Abstract
Characterizing microorganisms and enzymes involved in lignin biodegradation in thermal ecosystems can identify thermostable biocatalysts. We integrated stable isotope probing (SIP), genome-resolved metagenomics, and enzyme characterization to investigate the degradation of high-molecular weight, 13C-ring-labeled synthetic lignin by microbial communities from moderately thermophilic hot spring sediment (52 °C) and a woody “hog fuel” pile (53 and 62 °C zones). 13C-Lignin degradation was monitored using IR-GCMS of 13CO2, and isotopic enrichment of DNA was measured with UHLPC-MS/MS. Assembly of 42 metagenomic libraries (72 Gb) yielded 344 contig bins, from which 125 draft genomes were produced. Fourteen genomes were significantly enriched with 13C from lignin, including genomes of Actinomycetes (Thermoleophilaceae, Solirubrobacteraceae, Rubrobacter sp.), Firmicutes (Kyrpidia sp., Alicyclobacillus sp.) and Gammaproteobacteria (Steroidobacteraceae). We employed multiple approaches to screen genomes for genes encoding putative ligninases and pathways for aromatic compound degradation. Our analysis identified several novel laccase-like multi-copper oxidase (LMCO) genes in 13C-enriched genomes. One of these LMCOs was heterologously expressed and shown to oxidize lignin model compounds and minimally transformed lignin. This study elucidated bacterial lignin depolymerization and mineralization in thermal ecosystems, establishing new possibilities for the efficient valorization of lignin at elevated temperature.
Similar content being viewed by others
Introduction
Lignin is the second most abundant terrestrial biopolymer after cellulose, and can comprise 10–30% of a plant’s dry weight [1]. While biological conversion of plant biomass to fuels and chemicals can reduce fossil fuel consumption, few processes exist to valorize lignin to value-added chemicals [2]. This is due in part to its recalcitrance and heterogeneity: lignin is a complex heteropolymer containing diverse ether and carbon-carbon bonds linking phenylpropanoid aromatic subunits. A promising approach to divert lignin from waste streams for production of valuable bio-products is biological lignin valorization, involving lignin depolymerization and biocatalysts that funnel lignin-derived aromatic compounds (LDACs) into commercial chemicals [2]. There is a need to develop thermotolerant biocatalysts for efficient conversion of lignin derivatives produced by industrial processes [3].
In nature, fungi are thought to be mainly responsible for lignin depolymerization, with white rot fungi utilizing lignin peroxidases (EC 1.11. 1.14) and laccases (benzenediol oxygen oxidoreductases, EC 1.10.3.2) to do so. However, bacteria are increasingly recognized for their contributions to this process [4,5,6]. Investigation of bacterial lignin depolymerization has focused on two enzyme classes: dye-depolymerizing peroxidases (DyPs) and laccase-like multi-copper oxidases (LCMOs). Bacterial LCMOs carry out myriad reactions. Importantly, LCMOs in two-domain super-families, including K-type small laccases (SLACs), are capable of efficient lignin depolymerization [7, 8]. Further, bacteria have catabolic pathways that funnel diverse LDACs into catabolic intermediates (e.g., protocatechuate and catechol). These diols are typically degraded via either meta or ortho ring-cleavage pathways [6, 9]. Characterization of novel thermotolerant lignin-degrading bacteria therefore requires (1) evidence for their involvement in lignin degradation, (2) identification of enzymes that depolymerize lignin, and (3) identification of pathways for catabolic funneling of LDACs.
To facilitate identification of novel lignolytic organisms and biocatalysts, we undertook genomic bioprospecting in thermal environments. Massive piles of wood residue, known as “hog fuel,” can reach temperatures sufficient for spontaneous combustion due to biological activity. We hypothesized that thermophilic microbes within hog fuel are adapted to use lignin and LDACs as carbon sources. Likewise, we hypothesized that geothermal hot springs with regular inputs of woody biomass harbor thermophilies capable of catabolizing lignin. To test these hypotheses, samples of hog fuel and hot spring microbial communities were incubated with a synthetic 13C-ring-labeled lignin dehydrogenation polymer (DHP) to facilitate stable-isotope probing (SIP). 13C-Enriched genomes resolved from metagenomic libraries encoded a variety of enzymes with the potential for depolymerization of lignin and catabolism of LDACs. Subsequently, we heterologously expressed a two-domain LMCO and characterized its ability to transform β-aryl ether lignin model compounds and Eucalyptus milled wood lignin.
Materials and methods
Sampling thermal environments
Lakelse hot spring (54°21′30.7″N, 128°32′28.0″W), near Terrace, Canada, is a concrete-enclosed pool 1–5 m deep, fed by geothermally warmed spring water (53.2 °C, pH 7.5) [10, 11]. About 500 ml of the top ~5 cm layer of organic-rich sediment was sampled at four equidistant locations on August 24th, 2017 using a manual pump. Sediment and spring water were placed in autoclaved 1 L Nalgene bottles and placed on ice. Two additional sediment samples per location for DNA extraction were placed in 5 ml screw-top vials and placed immediately on dry-ice. A 1.93 ha hog fuel pile in Crofton, Canada (48°52′31.9″N, 123°39′08.1″W) containing sub-boreal spruce, western redcedar and Douglas-fir reside was sampled on September 27, 2017. Three 1 m pits were dug at 40 m intervals along the perimeter of the pile and ~500 ml samples were removed from 20 and 80 cm depths (52.9 °C and 58.7 °C, respectively) with an ethanol- and distilled H2O-washed trowel. Bulk samples were stored on ice and 5 ml aliquots were stored on dry ice.
13C-DHP lignin microcosms
Sediment was separated from spring water using Steritop Filter bottles (Sigma-Aldrich, St. Louis, U.S.A.). Two sets of three replicate sediments from Lakelse, and hog fuel samples from 20 and 80 cm equivalent to 1 g dry weight were added to autoclaved 50 ml serum bottles with 0.1 g of 10% 13C-DHP lignin or 12C-DHP lignin plus 5 ml M9 buffer [12], and the bottles were crimp-sealed. 13C-DHP lignin was synthesized as in [4] and in the Supplementary Methods. Lakelse sediment and 20 cm hog fuel were incubated at 53 °C, while 80 cm hog fuel was incubated at 62 °C. Incubations were in rotary shakers at 150 rpm.
13C-CO2 respiration analysis
We monitored 13C-CO2 production as an indicator of 13C-DHP lignin mineralization. In total, 0.5 ml of serum bottle headspace air was manually-injected into an Isoprime gas chromatograph isotope ratio mass spectrometer (GV Instruments, Wythenshave, U.K.) using a 1.0-ml glass syringe. Headspace CO2 concentrations were calculated using a standard curve of 0.05, 0.5, 5, and 10% of 12C-CO2 (~1.2% atom 13C Praxair Inc., Danbury, U.S.A.) in N2, and 99.0 atom % 13C CO2 (Sigma-Aldrich) in N2 to 0.01, 0.05, 0.1 and 0.5% 13C-CO2. Control microcosms without sediment were monitored to test stability of 13C-DHP lignin at elevated temperature.
DNA extractions and fractionation
DNA was extracted from 0.5 g of three replicate in situ thermal hot spring sediment and hog fuel samples using NucleoSpin Soil kits (Macherey-Nagel, Düren, Germany). After 24 days incubation, three replicate microcosms with each of the three inocula were emptied into sterile 15-ml Falcon tubes and centrifuged for 10 min at 4000 rpm at 4 °C. DNA was extracted 4 times from 0.5 g sediment or hog fuel using the above kit to achieve ≥5.0 µg recovered DNA. Cesium chloride density gradient centrifugation and fractionation was conducted according to published protocols [13, 14]. The level of 13C enrichment in each purified DNA fraction was quantified using ultrahigh-performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS). Details are provided in [15] and in the Supplementary Methods.
Fraction selection and shotgun sequencing
Fraction four (F4, ~1.737 g ml−1) was selected as the “heavy” fraction, based on density measurement, % atom 13C-DNA, and absence of DNA recovered in this fraction from 12C-DHP lignin microcosms. Fraction six (F6, ~1.727 g ml−1) was used as the heavy fraction for 12C-DHP lignin samples. Fraction ten (F10, ~1.717 g ml−1) was used as the “light” fraction for both. One ng DNA from light and heavy fractions were used to generate metagenomic sequencing libraries using the Nextera XT DNA Library Prep Kit (Illumina, San Diego, U.S.A.). In total, 12 libraries from hog fuel microcosms and 12 libraries from hot spring microcosms (Supplementary Table 1) were multiplexed separately on two runs of NextSeq (Illumina) using 150-bp paired-end sequencing in High Output mode at the UBC Sequencing and Bioinformatics Consortium (Vancouver, CAN).
Metagenome assembly, binning and annotation
Trimmomatic 0.36 [16] was used to quality filter reads and trim Illumina adapters using default parameters. Reads from hot spring and hog fuel libraries were assembled separately with metaSPAdes v3.11.1 [17] using kmers = [21,33,55,77,99,127]. Contigs were binned with MyCC [18], MetaBAT2 2.12.1 [19], CONCOCT 1.0 [20] and MaxBin2.2.7 [21], and a dereplicated set of metagenome-assembled-genomes (MAGs) was generated with DASTool 1.1.2 [22]. MAGs were assessed for completeness and redundancy using CheckM 1.0.1 [23]. Taxonomic classification used GTDB-Tk [23] (github.com/Ecogenomics/GTDBTk). Coding sequences in binned and unbinned contigs were predicted using Prodigal 2.6.3 [24], and annotated with (1) DIAMOND 0.9.22.123 [25] blastp against the RefSeq 94 non-redundant (nr) database with a cut-off of e ≤ 1E−50, (2) hidden Markov models (HMMs) against the carbohydrate-active enzymes (CAZy) [26] database with dbCAN2 [27], Pfam/TIGRfam [28, 29], and (3) the KEGG database using the HMM-based KOFAMSCAN [30].
Statistical analysis
Base-2 logarithmic fold change (L2FC) of MAG abundance (sequencing depth) between 13C heavy (“enriched”) and light fractions, as well as between 13C heavy and 12C heavy fractions, was determined with DeSeq2 [31] in R 3.5.1 [32]. Phylogenetic trees were visualized using iTol [33] and ggtree [34].
Laccase preparation and characterization
LacOST51, identified in an enriched MAG from hog fuel, was produced heterologously as an N-terminal polyHis-tagged (Ht-) protein using E. coli BL-21 λ (DE3) containing pET_LacOST51 (details in Supplementary Methods). The molecular weight and purity of the protein were analyzed using SDS-PAGE. The copper content of LacOST51 was quantified using 2,2′-bicinchoninic acid after reduction of copper ions released from the holoenzyme [35]. Laccase activity was measured spectrophotometrically at 436 nm (ε = 36,000 M−1 cm−1) and 468 nm (ε = 49,600 M−1 cm−1) for assays performed using 3 mM 2,2’-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) (20 mM sodium acetate, pH 5) or 1 mM DMP (20 mM sodium phosphate, pH 8), respectively. One unit of activity (U) is defined as the amount of enzyme required to transform 1 μmol of substrate to product per minute at 25 °C. The specific activity of sLac from Amycolatopsis sp. 75iv3, a SLAC (Singh et al. [7]), was determined in parallel under the same conditions as a positive control. The optimal pH of LacOST51 for DMP was evaluated over a range between pH 6 and 9 using 20 mM sodium phosphate (I = 0.1 M, pH 6–8) and 20 mM Tris-HCl (I = 0.1 M, pH 9) buffers. The thermostability of the enzyme was analyzed by measuring the residual activity on DMP at pH 8, after incubating the enzyme at 45, 55, 65 and 75 °C for up to 24 h.
The ability of LacOST51 to transform guaiacylglycerol-β-guaiacyl ether and veratrylglycerol-β-guaiacyl ether was performed as described elsewhere [36]. Briefly, 1 mM of β-O-4 biaryl ether was incubated with 1 μM LacOST51 in 20 mM sodium phosphate, pH 8. The reactions were incubated at 55 °C with stirring, and quenched after 6 h by adding acetic acid to 10% final concentration. The quenched reaction was centrifuged at 16,000 × g for 5 min, and the cleared solution was analyzed by reverse-phase HPLC.
To characterize activity with a minimally transformed lignin, enzymatic mild acidolysis lignin (EMAL) from Eucalyptus wood [37] was dissolved in DMSO (100 mg ml−1) and used at 0.5% (w/v) for assays. Reactions were in performed in 10 ml 12.5 mM potassium phosphate, pH 8, containing 10% DMSO, incubated with or without 6 μM laccase at 30 °C and 200 rpm for 6 days. Reactions were performed using either LacOST51 or sLac from Amycolatopsis sp. 75iv3. To analyze the release of monomers after incubation, 100 μl of each reaction was quenched by adding acetic acid to 10% final concentration and analyzed by reverse-phase HPLC. The dried lignin was further analyzed by HSQC NMR and gel permeation chromatography (GPC). Full assay details are provided in Supplementary Methods.
Results
13C-lignin catabolism
In this study, we investigated the ability of thermophilic bacteria to mineralize synthetic lignin and assimilate lignin derivatives via stable isotope probing. We first synthesized ~3.5 g 13C-DHP with a mean molecular weight of 19.3 ± 0.2 kDa, approximately equivalent to 100 aromatic nuclei per DHP polymer. This synthetic lignin contained 13 β-O-4, 20 β-β, and 18 β-5 bonds per 100 guaiacyl subunits (Fig. 1A).
A Detail of DHP lignin structure supported by GPC-MALS, 2D HSQC NMR and 13C NMR analysis. Mn number average molecular weight (average molecular weight of all the polymer chains), PDI polydispersity index or Mw/Mn ratio. Mw, Mn and PDI determined by GPC-MALS. Red dots show possible positions of 13C-isotopes in coniferyl subunit rings. B 13C-CO2 measured in the headspace of 50-ml sealed serum bottles containing 1 g (dw) of each sample incubated with 100 mg (10%) 13C-DHP lignin, determined by IR-GCMS. 13C-CO2 evolution for control bottles with no inoculum (NI) or 100 mg unlabeled 12C-DHP lignin (NC) are provided. Each point is the mean of n = 3, and error bars represent standard error. C Post-ultra-centrifugation DNA gradient in a 5 ml centrifuge tube with 13C-Low (F10), 12C-Low (F10), 13C-High (F4) and 12C-High (F6) demarcated with dashed gray lines. Graphs show DNA concentration vs. fractional density and %13C-DNA for Hog Fuel 20 cm and Lakelse samples. %13C-DNA measured using UPLC-MS/MS.
Approximately 0.75% (w/w) 13C-DHP or 12C-DHP (control) was incubated with 2 g hot spring sediment or ground hog fuel for up to 24 days. We monitored the mineralization of 13C-DHP to 13C-CO2 to determine if the lignin was mineralized (Fig. 1B). Measuring the incubation headspace using IR-GC-MS showed that, after 24 days, hog fuel from 20 and 80 cm depths evolved about 473 ± 18 and 681 ± 82 µmol 13C-CO2 per gram of hot spring sediment or ground hog fuel, respectively, while Lakelse sediment evolved about 245 ± 45 µmol 13C-CO2 g−1. For comparison, 13C-DHP incubated without inoculum evolved 12 µmol 13C-CO2 g−1, and sediment incubated without 13C-DHP evolved 36 µmol 13C-CO2 g−1.Thus, microbial communities from thermal environments mineralized lignin at in situ temperatures.
Density fractionation was used to isolate DNA from microbes that incorporated 13C from the labeled synthetic lignin during incubation. To verify isotopic-labeling of DNA, UPLC-MS/MS was used to calculate atom% 13C in each of the 12 recovered fractions (Fig. 1C). Fractions 1–4 (F1–4) from 13C-DHP microcosms had a mean density of 1.738–1.745 g ml−1 and contained about 20–60 atom% 13C. All fractions from 12C-DHP microcosms had a baseline of about 6 atom% 13C DNA. Thus, fractions 1–4 were pooled to recover sufficient 13C-DNA for shotgun metagenome sequencing. Detectable DNA was not recovered from fractions 1–4 of the 12C-DHP microcosms. Therefore, fraction 6 (F6; 1.72 g ml−1) was used for the “high-density” fraction for these control microcosms.
Resolution of genomes from 13C-enriched metagenomes
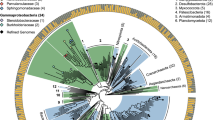
To facilitate the identification and characterization of putatively lignolytic bacteria in thermal environments, we focused our investigation on genome assemblies resolved from shotgun sequencing of fractionated DNA. Fifty-two of these MAGs passing quality thresholds (>80% completion, <5% contamination) were assembled from hog fuel and 72 from hot spring sediment. DESeq2 was used to statistically compare MAG abundance between high-density fractions from 13C-DHP (F1–4) and 12C-DHP (F6) microcosms (Figs. 2 and 3), as well as between high-density and low-density (F10) fractions from 13C-DHP microcosms (Figs. S2 and S3). MAGs with significantly higher (pHDR < 0.05) abundance in 13C F1–4 relative to 12C F6 were considered 13C-enriched. There were four 13C-enriched MAGs in hog fuel 20 cm libraries, another three in the 80 cm libraries, and two in libraries from both depths. There were five 13C-enriched MAGs in hot spring sediment libraries. One gammaproteobacterial MAG enriched in hog fuel (MB2.51) was placed in family Steroidobacteraceae (Fig. 2). The remainder of enriched MAGs were Gram-positive bacteria, including the phyla Chloroflexi, Actinobacteria and Firmicutes. MB2.64 from hog fuel was placed in the thermophilic actinobacterial family Solirubrobacteraceae, and was enriched over 300-fold in both 20 and 80 cm libraries.
Phylogenetic tree MAG placement using GTDB-TK v1.0.2 based on 120 bacterial single-copy genes. 13C-enriched genomes shown in bold. Scale shows length equivalent to 0.1 substitutions. Heatmap shows square-root transformed mean MAG abundance following DeSeq2 normalization in triplicate libraries. Abundance calculated by mapping quality-filtered reads to MAG nucleotide sequences with bbmap 38.22. Bar plot shows log2 fold change (L2FC) between 13C-High and 12C-High for each genome with >L2FC 0 indicating enrichment in 13C-High libraries. Error bars represent standard error of L2FC. Bar plot provides cut-off estimates for significance at αadj = 0.05 (individual padj values <0.05 provided).
Phylogenetic tree, heatmaps, and bar plot as in Fig. 2.
While we were able to recover 125 MAGs with an average single-copy gene completeness of 87% (Supplementary Data 2), the full suite of metabolism-encoding genes was likely not recovered for all, potentially resulting in incomplete annotation of aromatic degradation pathways. Of the hog fuel MAGs, MB2.64 (99% completeness), encoded catechol and protocatechuate ortho-cleavage pathways, and 4-hydroxybenzoate monooxygenase (Fig. 4). A Thermoleophilaceae MAG, MB2.39 was also highly enriched with 13C in the 80 cm hog fuel microcosms, and like MB2.64, is a member of the thermophilic order Solirubrobacterales. Other 13C-enriched Actinobacteria include CON22 (Actinomadura rubrobrunea), MAX.045 (Frankia sp.), MAX.030 (Micromonosporaceae), CON124 (Rubrobacter sp.) and MB2.74 (Rubrobacter sp.) (Fig. 3). These MAGs all encoded protocatechuate degradation. In addition, A. rubrobrunea encoded a two-component vanillate O-demethylase, while the Micromonosporaceae MAG appeared to encode a LigM-type (aminomethyltransferase) vanillate O-demethylase based on pHMM results (Fig. 4). To account for incomplete annotation of our MAGs, their aromatic degradation pathways were compared with those annotated in closely related strains (Fig. 4), revealing that vanillate O-demethylation and protocatechuate ortho-cleavage are encoded widely in thermophilic Actinobacteria.
Phylogeny of MAGs as in Figs. 2 and 3. Aromatic degradation pathway genes were annotated using profile HMMs for KEGG orthologs (KO) with e < 0.01 and HMM scores above KO-specific thresholds. Syringate O-demethylase (LigM) orthologs annotated using the TreeSAPP reference package are shown with 50% opacity (i.e., light pink). Individual orders containing enriched MAGs are highlighted. H4F Tetrahydrofolate, PCA Protocatechuic acid, HB Hydroxybenzoate, BKA beta-ketoadipate.
The 13C-enriched Lakelse hot spring MAGs represented a higher proportion of Firmicutes than the hog fuel MAGs (Figs. 2 and 3). These included Kyrpidia sp. (CON.60) and Alicyclobacillus sp. (MB2.88, CON.104). While some Kyrpidia and Alicyclobacillus reference genomes encode catechol meta-cleavage (Fig. 4), only CON.60 was found to encode this pathway in our MAG dataset. Three Alicyclobacillus genomes (MB2.88, CON25, and MB2.97) encoded LMCOs with high amino acid identity (>85%) to a homolog in phenolic- and polyphenolic-oxidizing Alicyclobacillus acidocaldarius DSM 446 [38]. Of these, MB2.88 contained two L-type 3-domain LCMOs with 100% amino acid identity to those encoded by DSM 446.
Identification of putative ligninases
Typically, DyPs and LCMOs are only broadly classified by existing pHMMs, or in sequence databases. We therefore applied phylogenetic profiling with TreeSAPP [39] to classify these enzymes into discrete sub-families (Fig. 5A). Sequences from MAGs were placed only into A and B DyP types. Few DyPs were recovered (Fig. 4), none of which contained secretion signals. Nevertheless, DyP2, a C-type DyP from Amycolatopsis sp. 75iv2 involved in lignin depolymerization has no detectable signal sequence [36]. In contrast to DyPs, 82 LCMOs were detected in hot spring MAGs, and 100 were detected in hog fuel MAGs. Of these, only four were detected in LCCED sub-family 11, corresponding to SLAC or K-type laccases, all of which contained TAT signal peptides (Fig. 5B). Additionally, an O-type two-domain LCMO was detected in MB2.51 (Steroidobacteraceae), also containing a leading secretion signal peptide (Fig. 5C). We named this enzyme LacOST51.
A Phylogeny of reference LCMOs. Protein sequences were aligned using MAFFT using the ginsi setting under 1000 iterations. Phylogeny was reconstructed using RAxML under the PROTGAMMAPMB model with 1000 iterations. Values beside labels show the number of reference sequences for each clade. B K-type (2dMCO SLAC) sequences from MAGs placed into reference tree. Presence of 5’ signal peptides shown using a green square. C Phylogenetic placement of LCMO sequences from MAGs in the O-type (2dMCO) clade. D Multiple alignment of reference and MAG LMCO sequences. Blue denotes 2dMCOs and red denotes 3dMCOs. Length of 5’ signal peptides shown in green. Yellow markings denote conserved copper-binding residues. Blue markings denote substrate-binding residues. Gray denotes strength of conservation. Yellow region shows possible active-site protecting fold in Type-K1 SLACs.
The classification of LCMOs into super-families, and comparison with enzymes of known function, may shed light on their functional roles. We applied structural alignment to assess the relationships between sequence, structure and function (Fig. 5D). Cu- and substrate-binding residues were highly conserved across K-type LMCOs including those recovered in this study. However, a 15-amino acid sequence hypothesized to act as a “flap” covering one of the channels leading to the trinuclear cluster [40], which was present in all known SLACs, was absent in the recovered thermophilic K-type LMCOs. Based on these sequence differences and the results of phylogenetic clustering, we categorize the LMCOs recovered from thermal systems as K2-type LCMOs, in contrast to the K1-type LCMOs found in mesophilic Actinobacteria such as Streptomyces coelicolor [40]. The lignin-degradation potential and thermotolerance of the K2 laccase clade remains uncharacterized.
Putative tetrahydrofolate-dependent O-demethylases
As our model lignin is comprised of 100% guaiacyl- subunits, we hypothesized that mineralization of DHP requires O-demethylation. While we identified a small number of vanillate O-demethylases (Fig. 4), which are Rieske-type oxygenases, we also investigated the potential for tetrahydrofolate-dependent O-demethylation of methoxylated aromatic compounds. To interrogate MAGs for tetrahydrofolate-dependent aryl O-demethylases, we once again used phylogenetic placement. We categorized aminomethyltransferases by putative function and taxonomic identity, with LigM and DesA sequences partitioning into distinct clusters (Fig. 6A). Assembled sequences placed into the tree formed a separate clade emerging from the DesA branch, which we have labeled “DesA-like aminomethyltransferases” (Fig. 6B). Specifically, MB2.64 (Solirubrobacterales), MB2.39 (Thermoleophilaceae), CON124 (Rubrobacter sp.), and MB2.51 (Steroidobacteraceae) all contained what appear to be DesA-like aminomethyltransferases, with conservation of a methyl-transferring tyrosine residue verified by structure-guided protein alignment (Fig. 6C). While the metabolic function of these enzymes requires validation, it is intriguing that they may facilitate the O-demethylation of LDACs in thermophilic bacteria.
A Reference tree produced by TreeSAPP with 50 amino acid sequences using RAxML under the PROTGAMMALG model and 1000 iterations. Tree includes experimentally-validated vanillate/3-O-methylgalate O-demethylase (LigM) and syringate O-demethylase (DesA) proteins. B Placement of predicted MAG-encoded aminomethyltransferases into the reference tree. C Multiple sequence alignment with MAFFT using the ginsi setting under 1000 iterations for select sequences. Aromatic-binding residues derived from LigM structural model are shown in pink, folate-binding residues are shown in green, and the primary methyl-transferring catalytic tyrosine residue shown in orange. D Pathway diagram of tetahydrofolate-dependent O-demethylation of methoxylated aromatic compounds.
Characterization of LacOST51
A key question that emerged from metabolic reconstruction of 13C-DHP-enriched MAGs following the microcosm study was the mechanism for the observed lignin depolymerization. In the above analysis we focused on the LCMOs as a possible answer. To test this hypothesis, and potentially identify novel biocatalysts, we selected four two-domain LCMOs (two K2-type, one O-type, one N-type) for heterologous expression, with the objective of evaluating their role in depolymerizing lignin (Table 1). Of these only “LacOST51,” the O-type LCMO from MB2.51, proved soluble when expressed in E. coli (Table 1). Expression in Rhodococcus jostii RHA1 did not improve the solubility of the other proteins. SDS-PAGE indicated that the LacOST51 protein was purified to >99% apparent homogeneity and had a molecular mass of ~36 kDa. Purified LacOST51 exhibited the blue color typical of laccases, and had an absorption band at 620 nm, characteristic of a T1 blue copper site. The preparation had a molar copper content of 4.0 ± 0.2, indicating that the purified LacOST51 was loaded with a full complement of copper.
LacOST51 utilized 2,2′-azino-di-(3-ethylbenzthiazoline sulfonate) (ABTS) and 2,6-dimethoxyphenol (DMP) as reducing substrates with specific activities of 1.46 and 0.03 U mg−1, respectively. The specific activity of LacOST51 for ABTS was of the same order of magnitude as that reported for other bacterial laccases (Table 1). However, specific activity for DMP was lower than the value for most other reported laccases. The oxidation of DMP was optimal at pH 8, and the enzyme retained ~90% activity at pH 9. These values are higher than the average reported for other bacterial laccases [41], and match the optimal values described for alkaline laccases (e.g. [42, 43]). LacOST51 retained >50% of its activity after 12 h when incubated at 55 °C. However, the half-life of LacOST51 dropped significantly at 65 °C. Generally, the high pH preference and the thermal stability indicate that the enzyme is suitable for industrial applications.
To evaluate the ability of LacOST51 to transform lignin, we initially tested the reactivity of LacOST51 toward guaiacylglycerol-β-guaiacyl ether (GGE) and veratrylglycerol-β-guaiacyl ether (VGE), phenolic and non-phenolic compounds, respectively, that contain the β-O-4 linkage prevalent in lignin. GGE was depleted upon incubation with LacOST51 for 6 h, and several products were detected by HPLC (Fig. 7A). The retention times of these compounds suggest that they are oligomerization products [44]. In contrast, the enzyme did not detectably transform VGE. These results indicate that LacOST51 can react with phenolic substrates in the absence of mediators.
A Reactivity of LacOST51with β-O-4 biaryl ethers. LacOST51 (1 μM) was incubated for 6 h with 1 mM guaiacylglycerol-β-guaiacyl ether (left) or veratrylglycerol-β-guaiacyl ether (right) with 20 mM sodium phosphate and pH 8 at 55 °C. HPLC traces are of reactions with (solid line) and without (dotted) enzyme. B Treatment of EMAL with LacOST51. LacOST51 (6 μM) was incubated for 6 days with 0.5% (w/v) EMAL (12.5 mM sodium phosphate, pH 8, 10% DMSO, at 30 °C). HPLC traces are of reactions with (solid line) and without (dotted) enzyme. The identities of the indicated compounds were confirmed using authentic standards. DMBQ: 2,6-Dimethoxy benzoquinone. Inset: Effect of laccase treatment on molar mass distribution of Eucalyptus EMAL, where EMAL was treated with either LacOST51, sLac or no enzyme (bsLac treatment generated insoluble material that was not analyzed using GPC). C HSQC NMR spectra of laccase-treated EMAL. EMAL was incubated with no enzyme (a) and (d), LacOST51 (b) and (e), or sLac (c) and (f). The top and the bottom panels show the aliphatic and aromatic regions, respectively, of the 2D-NMR spectra. Linkages and units are expressed as per 100 aromatic units (100 Ar), which represented the integration of the G2 + 1/2S2. Structures of the regions are shown to the right.
Transformation of enzymatic mild acidolysis lignin by LacOST51
We tested the ability of LacOST51 to transform EMAL, a minimally altered form of lignin that contains little residual cellulose or hemicellulose from Eucalyptus wood [45]. A solution of EMAL incubated without enzyme for 6 days contained a significant quantity of vanillin (tR = 10.6 min) and oligomeric material that eluted as a broad band (tR = 13–22 min) (Fig. 7B). Incubation with 6 µM LacOST51 additionally resulted in the production of syringaldehyde and 2,6-dimethoxy benzoquinone (DMBQ) as well as a reduction in the amount of oligomeric material. To provide insight into how LacOST51 modified the lignin, the transformed lignin was isolated and characterized using NMR and GPC-MALS spectrometry. In these experiments, sLac was used as a positive control as it had previously been shown to transform lignin [7]. HSQC NMR spectra from laccase-treated lignin samples differed significantly from those of the no-enzyme controls (Fig. 7C). Specifically, in samples treated with either LacOST51 or sLac, the secondary aliphatic hydroxyl groups of the lignin were oxidized to their benzylic ketone, as reflected by the increased in signals S’, G,’ A’, and A”. This oxidization process leads to the ability to cleave the propyl side chain with oxidative reagents resulting in rupture of native linkages (β-O-4, A) as illustrated with model compounds [46]. The slightly modified reaction on the C-C linkages including β-5 (B) and β-β (C) in our study and elsewhere [47] require more investigation. Based on the amount of linkages modified, sLac modified the lignin more efficiently than LacOST51.
GPC analysis indicated that treatment of EMAL with either LacOST51 or sLac yielded lignin with a higher apparent molar mass (Mw and Mn) and increased the range of fragment size (Fig. 7B). However, sLac treatment resulted in a significant amount of insoluble material that was not included in this analysis. The higher molecular weight for both treated materials is presumably due to condensation reactions between the aromatic radicals [48]. The observed polymerization activity of LacOST51 and sLac is consistent with studies of other laccases (e.g. [49]) and depends on the reaction conditions, particularly the relative concentrations of lignin species of different molecular weight. Indeed, sLac catalyzes the depolymerization of lignin in steam-pretreated poplar in the presence of natural mediators [7]. Overall, these data demonstrate that LacOST51 oxidatively transforms lignin in the same manner as other laccases.
Discussion
We hypothesized that lignin-degrading microorganisms occur in thermal environments that receive woody biomass inputs and would assimilate carbon from a synthetic lignin. The synthesis of 13C-DHP lignin in our laboratory was essential for accurate assessment of such assimilation, as 13C-labeled lignin of the purity used in this study is not commercially available. We verified lignin degradation in microcosms by monitoring 13C-CO2 production and 13C-incoropration into recovered DNA. The incorporation of 13C molecules from labeled, high-molecular-weight synthetic lignin into bacterial DNA provides direct evidence for bacterial catabolism of LDACs, and indirect support for bacterial lignin depolymerization. While it is evident that lignin was degraded, we cannot completely rule out other mechanisms of lignin depolymerization undetected by our methods. For example, a stable pool of extracellular fungal lignin peroxidases in our inoculum could have contributed to lignin depolymerization. However, genes putatively encoding lignin depolymerization were found in the MAGs of organisms that assimilated lignin in the microcosms, suggesting that thermophilic bacteria contributed to lignin depolymerization.
Several bacterial taxa were enriched with 13C from synthetic lignin. In hog fuel, these bacteria primarily belong to the proposed actinobacterial class Thermoleophilia—members of which are abundant in geothermal environments and soil but have poorly characterized metabolic potential [50]. Specifically, the genus Rubrobacter includes known thermophiles [51, 52], and Rubrobacter OTUs were strongly associated with lignocellulose degradation and tolerance of phenolic lignin metabolites at 55 °C [53]. Related actinobacterial MAGs, including from an Actinomadura rubrobrunea strain (CON22), were also 13C-enriched in hog fuel microcosms. CON22 contained one of few Rieske vanillate O-demethylases found in this study and encoded complete protocatechuate ortho-cleavage and partial meta-cleavage pathways (Fig. 4). While the lignin-degradation potential of A. rubrobrunea has yet to be characterized, other Actinomadura strains have been found to solubilize lignocellulose [54], and contribute to the degradation of the cellulose [55] or lignin [56]. The Rubrobacter and A. rubrobrunea strains identified in this study make compelling targets for further investigation. The MB2.64 MAG was placed in the actinobacterial family Solirubrobacteraceae. We previously identified putatively lignolytic Solirubrobacterales OTUs in forest soil through a similar stable isotope probing approach [14], but we were unable to resolve MAGs from lignin-assimilating Actinobacteria or identify enzymatic mechanisms that would explain their involvement in lignin degradation. We propose MB2.64 from hog fuel has robust lignin degradation potential. In contrast, in hot spring communities, 13C-enriched taxa were predominantly Firmicutes such as Alicyclobacillus ssp. Therefore, this study expands the taxonomic range of bacteria associated with lignin degradation to include other thermophilic Actinobacteria and Firmicutes.
In addition to incorporation of 13C from synthetic lignin and presence of putative ligninases, we used the presence of genes encoding catabolism of aromatic compounds to evaluate each MAG for its lignin degradation potential. A key difference between MAGs from hog fuel and hot springs was not only taxonomy, but also the capacity for aromatic catabolism. Hog fuel MAGs encoded vanillate O-demethylation and the protocatechuate ortho-cleavage pathway, suggesting that LDACs liberated from lignin were funneled into specific degradation pathways. Although several cultured representatives of Solirubrobacteriales encode a two-component Rieske vanillate O-demethylase (Fig. 4), none were found in the genome of MB2.64 from the hot spring. This was at odds with our hypothesis that O-demethylation is critical to catabolism of LDACs. However, a methyltransferase with full-length amino acid identity of 69.5% to syringate O-demethylase (DesASYK-6), and with conserved substrate-binding residues, was identified through phylogenetic placement and sequence alignment. Accordingly, the 5,10-methylene-tetrahydrofolate reductase (metF) and formate-tetrahydrofolate ligase (ligH) genes, encoding the tetrahydrofolate-mediated C1 metabolic pathway [57, 58], were also found in the MB2.64 genome (Fig. 5D). Genomes of other Gram-positive bacteria such as Rubrobacter xylanophilus DSM9941 [57], Acetobacterium dehalogenans and Desulfitobacterium hafniense [59] (both Firmicutes) encode vanillate-demethylating methyltransferases. Thus, the MB2.64 genome provides strong evidence for lignin degradation mediated by bacterial thermophiles, facilitated by a novel one-component actinobacterial O-demethylase.
The 13C-enriched MAG, MB2.51, from hog fuel was placed in the gammaproteobacterial family, Steroidobacteraceae. As the name suggests, bacteria in this family can degrade steroidal hormones [60], but also polyvinyl alcohol [61] and rubber [62]. MB2.51 encoded full catechol and protocatechuate meta-cleavage pathways, as well as monooxygenases involved in 4-hydroxybenzoate and phenol hydroxylation. A methyltransferase from MB2.51 contained all conserved LigMSYK-6 residues involved in methyl-transfer, as well as aromatic substrate and folate binding, suggesting a role for this gene in degradation of methoxylated aromatic compounds. No thermophilic Steroidobacteraceae strains have been previously reported. Yet this MAG yielded the only soluble, thermotolerant laccase found in this study, discussed below.
Lignin degradation mechanisms in 13C-enriched hot spring MAGs are less clear than in their hog fuel counterparts. One possible explanation for this is cross-feeding, the catabolism by one organism of LDACs produced by a different lignin depolymerizing organism. However, the two 13C-enriched Alicyclobacillus sp. MAGs from hot spring sediment encoded a suite of L- and O-type LCMOs that bore high sequence identity to LCMOs found in A. acidocaldarius capable of non-specific cleavage of lignin-derived polyphenols [38]. The catechol meta-cleavage pathway genes in Alicyclobacillus sp. MAGs are incomplete, although the complete pathway is encoded in closely related Alicyclobacillus genomes (Fig. 4). We propose that, similar to A. acidocaldarius, the Alicyclobacillus sp. MAGs recovered herein encode non-specific oxidative degradation of lignin-derived polyphenols, which can serve as a source of carbon in carbon and nutrient limited oligotrophic ecosystems such as geothermal hot springs. This may also serve to as a detoxification mechanism.
Lignin-degrading organisms can serve as a source of novel ligninases, including laccases. Laccases are multi-copper oxidases that oxidize a broad range of compounds including substituted phenols, arylamines and aromatic thiols [63]. Bacterial laccases are appealing and versatile catalysts due to their thermal stability [64], use of molecular oxygen as the final electron acceptor and production of only water as a by‐product [65]. Here, we used newly published software, TreeSAPP [39], which places novel sequences into reference phylogenies. We designed a multi-copper oxidase reference phylogeny based on a model of 16 sub-families [66]. Specifically, we identified a possible thermophilic clade of the lignolytic two-domain K-type SLACs found in 13C-enriched Solirubrobacteraceae and Thermoleophilales, which we refer to as K2-type laccases. We also annotated a number of two- and three-domain LMCOs in 13C-enriched Steroidobacteraceae and Alicyclobacillus MAGs from sub-families O and L, which contain members capable of phenolic oxidation [67, 68]. Together, these results suggest that bacterial laccases are involved in lignin degradation in thermal environments.
To validate putatively lignin-degrading LCMOs identified with 13C-lignin SIP, we heterologously expressed a selection of these enzymes. Specifically, we attempted to express two K2-type, one O-type and one N-type laccase in E. coli. The two K2-type enzymes were expressed as insoluble forms, and expression of the N-type was not detected in either soluble or insoluble form. We also attempted to express the actinobacterial K2-type laccases in Rhodococcus jostii RHA1, but they were again insoluble—thus, the lignin degradation potential of the K2- and N-type laccases remains uncharacterized. The O-type laccase originating from the gammaproteobacterial MB2.51 MAG was expressed in soluble form in E. coli and further purified and characterized. This enzyme (LacOST51) transformed a minimally transformed Eucalyptus lignin, liberating LDACs, including DMBQ and syringaldehyde. We previously demonstrated that syringaldehyde is a major degradation product of Eucalyptus lignin, and can be funnelled into the syringic acid meta-cleavage pathway in thermophilic Alphaproteobacteria [37]. Our results herein demonstrate that bacterial laccases, such as LacOST51 or the previously-characterized sLac, can generate lignin-derived mono-aromatic compounds at elevated temperature. We propose that that these thermostable biocatalysts can be employed for bacterial bio-product production.
In this paper we characterized 14 genomes from bacteria in thermal hog fuel and hot spring sediment environments that incorporated synthetic 13C-labeled lignin. The results supported our hypothesis that these communities harbor thermophilic, lignin-degrading bacteria. These bacteria include members of the actinobacterial families Solirubrobacterales and Thermoleophilaceae that have a distinct clade of K2-type SLACs. These bacteria also included a Gammaproteobacteria (Steroidobacteraceae) from which we expressed a lignin-transforming O-type laccase. Overall, this study advanced our knowledge of how thermophilic bacteria can degrade lignin and LDACs and identifies enzymes potentially useful in biocatalysts for lignin valorization.
References
Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–46. https://doi.org/10.1146/annurev.arplant.54.031902.134938.
Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, et al. Lignin valorization: improving lignin processing in the biorefinery. Science. 2014;344:1246843. https://doi.org/10.1126/science.1246843.
Hildén K, Hakala TK, Lundell T. Thermotolerant and thermostable laccases. Biotechnol Lett. 2009;31:1117. https://doi.org/10.1007/s10529-009-9998-0.
Wilhelm RC, Singh R, Eltis LD, Mohn WW. Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. 2018;1. https://doi.org/10.1038/s41396-018-0279-6.
Bugg TDH, Ahmad M, Hardiman EM, Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol. 2011;22:394–400. https://doi.org/10.1016/j.copbio.2010.10.009.
Kamimura N, Takahashi K, Mori K, Araki T, Fujita M, Higuchi Y, et al. Bacterial catabolism of lignin-derived aromatics: new findings in a recent decade: update on bacterial lignin catabolism. Environ Microbiol Rep. 2017;9:679–705. https://doi.org/10.1111/1758-2229.12597.
Singh R, Hu J, Regner MR, Round JW, Ralph J, Saddler JN, et al. Enhanced delignification of steam-pretreated poplar by a bacterial laccase. Sci Rep. 2017;7:42121. https://doi.org/10.1038/srep42121.
Perna V, Meyer AS, Holck J, Eltis LD, Eijsink VGH, Wittrup Agger J. Laccase-catalyzed oxidation of lignin induces production of H2O2. ACS Sustain Chem Eng. 2020;8:831–41. https://doi.org/10.1021/acssuschemeng.9b04912.
Johnson CW, Salvachúa D, Rorrer NA, Black BA, Vardon DR, St. John PC, et al. Innovative chemicals and materials from bacterial aromatic catabolic pathways. Joule. 2019;3:1523–37. https://doi.org/10.1016/j.joule.2019.05.011.
Brady AL, Sharp CE, Grasby SE, Dunfield PF. Anaerobic carboxydotrophic bacteria in geothermal springs identified using stable isotope probing. Front Microbiol. 2015;6. https://doi.org/10.3389/fmicb.2015.00897.
Grasby SE, Hutcheon I, Krouse HR. The influence of water–rock interaction on the chemistry of thermal springs in western Canada. Appl Geochem. 2000;15:439–54. https://doi.org/10.1016/S0883-2927(99)00066-9.
Bauchop T, Elsden SR. The growth of micro-organisms in relation to their energy supply. Microbiology. 1960;23:457–69. https://doi.org/10.1099/00221287-23-3-457.
Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, et al. DNA stable-isotope probing. Nat Protoc. 2007;2:860–6. https://doi.org/10.1038/nprot.2007.109.
Wilhelm RC, Singh R, Eltis LD, Mohn WW. Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. 2019;13:413–29. https://doi.org/10.1038/s41396-018-0279-6.
Wilhelm R, Szeitz A, Klassen TL, Mohn WW. Sensitive, efficient quantitation of 13C-enriched nucleic acids via ultrahigh-performance liquid chromatography-tandem mass spectrometry for applications in stable isotope probing. Appl Environ Microbiol. 2014;80:7206–11. https://doi.org/10.1128/AEM.02223-14.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma Oxf Engl. 2014;30:2114–20. https://doi.org/10.1093/bioinformatics/btu170.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. https://doi.org/10.1089/cmb.2012.0021.
Lin H-H, Liao Y-C. Accurate binning of metagenomic contigs via automated clustering sequences using information of genomic signatures and marker genes. Sci Rep. 2016;6:24175. https://doi.org/10.1038/srep24175.
Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7:e7359. https://doi.org/10.7717/peerj.7359.
Alneberg J, Bjarnason BS, Bruijn ID, Schirmer M, Quick J, Ijaz UZ, et al. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11:1144–6. https://doi.org/10.1038/nmeth.3103.
Wu Y-W, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinforma Oxf Engl. 2016;32:605–7. https://doi.org/10.1093/bioinformatics/btv638.
Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 2018;3:836–43. https://doi.org/10.1038/s41564-018-0171-1.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. https://doi.org/10.1101/gr.186072.114.
Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119. https://doi.org/10.1186/1471-2105-11-119.
Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. https://doi.org/10.1038/nmeth.3176.
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–5. https://doi.org/10.1093/nar/gkt1178.
Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–101. https://doi.org/10.1093/nar/gky418.
El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–32. https://doi.org/10.1093/nar/gky995.
Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, et al. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 2001;29:41–3.
Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020. https://doi.org/10.1093/bioinformatics/btz859.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15. https://doi.org/10.1186/s13059-014-0550-8.
R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. https://www.R-project.org.
Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5. https://doi.org/10.1093/nar/gkw290.
Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. https://doi.org/10.1111/2041-210X.12628.
Brenner AJ, Harris ED. A quantitative test for copper using bicinchoninic acid. Anal Biochem. 1995;226:80–4. https://doi.org/10.1006/abio.1995.1194.
Brown ME, Barros T, Chang MCY. Identification and characterization of a multifunctional dye peroxidase from a lignin-reactive bacterium. ACS Chem Biol. 2012;7:2074–81. https://doi.org/10.1021/cb300383y.
Levy-Booth DJ, Hashimi A, Roccor R, Liu L-Y, Renneckar S, Eltis LD, et al. Genomics and metatranscriptomics of biogeochemical cycling and degradation of lignin-derived aromatic compounds in thermal swamp sediment. ISME J. 2021;15:879–93. https://doi.org/10.1038/s41396-020-00820-x.
Aston JE, Apel WA, Lee BD, Thompson DN, Lacey JA, Newby DT, et al. Degradation of phenolic compounds by the lignocellulose deconstructing thermoacidophilic bacterium Alicyclobacillus Acidocaldarius. J Ind Microbiol Biotechnol. 2016;43:13–23. https://doi.org/10.1007/s10295-015-1700-z.
Morgan-Lang C, McLaughlin R, Armstrong Z, Zhang G, Chan K, Hallam SJ. TreeSAPP: the tree-based sensitive and accurate phylogenetic profiler. Bioinformatics. 2020. https://doi.org/10.1093/bioinformatics/btaa588.
Machczynski MC, Vijgenboom E, Samyn B, Canters GW. Characterization of SLAC: a small laccase from streptomyces coelicolor with unprecedented activity. Protein Sci Publ Protein Soc. 2004;13:2388–97. https://doi.org/10.1110/ps.04759104.
Berini F, Verce M, Ausec L, Rosini E, Tonin F, Pollegioni L, et al. Isolation and characterization of a heterologously expressed bacterial laccase from the anaerobe Geobacter metallireducens. Appl Microbiol Biotechnol. 2018;102:2425–39. https://doi.org/10.1007/s00253-018-8785-z.
Yin Q, Zhou G, Peng C, Zhang Y, Kües U, Liu J, et al. The first fungal laccase with an alkaline pH optimum obtained by directed evolution and its application in indigo dye decolorization. AMB Express. 2019;9:151. https://doi.org/10.1186/s13568-019-0878-2.
Kumar D, Kumar A, Sondhi S, Sharma P, Gupta N. An alkaline bacterial laccase for polymerization of natural precursors for hair dye synthesis. 3 Biotech. 2018;8:182. https://doi.org/10.1007/s13205-018-1181-7.
Hilgers R, Vincken J-P, Gruppen H, Kabel MA. Laccase/mediator systems: their reactivity toward phenolic lignin structures. ACS Sustain Chem Eng. 2018;6:2037–46. https://doi.org/10.1021/acssuschemeng.7b03451.
Wu S, Argyropoulos D. An improved method for isolating lignin in high yield and purity. J Pulp Pap Sci. 2003;29:235–40.
Gao R, Li Y, Kim H, Mobley JK, Ralph J. Selective oxidation of lignin model compounds. ChemSusChem. 2018;11:2045–50. https://doi.org/10.1002/cssc.201800598.
Rahimi A, Azarpira A, Kim H, Ralph J, Stahl SS. Chemoselective metal-free aerobic alcohol oxidation in lignin. J Am Chem Soc. 2013;135:6415–8. https://doi.org/10.1021/ja401793n.
Schutyser W, Renders T, Bosch SV, den, Koelewijn S-F, Beckham GT, Sels BF. Chemicals from lignin: an interplay of lignocellulose fractionation, depolymerisation, and upgrading. Chem Soc Rev. 2018;47:852–908. https://doi.org/10.1039/C7CS00566K.
Sun X, Bai R, Zhang Y, Wang Q, Fan X, Yuan J, et al. Laccase-catalyzed oxidative polymerization of phenolic compounds. Appl Biochem Biotechnol. 2013;171:1673–80. https://doi.org/10.1007/s12010-013-0463-0.
Hu D, Zang Y, Mao Y, Gao B. Identification of molecular markers that are specific to the class Thermoleophilia. Front Microbiol. 2019;10. https://doi.org/10.3389/fmicb.2019.01185.
Chen M-Y, Wu S-H, Lin G-H, Lu C-P, Lin Y-T, Chang W-C, et al. Rubrobacter taiwanensis sp. nov., a novel thermophilic, radiation-resistant species isolated from hot springs. Int J Syst Evol Microbiol. 2004;54:1849–55. https://doi.org/10.1099/ijs.0.63109-0.
Tomariguchi N, Miyazaki K. Complete genome sequence of Rubrobacter xylanophilus strain AA3-22, isolated from Arima Onsen in Japan. Microbiol Resour Announc. 2019;8. https://doi.org/10.1128/MRA.00818-19.
Ceballos SJ, Yu C, Claypool JT, Singer SW, Simmons BA, Thelen MP, et al. Development and characterization of a thermophilic, lignin degrading microbiota. Process Biochem. 2017;63:193–203. https://doi.org/10.1016/j.procbio.2017.08.018.
Clark Mason J, Richards M, Zimmermann W, Broda P. Identification of extracellular proteins from actinomycetes responsible for the solubilisation of lignocellulose. Appl Microbiol Biotechnol. 1988;28:276–80. https://doi.org/10.1007/BF00250455.
Yin Y-R, Sang P, Xian W-D, Li X, Jiao J-Y, Liu L, et al. Expression and characteristics of two glucose-tolerant GH1 β-glucosidases from Actinomadura amylolytica YIM 77502T for promoting cellulose degradation. Front Microbiol. 2018;9. https://doi.org/10.3389/fmicb.2018.03149.
Zimmermann W, Broda P. Utilization of lignocellulose from barley straw by actinomycetes. Appl Microbiol Biotechnol. 1989;30:103–9. https://doi.org/10.1007/BF00256005.
Abe T, Masai E, Miyauchi K, Katayama Y, Fukuda M. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J Bacteriol. 2005;187:2030–7. https://doi.org/10.1128/JB.187.6.2030-2037.2005.
Varman AM, He L, Follenfant R, Wu W, Wemmer S, Wrobel SA, et al. Decoding how a soil bacterium extracts building blocks and metabolic energy from ligninolysis provides road map for lignin valorization. Proc Natl Acad Sci USA. 2016;113:E5802–11. https://doi.org/10.1073/pnas.1606043113.
Studenik S, Vogel M, Diekert G. Characterization of an O-demethylase of Desulfitobacterium hafniense DCB-2. J Bacteriol. 2012;194:3317–26. https://doi.org/10.1128/JB.00146-12.
Fahrbach M, Kuever J, Remesch M, Huber BE, Kämpfer P, Dott W, et al. Steroidobacter denitrificans gen. nov., sp. nov., a steroidal hormone-degrading gammaproteobacterium. Int J Syst Evol Microbiol. 2008;58:2215–23. https://doi.org/10.1099/ijs.0.65342-0.
Nogi Y, Yoshizumi M, Hamana K, Miyazaki M, Horikoshi K. Povalibacter uvarum gen. nov., sp. nov., a polyvinyl-alcohol-degrading bacterium isolated from grapes. Int J Syst Evol Microbiol. 2014;64:2712–7. https://doi.org/10.1099/ijs.0.062620-0.
Sharma V, Siedenburg G, Birke J, Mobeen F, Jendrossek D, Prakash T. Metabolic and taxonomic insights into the Gram-negative natural rubber degrading bacterium Steroidobacter cummioxidans sp. nov., strain 35Y. PLoS ONE. 2018;13:e0197448. https://doi.org/10.1371/journal.pone.0197448.
Reiss R, Ihssen J, Richter M, Eichhorn E, Schilling B, Thöny-Meyer L. Laccase versus laccase-like multi-copper oxidase: a comparative study of similar enzymes with diverse substrate spectra. PLoS ONE. 2013;8:e65633. https://doi.org/10.1371/journal.pone.0065633.
Christopher LP, Yao B, Ji Y. Lignin biodegradation with laccase-mediator systems. Front Energy Res. 2014;2. https://doi.org/10.3389/fenrg.2014.00012.
Mate DM, Alcalde M. Laccase: a multi‐purpose biocatalyst at the forefront of biotechnology. Micro Biotechnol. 2016;10:1457–67. https://doi.org/10.1111/1751-7915.12422.
Sirim D, Wagner F, Wang L, Schmid RD, Pleiss J. The Laccase Engineering Database: a classification and analysis system for laccases and related multicopper oxidases. Database J Biol Databases Curation. 2011;2011. https://doi.org/10.1093/database/bar006.
Fang Z, Li T, Wang Q, Zhang X, Peng H, Fang W, et al. A bacterial laccase from marine microbial metagenome exhibiting chloride tolerance and dye decolorization ability. Appl Microbiol Biotechnol. 2011;89:1103–10. https://doi.org/10.1007/s00253-010-2934-3.
Komori H, Miyazaki K, Higuchi Y. X-ray structure of a two-domain type laccase: a missing link in the evolution of multi-copper proteins. FEBS Lett. 2009;583:1189–95. https://doi.org/10.1016/j.febslet.2009.03.008.
Sherif M, Waung D, Korbeci B, Mavisakalyan V, Flick R, Brown G, et al. Biochemical studies of the multicopper oxidase (small laccase) from Streptomyces coelicolor using bioactive phytochemicals and site-directed mutagenesis. Microb Biotechnol. 2013;6:588–97. https://doi.org/10.1111/1751-7915.12068
Gunne M, Urlacher VB. Characterization of the alkaline laccase Ssl1 from Streptomyces sviceus with unusual properties discovered by genome mining. PLOS ONE. 2012;7:e52360 https://doi.org/10.1371/journal.pone.0052360
Dubé E, Shareck F, Hurtubise Y, Beauregard M, Daneault C. Decolourization of recalcitrant dyes with a laccase from Streptomyces coelicolor under alkaline conditions. J Ind Microbiol Biotechnol. 2008;35:1123–9. https://doi.org/10.1007/s10295-008-0391-0
Koschorreck K, Richter SM, Ene AB, Roduner E, Schmid RD, Urlacher VB. Cloning and characterization of a new laccase from Bacillus licheniformis catalyzing dimerization of phenolic acids. Appl Microbiol Biotechnol. 2008;79:217–24. https://doi.org/10.1007/s00253-008-1417-2
Mohammadian M, Fathi-Roudsari M, Mollania N, Badoei-Dalfard A, Khajeh K. Enhanced expression of a recombinant bacterial laccase at low temperature and microaerobic conditions: purification and biochemical characterization. J Ind Microbiol Biotechnol. 2010;37:863–9. https://doi.org/10.1007/s10295-010-0734-5
Ausec L, Berini F, Casciello C, Cretoiu MS, van Elsas JD, Marinelli F, et al. The first acidobacterial laccase-like multicopper oxidase revealed by metagenomics shows high salt and thermo-tolerance. Appl Microbiol Biotechnol. 2017;101:6261–76. https://doi.org/10.1007/s00253-017-8345-y
Ausec L, Črnigoj M, Šnajder M, Ulrih NP, Mandic-Mulec I. Characterization of a novel high-pH-tolerant laccase-like multicopper oxidase and its sequence diversity in Thioalkalivibrio sp. Appl Microbiol Biotechnol. 2015;99:9987–99. https://doi.org/10.1007/s00253-015-6843-3
Acknowledgements
This study was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (STPGP 506595-17) and a research contract from Genome BC (SIP004). LDE is the recipient of Canada Research Chair. Sequencing was performed by Dr. Sunita Sinha at The University of British Columbia Sequencing and Bioinformatics Consortium (Vancouver, CAN). We thank Dr. Peter Dunfield (University of Calgary) for assistance with sampling, and Carlo Dal Monte and Mark Wunderlich (Catalyst Paper) for access to Crofton Mill and assistance with sampling.
Author information
Authors and Affiliations
Contributions
DJLB conducted sampling, SIP experiments, genomic analyses, and wrote the paper. LEN performed enzymology experiments and co-wrote the paper. MMF synthesized DHP lignin substrates. LYL and SR performed lignin chemistry analysis and co-wrote the paper. TD performed SIP experiments. LDE and WWM designed experiments and co-wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Levy-Booth, D.J., Navas, L.E., Fetherolf, M.M. et al. Discovery of lignin-transforming bacteria and enzymes in thermophilic environments using stable isotope probing. ISME J 16, 1944–1956 (2022). https://doi.org/10.1038/s41396-022-01241-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-022-01241-8
This article is cited by
-
Two Paenibacillus spp. strains promote grapevine wood degradation by the fungus Fomitiporia mediterranea: from degradation experiments to genome analyses
Scientific Reports (2024)
-
Lignocellulosic fraction-induced niche differentiation within dissimilatory iron reducing bacterial groups in a paddy soil
Soil Ecology Letters (2024)
-
Enhanced biodegradation of lignin and lignocellulose constituents in the pulp and paper industry black liquor using integrated magnetite nanoparticles/bacterial assemblage
Applied Water Science (2024)