 days to eliminate residual antibiotics. All flies starved for 4 h before fed on bacteria in vials (10 flies per vial). To determine the number of viable bacterial cells inside the flies, the surfaces of the flies were sterilized with 70% ethanol for 30 s and washed three times with sterile PBS. Flies were then crushed in an Eppendorf tube with 1 mL of culture medium. To enumerate CFU of bacteria in flies, serial dilutions of the homogenates were plated on the following agar plates: Staphylococcus Medium 110 for S. aureus and Columbia Blood agar for S. parasanguinis.
Screening of S. aureus transposon library
A two-step screening was conducted. First, each transposon mutant from the Nebraska Transposon Mutant Library of S. aureus USA 300 LAC was co-cultured with S. parasanguinis FW213 using the dual-species biofilm conditions described above. Second, S. aureus defective mutants identified from the dual-species biofilm formation were further evaluated in the presence of rBapA1, as rBapA1 alone promoted S. aureus biofilm formation. In brief, the middle-log phase wild-type USA300 and selected mutants were 1000-fold diluted in TSBYG with or without 0.04 mg/mL rBapA1-C. Biofilms were grown and measured. Each strain was repeated in triplicate.
Construction of S. aureus deletion mutants and their complement strains
To ensure mutant phenotypes are related to transposon-inactivated genes, we constructed corresponding in-frame deletion mutants as described previously [35]. Briefly, 1000 bp of upstream and downstream fragments of codY, sarA, atl, and nuc were amplified by PCR with appropriate primers (Supplementary Table S2) from the USA300 LAC chromosome, respectively. These PCR products were cloned into pJB38 to construct in-frame deletion plasmids. The ligation mixtures were first introduced into S. aureus RN4220 to obtain corresponding recombinant plasmids, which were purified and validated by DNA sequencing before being electro-transformed into USA300 LAC or its mutants. In-frame deletion mutants were selected by appropriate antibiotics and confirmed by PCR and DNA sequencing.
Complementation plasmids were constructed as follows. DNA fragments containing the promoter and its corresponding open reading frame of codY, sarA, and atl were amplified using primer sets (Supplementary Table S2). The PCR products were ligated into pLI50 to generate pLI50-codY, pLI50-atl, and pLI50-sarA, respectively. The resultant plasmids were validated by DNA sequencing, transformed into corresponding mutants by electroporation, and selected by appropriate antibiotics. Due to the toxic nature of atl in E. coli, the construction of the atl complementation plasmid was unsuccessful.
Cloning, expression, and purification of various recombinant BapA1 proteins
Recombinant 6xHis-tagged, GFP-tagged, and GST-fused BapA1 were produced from E. coli BL21 (DE3) and purified, respectively, as described elsewhere [29]. Briefly, BapA1 domains, including N-terminal BapA1(BapA1-N, 40-1032aa) and C-terminal antigen domain (BapA1-C, 1430-1571aa), were amplified with their corresponding primer sets (Supplementary Table S2) from S. parasanguinis. Next, PCR products were digested by Nhe1 and Xho1, purified by gel extraction, and cloned into pET-21a.
Likewise, the GST and GFP genes were amplified from pGEX-6P-1 and pVPT-GFP using corresponding primer sets (Supplementary Table S2). Subsequently, the PCR products were digested by Sac1 and Xho1 and cloned into pET-21a to obtain pET-21a-GST and pET-21a-GFP, respectively. Then, the BapA1-C was amplified, digested, and cloned into resultant pET-21a-GST and pET-21a-GFP to generate GST-fused and GFP-tagged rBapA1-C in E. coli BL21 (DE3).
Pull-down of rBapA1-C by S. aureus cells
The wide-type S. aureus and its mutants were grown for 24 h in TSBYG with or without 0.1 mg/mL of rBapA1-C. Subsequently, bacterial cells were collected and lysed with lysis buffer (100 µg/mL of lysostaphin in PBS). Finally, total proteins were separated onto 10% Tris-glycine SDS-PAGE gel and stained with Coomassie Brilliant Blue R-250 to quantify rBapA1-C captured by S. aureus cells. In addition, rBapA1-C was confirmed by western blotting analysis.
Pull-down of DNA by GST-fused rBapA1-C
To determine the interaction of rBapA1 and eDNA, we performed a pull-down of DNA by GST-fused rBapA1-C. First, the chromosomal DNA of S. aureus USA 300 was prepared and then sonicated into small fragments of approximately 250 bp. Second, the GST-fused rBapA1-C conjugated to glutathione beads (GE, USA) was incubated with DNA fragments (10, 25, and 50 µg/mL) in the binding buffer (culture supernatant of middle log phase of USA 300), rotating for 2 h at 30 °C. The same amount of GST conjugated to glutathione beads was used as a negative control. Next, the beads were washed once with the binding buffer. Finally, the bound DNA was eluted by elution buffer (1% SDS in PBS), purified by phenol/chloroform, and subjected to 1% agarose gel analysis and ethidium bromide staining.
Results
S. parasanguinis promotes S. aureus biofilm formation
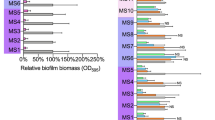
Because S. parasanguinis is a representative commensal streptococcus that colonizes the same upper respiratory niche as S. aureus, we attempted to determine whether S. parasanguinis affects S. aureus biofilm formation using S. parasanguinis strain FW213 and three S. aureus strains, including a pandemic methicillin-resistant S. aureus (MRSA) strain USA300 LAC, methicillin-sensitive (MSSA) clinical isolates Sau10 and Sau65. Co-culture of FW213 with each S. aureus strain significantly increased the formation of dual-species biofilms. About a 3-fold increase in dual-species biofilm biomasses was observed when compared with single-species biofilms (Fig. 1A). The relative contribution of each microorganism within the biofilms was further determined. The maximal number of single S. aureus biofilms was about 109 CFU/ml (Fig. 1B), similar to the highest densities in the planktonic cultures. However, the extraordinarily high cell densities of S. aureus (up to 1 × 1012 CFU/ml) were observed from the 24 h dual-species biofilms (Fig. 1B). In addition, the number of streptococcal cells from the dual-species biofilm also increased significantly (Fig. 1C). The S. aureus cells greatly outnumbered S. parasanguinis cells, so S. aureus is the major biofilm constituent in dual-species biofilms. These data demonstrate a strong polymicrobial synergy between commensal streptococcus and S. aureus.
A The 24 h dual-species biofilms formed by three different S. aureus strains (USA300, Sau10, and Sau65) with either wild-type S. parasanguinis FW213 or its variants (srtA, bapA1, and fap1). Biofilms were determined by crystal violet staining. B CFUs of three S. aureus strains within single and dual-species biofilms. C CFUs of wild-type FW213 and the bapA1 mutant within single and dual-species biofilms. D Representative CLSM images of single and dual-species biofilms of S. aureus with S. parasanguinis and its bapA1 mutant. S. aureus cells (green) were probed with a specific polyclonal antibody PA1-7246 and stained with goat anti-rabbit Alex-Fluor 488 secondary antibody. S. parasanguinis cells (red) were probed with a monoclonal antibody F51 and stained with goat anti-mouse Alex-Fluor 594 secondary antibody. Images were examined at 60× objective magnification. Scale bar: 20 µm. E Biofilm thickness analysis of single or dual-species biofilms using NIS Elements imaging software. F Biovolumes analysis of single or dual-species biofilms using BiofilmQ software. Data represent the means of three independent experiments. Error bars denote SEM. ns means no significance, *p < 0.05, **p < 0.01 and ***p < 0.001 (t-test).
BapA1 of S. parasanguinis mediates the increased dual-species biofilms
To determine what streptococcal surface components are required, we examined the role of cell wall-anchored surface proteins of S. parasanguinis. Sortase A (SrtA) [36] plays an important role in displaying numerous surface adhesins, such as Fap1 [37] and BapA1 [29]. We thus evaluated whether the loss of those genes would have any effect. Both ∆srtA and ∆bapA1 significantly decreased the dual-species biofilms, whereas ∆fap1 exhibited the same phenotype as the wild-type strain (Fig. 1A), although ∆fap1 reduced its single-species biofilm. To further characterize the biofilms, we determined CFUs of each organism. The ∆bapA1 failed to increase CFUs of all three S. aureus strains (Fig. 1B) compared to the wild-type FW213 within the dual-species biofilms. Albeit, ∆bapA1 cells slightly increased in the dual-species biofilms (Fig. 1C). These results suggested the importance of BapA1 to S. aureus within the dual-species biofilms.
Because identical biofilm phenotypes were observed for all three S. aureus strains tested, further experiments were performed only using the USA300 strain. CLSM images of single- and dual-species biofilms validated the synergistic association between the two wild-type strains, albeit ∆bapA1 failed to support the biofilms formation with USA300 (Fig. 1D). The thickness and biovolume of the dual-species biofilm formed by ∆bapA1 were significantly lower than that formed by wildtype FW213 (Fig. 1E, F). These data demonstrate that BapA1 plays a vital role in supporting the dual-species biofilm.
To validate the role of BapA1, we hypothesized that rBapA1 alone would promote the dual-species biofilm. Given that the gene encoding BapA1 is large, consisting of 3400 amino-acid residues, it is challenging to conduct a stable genetic complementation using the full-length bapA gene of 10.2 kb with an extensive repeat region. Thus, rBapA1 fragments from the N-terminal domain to the C-terminal region were produced and used to evaluate their roles. The rBapA1-C, not rBapA1-N, greatly enhanced the S. aureus biofilm formation (p < 0.01) (Fig. 2A). Further, the effect of the rBapA1-C is dose-dependent (Fig. 2B). These results were also supported by fluorescent microscopy studies (Supplementary Fig. S1), demonstrating that streptococcal BapA1 is crucial for the enhanced biofilm formation by S. aureus in vitro.
A Biofilms of S. aureus in the presence of rBapA1-C (0.04 mg/mL) or rBapA1-N (0.8 mg/mL). The binding of rBapA1-N, and C fragments to the substratum were used as controls. B rBapA1-C dose-dependently promoted S. aureus USA300 biofilm formation. Biofilms were determined by crystal violet staining. Data represent the means of three independent experiments. Error bars denote SEM. ns means no significance, *p < 0.05 and ***p < 0.001 (t-test or one-way ANOVA).
S. parasanguinis promotes S. aureus colonization in a Drosophila melanogaster model
We tested whether observed in vitro effects were also relevant in vivo for a D. melanogaster model, which was accepted as a general model for studying S. aureus colonization [25, 38]. The presence of S. parasanguinis significantly enhanced the staphylococcal colonization approximately 10-fold in flies (e.g., about 9.2 × 105 versus 2.5 × 104 for USA300) (Fig. 3A). In contrast, S. parasanguinis colonization was not altered (Fig. 3B). Because the flies’ immunity and microbiota would likely affect bacterial colonization [38,39,40], it seems reasonable that the magnitude of increase differed dramatically from a 4-5 log increase observed in vitro. Likewise, ∆bapA1 failed to promote S. aureus colonization in flies (Fig. 3A). These in vivo results are consistent with the in vitro findings, further demonstrating that BapA1 mediates the dual-species biofilms.
Flies were fed with single or dual species of S. parasanguinis and S. aureus using the following strains: FW213, bapA1 mutant, USA300, Sau10, and Sau65. After a 24 h infection, flies were crushed and enumerated CFUs for S. aureus (A) and S. parasanguinis (B). Data represent the means of three independent experiments. Error bars denote SEM. ns means no significance, *p < 0.05 and **p < 0.01 (t-test).
Identification of S. aureus virulence factors in BapA1-mediated dual-species biofilms
To identify S. aureus factors that mediate the dual-species biofilm, we screened an ordered USA300 Nebraska Transposon Mutant Library for mutants with defective dual-species biofilm formation. First, each USA300 mutant from the library was co-cultured with FW213, and a crystal violet staining assay assessed biofilm formation [25, 27, 41]. A total of 100 mutants that reduced the dual-species biofilms by at least 25% were initially identified. These mutants were subsequently subjected to a secondary screen using rBapA1-C because the rBapA1-C promoted S. aureus biofilm, and we were interested in studying BapA1-mediated staphylococcal biofilm formation. Only a few exhibited reproducible biofilm defects, including ∆codY, ∆sarA, and ∆atl (Fig. 4A). Fluorescent microscopy studies further validated the biofilm defect using a representative mutant ∆codY (Fig. 4B). These results suggested that the association of S. aureus with BapA1 is critical. For further validation, pull-down assays using staphylococcal cells were employed to test whether S. aureus cells bind to rBapA1-C directly. Wild-type S. aureus captured rBapA1-C, whereas the sarA mutant failed to pull down rBapA1-C. The ∆codY and ∆atl significantly reduced the amount of rBapA1-C captured, exhibiting an intermediate phenotype (Fig. 5A). As a control, the spa mutant did not exhibit any defect. The mutant phenotypes were readily complemented (Fig. 5B, C), demonstrating the importance of each gene in modulating BapA1-mediated biofilm activities. It is worth noting that only S. aureus biofilm cells captured rBapA1-C, whereas planktonic cells failed (Supplementary Fig. S2), further highlighting the importance of BapA1 in modulating staphylococcal biofilms.
A The ability of biofilm formation of three S. aureus mutants (ΔcodY, ΔsarA, and Δatl) was determined in both dual-species biofilms and rBapA1-C-mediated biofilms, and a Δspa was used as a negative control. Biofilms were determined by crystal violet staining, and biomasses of biofilms were normalized to the growth of cells within the biofilms (OD562/OD600). Data represent the means of three independent experiments. Error bars denote SEM. ns means no significance, ***p < 0.001 and ****p < 0.0001 (t-test or one-way ANOVA). B Representative SYTO 9 staining of wild-type S. aureus USA300 or ΔcodY cells within biofilms with the absence and presence of rBapA1-C (0.04 mg/mL). Images were examined at 10 × objective magnification. Scale bar: 100 µm.
The ability of wild-type S. aureus USA300 and its mutants (ΔcodY, ΔsarA, Δatl, and Δspa) (A), as well as complement strains of two mutants codYcomp and sarAcomp (B), to capture rBapA1-C under the biofilm formation conditions. The Δspa was used as a control. Black arrows denote the rBapA1-C. Bacterial cells were harvested after 24 h incubation in the absence or presence of rBapA1-C (0.1 mg/mL) and lysed by lysis buffer. Total proteins were then separated onto 10% SDS-PAGE gel and stained with Coomassie Brilliant Blue R-250. C Quantification of rBapA1-C from A and B with densitometry. Error bars denote SEM. *p < 0.05, ****p < 0.0001, ns no significance (t-test).
Reduced eDNA amounts are evident in S. aureus mutants
We hypothesized that S. aureus biofilm proteins might be involved in interactions because BapA1 is only bound to S. aureus biofilm cells. We carried out extensive pull-down studies using rBapA1-C and failed to identify any staphylococcal proteins (data not shown), suggesting biofilm matrix may be required.
Extracellular polysaccharides are a vital biofilm matrix required for S. aureus biofilms [42]. We re-examined transposon mutants that inactivate genes responsible for the biosynthesis of staphylococcal capsules and polysaccharides, including SAUSA300_0152 and 17 other relevant genes. None of these mutants exhibited biofilm defects when co-cultured with S. parasanguinis (data not shown), indicating that those polysaccharides were not responsible.
Given that eDNA is another well-recognized S. aureus biofilm matrix [43, 44], we hypothesized that the biofilm defects observed are related to decreased eDNA levels in those mutants. We first evaluated the role of eDNA in the development of the dual-species biofilm and found DNase I inhibited the biofilm formation, and the inhibition was dose-dependent (Fig. 6A). Further, DNase I was able to disperse the mature dual-species biofilms (Fig. 6B). These data suggest that eDNA is critical for the dual-species biofilms. We then tested whether the three S. aureus mutants exhibited any defect in the production of eDNA. No detectable eDNA was found in ∆sarA supernatant, while the eDNA level was significantly lower in both ∆codY and ∆atl (Fig. 6C). Consistently, ∆sarA, ∆codY, and ∆atl exhibited reduced PI staining (eDNA or dead cells) within their single biofilms (Supplementary Fig. S3). These data suggest that eDNA reduction is linked to the biofilm defects observed in those mutants.
A Effect of DNase I on developing dual-species biofilms. The 24 h dual-species biofilms were developed, and DNase I at different concentrations was added at the beginning of the co-culture of S. aureus and FW213 (0 h). B Effect of DNase I on pre-formed dual-species biofilms. The 24 h dual-species biofilms were treated with DNase I for an additional 6 h and further determined. C The eDNA amounts of planktonic S. aureus cells were quantified by qPCR using S. aureus 16s rDNA. D The 24 h dual-species biofilm biomass of FW213 and wild-type S. aureus or its mutants (codY, codY/nuc, sarA, sarA/nuc) was denoted as OD562/OD600. Biofilms were determined by crystal violet staining, measured at OD562nm, and normalized by bacterial growth at OD600nm. Error bars denote SEM. **p < 0.01, ***p < 0.001, ****p < 0.0001, ns no significance (t-test or one-way ANOVA).
Consistent with the reduced eDNA levels, ∆sarA overproduced thermonuclease to degrade DNA [45, 46], which may explain why ∆sarA had diminished eDNA and exhibited the biofilm defect. To further validate this, the gene nuc encoding the thermonuclease was inactivated in ∆sarA. Compared to ∆sarA, the resultant double mutant ∆sarA∆nuc partially restored the diminished biofilm (Fig. 6D), indicating the contribution of eDNA/nuclease to the dual-species biofilm. The double mutant ∆codY∆nuc did not rescue the ∆codY biofilm defect, suggesting the overexpressed nuclease is not responsible for ∆codY dual-species biofilm defect.
eDNA and BapA1 colocalized within the BapA1-mediated S. aureus biofilms
We hypothesized that BapA1 and eDNA associate within biofilms because S. parasanguinis BapA1 is required for the dual-species biofilms, and eDNA is a fundamental staphylococcal biofilm matrix. First, we validated a GFP-tagged rBapA1-C capable of promoting S. aureus biofilms (Supplementary Fig. S4). Second, the 24 h S. aureus biofilms in the presence of GFP-tagged rBapA1-C were developed. Next, the biofilms were stained with either 1 µM of PI to stain eDNA and dead cells (Fig. 7A) or 1 µM of CF®594 conjugated WGA to stain the polysaccharide matrix (Fig. 7B). The quantification of colocalization suggested that rBapA1-C (green) was strongly colocalized with the eDNA (red) (Pearson correlation coefficient = 0.87). In contrast, rBapA1-C was weakly colocalized with polysaccharide matrix (Pearson correlation coefficient = 0.15) (Fig. 7C). These results suggest that BapA1 selectively binds to eDNA within the biofilms.
The biofilms of S. aureus USA300 in the presence of GFP-tagged rBapA1-C were stained with PI (1 µM) for eDNA (A), and CF®594 conjugated WGA (1 µM) for extracellular polysaccharides (B). Images were examined at 40× objective magnification using a BioTek Cytation 5. Scale bar: 20 µm. The colocalization of rBapA1-C with eDNA or WGA was quantified by ImageJ and presented as a Pearson correlation coefficient. C Pearson correlation coefficient calculated from images of GFP-rBapA1-C and PI-eDNA in A or CF®594 red- WGA in B. D rBapA1-C bound to chromosomal DNA of USA 300 LAC in vitro. The GST-fused rBapA1-C conjugated to glutathione beads was used to pull down chromosomal DNA fragments of USA 300 LAC (10, 25, and 50 µg/mL). DNA fragments bound to GST-fused rBapA1-C were then eluted, purified, and subjected to 1% agarose gel, followed by ethidium bromide staining. Lane 1, DNA marker; lane 2, input control, 100 µg/mL of DNA fragments; lane 3–5, incubation of GST-fused rBapA1-C with DNA fragments (10, 25, and 50 µg/mL); lane 6–8, incubation of GST control with DNA fragments (10, 25, and 50 µg/mL). Error bars denote SEM. ***p < 0.001. GST and GST-fused rBapA1-C input were examined by SDS-PAGE (Supplementary Fig. S7).
As BapA1 colocalized with eDNA in the dual-species biofilms, we investigated whether BapA1 directly binds to eDNA. S. aureus DNA fragments dose-dependently bound to GST-fused rBapA1-C (Fig. 7D), demonstrating the direct interaction between rBapA1 and staphylococcal DNA.
Discussion
Recent studies of the integrative Human Microbiome Project demonstrated indigenous microbiota changes at an individual level and their significant contributions to health and disease [47,48,49]. Dynamic interactions among microorganisms occur in every known microbiome community. However, how microbe-microbe interactions develop and how they determine microbial fitness and pathogenesis have not been explored systematically. This is certainly the case for polymicrobial interactions between commensal streptococci and prevalent pathogenic microorganisms, such as S. aureus, where they often colonize a same ecological niche in the upper respiratory tract [24]. To date, the influence of commensal streptococci on S. aureus biofilm life cycle has not been fully appreciated. Our current study indicates that an oral streptococcus utilizes its unique major surface protein BapA1 to capture eDNA produced by S. aureus to build a biofilm matrix network, thereby enhancing the biofilm formation of S. aureus, a previously unappreciated molecular interaction that shapes polymicrobial interactions.
Previous studies demonstrated that both C. albicans [44] and Fusobacterium nucleatum [50] could increase the S. aureus biofilm formation through hypha formation and coaggregation, respectively. Coaggregation among diverse oral microbes is a hallmark of polymicrobial interactions in the oral cavity [51, 52]. However, we did not observe the coaggregation between S. parasanguinis and S. aureus (Supplementary Fig. S5), even though physical contact between the two species is required (Supplementary Fig. S6). These findings suggest a unique interaction between the two organisms.
Biofilm matrix often comprises eDNA, polysaccharides, lipids, and secreted or surface-exposed proteins. Indeed, eDNA has been implicated in supporting the biofilm structural integrity in various organisms, including S. aureus [43, 53,54,55,56]. Within single-species biofilms, it is documented that eDNA associates with other biofilm matrix components and builds an integral biofilm scaffold [55, 57,58,59]. For example, a family of “moonlighting” lipoproteins have been recently identified as eDNA binding proteins [60, 61]. In addition, the interaction between eDNA and secreted polysaccharides Pel is crucial for the integrity of P. aeruginosa biofilm matrix [57, 62]. However, the framework that eDNA from one microorganism might directly interact with distinct matrix components from a different microorganism has not been appreciated [63].
In this study, we demonstrated that oral commensal S. parasanguinis FW213 enhanced biofilm formation of S. aureus via its cell surface anchored protein BapA1. Using a transposon mutant library screening, we identified three known staphylococcal virulence factors, CodY, SarA, and Atl, that are required for BapA1-enhanced biofilm formation. The mutants defective in distinct proteins share one common feature: they all possessed significantly lower amounts of eDNA in their culture supernatants, thus diminishing the eDNA matrix. These findings imply the potential interaction between BapA1 and eDNA. Several additional lines of evidence support this conclusion. First, DNase I inhibited the dual-species formation. Second, rBapA1-C itself dose-dependently promoted biofilm formation of S. aureus and colocalized with eDNA within the dual-species biofilm. Third, rBapA1-C directly bound to staphylococcal DNA. Collectively, we demonstrate that the building of the integral interspecies biofilm matrix is shaped by the close association between S. parasanguinis BapA1 and S. aureus eDNA.
As far as we know, BapA1 does not belong to any eDNA binding proteins family reported previously. It is known that a binding domain (BR) of a streptococcal serine-rich repeat protein PsrP forms a split β-barrel-like structure to enable binding with eDNA and promotes bacterial aggregation [64]. Whether BapA1 possesses a BR structure and an interaction between eDNA and BapA1 represents a general mechanism for stabilizing and strengthening biofilms is unknown and awaits future studies. Interestingly, a PsrP homolog Fap1 of S. parasanguinis is not engaged in the formation of dual-species biofilms, albeit it is critical for single-species biofilm formation [65]. It is known that Fap1 makes the fibril that covers the bacterial surface. In the bald fap1 mutant, more BapA1 would be exposed that should enable it to interact more with staphylococcal eDNA, which would support the incorporation of S. parasanguinis into the S. aureus biofilm. In turn, the close association between two organisms and continuous secretion of BapA1 by S. parasanguinis within the micro-niche established by the BapA1/eDNA network should further enhance the dual-species biofilm formation. This again suggests the importance of the binding of BapA1 to eDNA and the development of the biofilm matrix network in dual-species biofilm formation.
On the side of S. aureus, three biofilm-associated factors, SarA, Atl, and CodY, were identified to play a critical role in the development of BapA1-enhanced biofilms. These seemingly disparate virulence factors are responsible for the same dual-species biofilm formation, implying the existence of a common operative mechanism. First, the staphylococcal accessory regulator SarA interacts with a complex regulatory network and mediates the single-species biofilm formation of S. aureus [66]. The sarA mutant degrades eDNA by its overproduced themonuclease [46]. Inactivation of a nuclease gene from the sarA mutant restored BapA1-mediated S. aureus biofilms, further demonstrating the role of eDNA and contribution of SarA. Second, the Atl has been implicated in releasing bacterial DNA from S. aureus cells [67,68,69]. Atl deficiency abolishes S. aureus biofilm formation [70]. However, there is no direct link between the expression of SarA and the activity of Atl, and additional studies are required to uncover the potential new association. Third, the CodY is a global regulator differentially regulating many genes responsible for a wide range of metabolic reprogramming [71,72,73,74,75,76] and positively modulates genes responsible for the production of extracellular adhesion components, such as cell wall anchored proteins. It also depresses genes whose products are harmful to the host, including nucleases, proteases, and lipases. This mechanism enables S. aureus to integrate metabolism and virulence [77], while the function of CodY in mediating S. aureus biofilm remains ill-defined [78]. However, deletion of a nuclease gene from ∆codY did not rescue any biofilm defect of ∆codY, suggesting that overproduced nuclease is not involved. It is possible that the codY deficiency led to overexpression of proteases and lipases that may degrade unknown eDNA binding moonlighting proteins and lipids, two additional biofilm matrix components, thereby attenuating the polymicrobial biofilm formation. Further studies are necessary to clarify the role of CodY. Altogether, all mutants significantly reduced the release of their eDNA. Unsurprisingly, these different virulence factors modulate BapA1-mediated S. aureus biofilms via a shared mechanism, eDNA binding. Whether and how SarA, CodY, and Atl are coordinately regulated to mediate this awaits further investigation.
In conclusion, this study has revealed a new interspecies link between a commensal streptococcus and a leading pathogen, S. aureus. Commensal bacteria’s ability to produce versatile adhesins that bind to key biofilm matrix from pathogens represents a new avenue to shape an integral polymicrobial biofilm matrix network to initiate the development of a pathogenic niche. Such a selective interaction between streptococcal adhesin and staphylococcal eDNA constitutes a potential therapeutic target that is amenable to the development of new precision anti-infection strategies.
Data availability
All data are available in the main text or Supplementary materials.
References
Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch Microbiol. 2018;200:525–40.
Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71:653–70.
Sudhakara P, Gupta A, Bhardwaj A, Wilson A. Oral dysbiotic communities and their implications in systemic diseases. Dent J. 2018;6:10.
Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the national healthcare safety network at the Centers for Disease Control and Prevention, 2011-2014. Infect Control Hosp Epidemiol. 2016;37:1288–301.
Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–32.
Bradley SF. Eradication or decolonization of methicillin-resistant Staphylococcus aureus carriage: what are we doing and why are we doing it? Clin Infect Dis. 2007;44:186–9.
Waters EM, Rowe SE, O’Gara JP, Conlon BP. Convergence of Staphylococcus aureus persister and biofilm research: Can biofilms be defined as communities of adherent persister cells? PLoS Pathog. 2016;12:e1006012.
Tognon M, Kohler T, Gdaniec BG, Hao Y, Lam JS, Beaume M, et al. Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J 2017;11:2233–43.
Khan F, Wu X, Matzkin GL, Khan MA, Sakai F, Vidal JE. Streptococcus pneumoniae eradicates preformed Staphylococcus aureus biofilms through a mechanism requiring physical contact. Front Cell Infect Microbiol. 2016;6:104.
Filkins LM, Graber JA, Olson DG, Dolben EL, Lynd LR, Bhuju S, et al. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol. 2015;197:2252–64.
Orazi G, O’Toole GA. Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. MBio 2017;8:e00873–17.
Radlinski L, Rowe SE, Kartchner LB, Maile R, Cairns BA, Vitko NP, et al. Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol. 2017;15:e2003981.
Ibberson CB, Stacy A, Fleming D, Dees JL, Rumbaugh K, Gilmore MS, et al. Co-infecting microorganisms dramatically alter pathogen gene essentiality during polymicrobial infection. Nat Microbiol. 2017;2:17079.
Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci USA 2013;110:1059–64.
Reiss-Mandel A, Regev-Yochay G. Staphylococcus aureus and Streptococcus pneumoniae interaction and response to pneumococcal vaccination: Myth or reality? Hum Vaccin Immunother. 2016;12:351–7.
Lewnard JA, Givon-Lavi N, Huppert A, Pettigrew MM, Regev-Yochay G, Dagan R, et al. Epidemiological markers for interactions among Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus in upper respiratory tract carriage. J Infect Dis. 2016;213:1596–605.
Reddinger RM, Luke-Marshall NR, Sauberan SL, Hakansson AP, Campagnari AA. Streptococcus pneumoniae modulates Staphylococcus aureus biofilm dispersion and the transition from colonization to invasive disease. MBio 2018;9:e02089–17.
Piewngam P, Zheng Y, Nguyen TH, Dickey SW, Joo HS, Villaruz AE, et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018;562:532–7.
Boldock E, Surewaard BGJ, Shamarina D, Na M, Fei Y, Ali A, et al. Human skin commensals augment Staphylococcus aureus pathogenesis. Nat Microbiol. 2018;3:881–90.
Smith AJ, Jackson MS, Bagg J. The ecology of Staphylococcus species in the oral cavity. J Med Microbiol. 2001;50:940–6.
Sands KM, Wilson MJ, Lewis MAO, Wise MP, Palmer N, Hayes AJ, et al. Respiratory pathogen colonization of dental plaque, the lower airways, and endotracheal tube biofilms during mechanical ventilation. J Crit Care. 2017;37:30–7.
Ohara-Nemoto Y, Haraga H, Kimura S, Nemoto TK. Occurrence of staphylococci in the oral cavities of healthy adults and nasal oral trafficking of the bacteria. J Med Microbiol. 2008;57:95–9. Pt 1
McCormack MG, Smith AJ, Akram AN, Jackson M, Robertson D, Edwards G. Staphylococcus aureus and the oral cavity: an overlooked source of carriage and infection? Am J Infect Control. 2015;43:35–7.
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14.
Scoffield JA, Duan D, Zhu F, Wu H. A commensal streptococcus hijacks a Pseudomonas aeruginosa exopolysaccharide to promote biofilm formation. PLoS Pathog. 2017;13:e1006300.
Fine DH, Markowitz K, Fairlie K, Tischio-Bereski D, Ferrendiz J, Furgang D, et al. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J Clin Microbiol. 2013;51:2850–61.
Duan D, Scoffield JA, Zhou X, Wu H. Fine-tuned production of hydrogen peroxide promotes biofilm formation of Streptococcus parasanguinis by a pathogenic cohabitant Aggregatibacter actinomycetemcomitans. Environ Microbiol. 2016;18:4023–36.
Accorsi EK, Franzosa EA, Hsu T, Joice Cordy R, Maayan-Metzger A, Jaber H, et al. Determinants of Staphylococcus aureus carriage in the developing infant nasal microbiome. Genome Biol. 2020;21:301.
Liang X, Chen YY, Ruiz T, Wu H. New cell surface protein involved in biofilm formation by Streptococcus parasanguinis. Infect Immun. 2011;79:3239–48.
Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 2013;4:e00537–12.
Wu H, Zeng M, Fives-Taylor P. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect Immun. 2007;75:2181–8.
Hartmann R, Jeckel H, Jelli E, Singh PK, Vaidya S, Bayer M, et al. Quantitative image analysis of microbial communities with BiofilmQ. Nat Microbiol. 2021;6:151–6.
Adler J, Parmryd I. Colocalization analysis in fluorescence microscopy. Methods Mol Biol. 2013;931:97–109.
Scoffield JA, Wu H. Oral streptococci and nitrite-mediated interference of Pseudomonas aeruginosa. Infect Immun. 2015;83:101–7.
Bose JL, Lehman MK, Fey PD, Bayles KW. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One. 2012;7:e42244.
Yamaguchi M, Terao Y, Ogawa T, Takahashi T, Hamada S, Kawabata S. Role of Streptococcus sanguinis sortase A in bacterial colonization. Microbes Infect. 2006;8:2791–6.
Wu H, Mintz KP, Ladha M, Fives-Taylor PM. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol. 1998;28:487–500.
Ramond E, Jamet A, Ding X, Euphrasie D, Bouvier C, Lallemant L, et al. Reactive oxygen species-dependent innate immune mechanisms control methicillin-resistant Staphylococcus aureus virulence in the Drosophila larval model. MBio 2021;12:e0027621.
Marra A, Hanson MA, Kondo S, Erkosar B, Lemaitre B. Drosophila antimicrobial peptides and lysozymes regulate gut microbiota composition and abundance. MBio 2021;12:e0082421.
Lee HY, Yoon CK, Cho YJ, Lee JW, Lee KA, Lee WJ, et al. A mannose-sensing AraC-type transcriptional activator regulates cell-cell aggregation of Vibrio cholerae. NPJ Biofilms Microbiomes. 2022;8:65.
Yang C, Scoffield J, Wu R, Deivanayagam C, Zou J, Wu H. Antigen I/II mediates interactions between Streptococcus mutans and Candida albicans. Mol Oral Microbiol. 2018;33:283–91.
Otto M. Staphylococcal biofilms. Microbiol Spectr. 2018;6:10.1128.
Sugimoto S, Sato F, Miyakawa R, Chiba A, Onodera S, Hori S, et al. Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus. Sci Rep. 2018;8:2254.
Kean R, Rajendran R, Haggarty J, Townsend EM, Short B, Burgess KE, et al. Candida albicans mycofilms support Staphylococcus aureus colonization and enhances miconazole resistance in dual-species interactions. Front Microbiol. 2017;8:258.
Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One. 2008;3:e3361.
Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822.
Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, et al. The vaginal microbiome and preterm birth. Nat Med. 2019;25:1012–21.
Zhou W, Sailani MR, Contrepois K, Zhou Y, Ahadi S, Leopold SR, et al. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature 2019;569:663–71.
Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62.
Lima BP, Hu LI, Vreeman GW, Weibel DB, Lux R. The oral bacterium fusobacterium nucleatum binds Staphylococcus aureus and alters expression of the staphylococcal accessory regulator sarA. Micro Ecol. 2019;78:336–47.
Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol. 2016;14:93–105.
Sanchez BC, Chang C, Wu C, Tran B, Ton-That H. Electron transport chain is biochemically linked to pilus assembly required for polymicrobial interactions and biofilm formation in the gram-positive Actinobacterium Actinomyces oris. MBio 2017;8:e00399–17.
Moormeier DE, Bayles KW. Staphylococcus aureus biofilm: a complex developmental organism. Mol Microbiol. 2017;104:365–76.
Dengler V, Foulston L, DeFrancesco AS, Losick R. An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J Bacteriol. 2015;197:3779–87.
Rostami N, Shields RC, Yassin SA, Hawkins AR, Bowen L, Luo TL, et al. A critical role for extracellular DNA in dental plaque formation. J Dent Res. 2017;96:208–16.
Okshevsky M, Meyer RL. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol. 2015;41:341–52.
Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci USA 2015;112:11353–8.
Rainey K, Michalek SM, Wen ZT, Wu H. Glycosyltransferase-mediated biofilm matrix dynamics and virulence of Streptococcus mutans. Appl Environ Microbiol. 2019;85:e02247–18.
Bowen WH, Burne RA, Wu H, Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018;26:229–42.
Graf AC, Leonard A, Schauble M, Rieckmann LM, Hoyer J, Maass S, et al. Virulence factors produced by Staphylococcus aureus biofilms have a moonlighting function contributing to biofilm integrity. Mol Cell Proteom. 2019;18:1036–53.
Kavanaugh JS, Flack CE, Lister J, Ricker EB, Ibberson CB, Jenul C, et al. Identification of extracellular DNA-binding proteins in the biofilm matrix. MBio 2019;10:e01137–19.
Passos da Silva D, Matwichuk ML, Townsend DO, Reichhardt C, Lamba D, Wozniak DJ, et al. The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nat Commun. 2019;10:2183.
Yang L, Liu Y, Markussen T, Hoiby N, Tolker-Nielsen T, Molin S. Pattern differentiation in co-culture biofilms formed by Staphylococcus aureus and Pseudomonas aeruginosa. FEMS Immunol Med Microbiol. 2011;62:339–47.
Schulte T, Mikaelsson C, Beaussart A, Kikhney A, Deshmukh M, Wolniak S, et al. The BR domain of PsrP interacts with extracellular DNA to promote bacterial aggregation; structural insights into pneumococcal biofilm formation. Sci Rep. 2016;6:32371.
Ramboarina S, Garnett JA, Zhou M, Li Y, Peng Z, Taylor JD, et al. Structural insights into serine-rich fimbriae from Gram-positive bacteria. J Biol Chem. 2010;285:32446–57.
Zielinska AK, Beenken KE, Mrak LN, Spencer HJ, Post GR, Skinner RA, et al. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol. 2012;86:1183–96.
Houston P, Rowe SE, Pozzi C, Waters EM, O’Gara JP. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun. 2011;79:1153–65.
Liu Y, Burne RA. The major autolysin of Streptococcus gordonii is subject to complex regulation and modulates stress tolerance, biofilm formation, and extracellular-DNA release. J Bacteriol. 2011;193:2826–37.
Jung CJ, Hsu RB, Shun CT, Hsu CC, Chia JS. AtlA mediates extracellular DNA release, which contributes to Streptococcus mutans biofilm formation in an experimental rat model of infective endocarditis. Infect Immun. 2017;85:e00252–17.
Chen C, Krishnan V, Macon K, Manne K, Narayana SV, Schneewind O. Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J Biol Chem. 2013;288:29440–52.
Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, et al. Direct targets of CodY in Staphylococcus aureus. J Bacteriol. 2010;192:2861–77.
Lei MG, Lee CY. Repression of capsule production by XdrA and CodY in Staphylococcus aureus. J Bacteriol. 2018;200:e00203–18.
Montgomery CP, Boyle-Vavra S, Roux A, Ebine K, Sonenshein AL, Daum RS. CodY deletion enhances in vivo virulence of community-associated methicillin-resistant Staphylococcus aureus clone USA300. Infect Immun. 2012;80:2382–9.
Rivera FE, Miller HK, Kolar SL, Stevens SM Jr, Shaw LN. The impact of CodY on virulence determinant production in community-associated methicillin-resistant Staphylococcus aureus. Proteomics 2012;12:263–8.
Waters NR, Samuels DJ, Behera RK, Livny J, Rhee KY, Sadykov MR, et al. A spectrum of CodY activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol Microbiol. 2016;101:495–514.
Kaiser JC, King AN, Grigg JC, Sheldon JR, Edgell DR, Murphy MEP, et al. Repression of branched-chain amino acid synthesis in Staphylococcus aureus is mediated by isoleucine via CodY, and by a leucine-rich attenuator peptide. PLoS Genet. 2018;14:e1007159.
Brinsmade SR. CodY, a master integrator of metabolism and virulence in Gram-positive bacteria. Curr Genet. 2017;63:417–25.
Atwood DN, Loughran AJ, Courtney AP, Anthony AC, Meeker DG, Spencer HJ, et al. Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. Microbiologyopen 2015;4:436–51.
Acknowledgements
This study was supported in part by NIH/NIDCR R01 DE017954 (HW). Nebraska Transposon Mutant Library (NTML) Screening Array, NR-48501, was provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources, NIAID, NIH.
Author information
Authors and Affiliations
Contributions
HW conceived and supervised the project. HW and LW designed the experiments. LW, HW, HZ performed experiments and analyzed data. LW and HW wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, L., Wang, H., Zhang, H. et al. Formation of a biofilm matrix network shapes polymicrobial interactions. ISME J 17, 467–477 (2023). https://doi.org/10.1038/s41396-023-01362-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-023-01362-8
This article is cited by
-
Staphylococcus aureus and biofilms: transmission, threats, and promising strategies in animal husbandry
Journal of Animal Science and Biotechnology (2024)
-
The soil plastisphere
Nature Reviews Microbiology (2024)