Abstract
Inoculating agricultural soils with nitrous oxide respiring bacteria (NRB) can reduce N2O-emission, but would be impractical as a standalone operation. Here we demonstrate that digestates obtained after biogas production are suitable substrates and vectors for NRB. We show that indigenous NRB in digestates grew to high abundance during anaerobic enrichment under N2O. Gas-kinetics and meta-omic analyses showed that these NRB’s, recovered as metagenome-assembled genomes (MAGs), grew by harvesting fermentation intermediates of the methanogenic consortium. Three NRB’s were isolated, one of which matched the recovered MAG of a Dechloromonas, deemed by proteomics to be the dominant producer of N2O-reductase in the enrichment. While the isolates harbored genes required for a full denitrification pathway and could thus both produce and sequester N2O, their regulatory traits predicted that they act as N2O sinks in soil, which was confirmed experimentally. The isolates were grown by aerobic respiration in digestates, and fertilization with these NRB-enriched digestates reduced N2O emissions from soil. Our use of digestates for low-cost and large-scale inoculation with NRB in soil can be taken as a blueprint for future applications of this powerful instrument to engineer the soil microbiome, be it for enhancing plant growth, bioremediation, or any other desirable function.
Similar content being viewed by others
Introduction
Nitrous oxide is an intermediate in the nitrogen cycle and a powerful greenhouse gas emitted in large volumes from agricultural soils, accounting for ~1/3 of total anthropogenic N2O emissions [1]. Reduced emissions can be achieved by minimizing the consumption of fertilizer nitrogen through improved agronomic practice and reduction of meat consumption [2, 3], but such measures are unlikely to do more than stabilize the global consumption of fertilizer-N [4]. This calls for more inventive approaches to reduce N2O emissions, targeting the microbiomes of soil [5], in particular the physiology and regulatory biology of the organisms involved in production and consumption of N2O in soil [6].
N2O turnover in soil involves several metabolic pathways, controlled by a plethora of fluctuating physical and chemical variables [7,8,9]). Heterotrophic denitrification is the dominant N2O source in most soils, while autotrophic ammonia oxidation may dominate in well drained calcareous soils and references therein [10]. Heterotrophic denitrifying organisms are both sources and sinks for N2O because N2O is a free intermediate in their stepwise reduction of nitrate to dinitrogen (NO3− →NO2− →NO → N2O → N2). Denitrification involves four enzymes collectively referred to as denitrification reductases: nitrate reductase (Nar/Nap), nitrite reductase (Nir), nitric oxide reductase (Nor) and nitrous oxide reductase (Nos), encoded by the genes nar/nap, nir, nor and nosZ, respectively. Oxygen is a strong repressor of denitrification, both at the transcriptional and the metabolic level [11, 12]. Many organisms have truncated denitrification pathways, lacking from one to three of the four reductase genes [13, 14], and truncated denitrifiers can thus act as either N2O producers (organisms without nosZ) or N2O reducers (organisms with nosZ only). The organisms with nosZ only, coined non-denitrifying N2O-reducers [15], have attracted much interest as N2O sinks in the environment [16]. Of note, organisms with a full-fledged denitrification pathway may also be strong N2O sinks depending on the relative activities and regulation of the various enzymes in the denitrification pathway [17, 18]. Despite their promise, feasible ways to utilize N2O-reducing organisms to reduce N2O emissions have not yet emerged.
A soil with a strong N2O-reducing capacity will emit less N2O than one dominated by net N2O producing organisms, as experimentally verified by Domeignoz-Horta et al. [19], who showed that soils emitted less N2O if inoculated with large numbers (107–108 cells g−1 soil) of organisms expressing Nos as their sole denitrification reductase. As a standalone operation, the large-scale production and distribution of N2O-respiring bacteria would be prohibitively expensive and impractical. However, the use of N2O-respiring bacteria could become feasible if adapted to an existing fertilization pipeline, such as fertilization with the nitrogen- and phosphate-rich organic waste (digestate) generated by biogas production in anaerobic digesters. Anaerobic digestion (AD) is already a core technology for treating urban organic wastes, and is expected to treat an increasing proportion of the much larger volumes of waste produced by the agricultural sector (Fig. 1), as an element of the roadmap towards a low-carbon circular economy [20]. This means that digestates from AD are likely to become a major organic fertilizer for agricultural soils, with a huge potential for reducing N2O emissions if enriched with N2O-respiring bacteria prior to application.
Solid arrows (top section) show streams of biomass available for anaerobic digestion (AD). Numbers indicate known estimates of currently used or potentially available amounts in Europe, in million tonnes dry-weight (DW) per year [50,51,52,53]. The arrow from anaerobic digestion to agricultural soil, indicates a credible pathway for digestate enriched with N2O-reducing bacteria (assuming enrichment after AD); fertilization with such enriched digestates strengthens the N2O sink capacity of the soil, hence reducing N2O emissions. N2O emissions from agricultural soil in Europe are estimated at 0.51 tG per year (min 0.33 – max 0.80), representing some 48% of total European N2O emissions [1], which account for ~3.5% of the global warming effect from European greenhouse gas emissions and 35% of the global warming effect from European agriculture [54]. The lower half of the picture shows the microbial nitrogen transformations underlying these N2O emissions, which are fed by fertilizers. Today, AD is primarily used for treating urban organic wastes, which comprise only ~10% of the biomass potentially available for AD. The amount of biomass treated by AD is expected to increase by an order of magnitude when adopted on a large scale in the agricultural sector. This would generate 70–135 Mt DW of digestate annually (assuming 50% degradation by AD), which is equivalent to 400–780 kg DW ha−1 y−1 if spread evenly on the total farmland of Europe (173 million ha).
Here we provide the first proof of this promising concept. Firstly, we demonstrate selective enrichment and isolation of fast-growing digestate-adapted N2O-respiring bacteria using a digestate from a wastewater treatment plant. Secondly, we demonstrate that the use of digestates enriched with such organisms as a soil amendment reduces the proportion of N leaving soil as N2O, confirming the suitability of such digestates for this purpose. Analysis of the enrichment process with multi-omics and in-depth monitoring of gas kinetics provides valuable insights into Nos-synthesis by the various enriched taxa, and the metabolic pathways of the anaerobic consortium providing substrates for these enriched N2O-respiring organisms.
Materials and methods
The digestates were taken from two anaerobic digesters, one mesophilic (37 °C) and one thermophilic (52 °C), which were running in parallel, producing biogas from sludge produced by a wastewater treatment plant. The sludge was a poly-aluminum chloride (PAX-XL61™, Kemira) and ferric chloride (PIX318™, Kemira) precipitated municipal wastewater sludge, with an organic matter content of 5.6% (w/w). Both digestors reduced the organic matter by approximately 60%, producing digestates containing ~2.1 % organic matter, 1.8–1.9 g NH4+-N L−1, ~16 and 32 Meq VFA L−1, pH = 7.6–7.8 and 8.2; mesophilic and thermophilic, respectively (see Supplementary Methods 1 for further details). The digestates were transported to the laboratory in 1 L insulated steel-vessels and used for incubation experiments 3–6 h after sampling.
The robotized incubation system developed by Molstad et al. [21, 22] was used in all experiments where gas kinetics was monitored. The system hosts 30 parallel stirred batches in 120 mL serum vials, crimp sealed with gas tight butyl rubber septa, which are monitored for headspace concentration of O2, N2, N2O, NO, CO2 and CH4 by frequent sampling. After each sampling, the system returns an equal volume of He, and elaborated routines are used to account for the gas loss by sampling to calculate the production/consumption-rate of each gas for each time interval between two samplings. More details are given in Supplementary Methods 2.
Enrichment culturing of N2O-respiring bacteria (NRB) in digestate was done as stirred (300 rmp) batches of 50 mL digestate per vial. Prior to incubation, the headspace air was replaced with Helium by repeated evacuation and He-filling [21] and supplemented with N2O. The N2O in the headspace was sustained by repeated injections in response to depletion. Liquid samples (1 mL) were taken by syringe, for metagenomic and metaproteomic analyses, and for quantification of volatile fatty acids (VFA) and 16S rRNA gene abundance. The samples were stored −80 °C before analyzed. The growth of NRB in the enrichments was modeled based on the N2O reduction kinetics. The modeling and the analytic methods (quantification of VFA and 16S rRNA gene abundance) are described in detail in Supplementary Methods 3.
Metagenomics and metaprotomics
Sequencing of DNA (Illumina HiSec4000), and the methods for Metagenome-Assembled Gemome (MAG) binning, and the phylogenetic placement of the MAGs is described in detail in Supplementary Methods 4. Proteins were extracted and digested to peptides, which were analyzed by nanoLC-MS/MS, and the acquired spectra were inspected, using the metagenome-assembled genomes (149 MAGs) as a scaffold (Supplementary Methods 5).
Isolation of N2O-respiring bacteria (NRB) (Supplementary Methods 6). NRB present in the enrichment cultures were isolated by spreading diluted samples on agar plates with different media composition, then incubated in an anaerobic atmosphere with N2O. Visible colonies were re-streaked and subsequently cultured under aerobic conditions, and 16s-sequenced. Three isolates, AS (Azospira sp), AN (Azonexus sp) and SP (Pseudomonas sp) (names based on their 16s sequence), were selected for genome sequencing, characterization of their denitrification phenotypes, and for testing their effect as N2O sinks in soil.
Genome sequencing and phenotyping of isolates
Three isolates were genome sequenced and compared with MAG’s of the enrichment culture (Supplementary Methods 7). The isolates’ ability to utilize various organic C substrates was tested on BiOLOG Phenotype MicroArray microtiter plates, and their characteristic regulation of denitrification was tested through a range of incubation experiments as in previous investigations [17, 18, 23, 24], by monitoring the kinetics of O2, N2, N2O, NO and NO2− throughout the cultures’ depletion of O2 and transition from aerobic to anaerobic respiration in stirred batch cultures with He + O2 (±N2O) in the headspace (Supplementary Methods 8). The kinetics of electron flow throughout the oxic and anoxic phase in these experiments were used to assess if the organisms were bet hedging, as demonstrated for Paracoccus denitrificans [17], i.e. that only a minority of cells express nitrate- and/or nitrite-reductase, while all express Nos, in response in response to oxygen depletion. Putative bet hedging was corroborated by measuring the abundance of nitrate-, nitrite- and nitrous oxide reductase (Supplementary Methods 9).
N2O mitigation experiments (Supplementary Methods 10). To assess the capacity of the isolates to reduce the N2O emission from soil, they were grown aerobically in sterilized digestate, which was then added to soil in microcosms, for measuring the NO-, N2O- and N2- kinetics of denitrification in the soil. For comparison, the experiments included soils amended with sterilized digestate, live digestate (no pretreatment), and digestate in which N2O-reducing bacteria had been enriched by anaerobic incubation with N2O (as for the initial enrichment culturing).
Results and discussion
Enrichment of indigenous nitrous oxide respiring bacteria (NRB) in digestates
We hypothesized that suitable organisms could be found in anaerobic digesters fed with sewage sludge, since such sludge contains a diverse community of denitrifying bacteria stemming from prior nitrification/denitrification steps [25]. We further hypothesized that these bacteria could be selectively enriched in digestates by anaerobic incubation with N2O. We decided to enrich at 20 °C, rather than at the temperatures of the anaerobic digesters (37 and 52 °C), to avoid selecting for organisms unable to grow within the normal temperature range of soils.
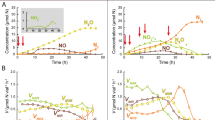
The digestates were incubated anaerobically as stirred batch cultures with N2O in the headspace (He atmosphere), and the activity and apparent growth of N2O reducers was assessed by monitoring the N2O-reduction to N2. Figure 2A shows the results for the first experiment, where liquid samples were taken at three time points (0, 115 and 325 h) for metagenomics, metaproteomics, and quantification of 16S rRNA gene copy abundance and volatile fatty acids (VFAs). N2O was periodically depleted (100–140 h) in this experiment, precluding detailed analysis of the growth kinetics throughout. This was avoided in the second enrichment, for which complete gas data are shown (Fig. 2B, C). Apart from the deviations caused by the temporary depletion of N2O in the first experiment, both experiments showed very similar N2 production rates (Fig. 2B and Supplementary Fig. S1B). The gas kinetics of the second enrichment are discussed in detail below.
Panel A shows results for the enrichment culture (triplicate culture vials) sampled for metagenomics, metaproteomics, quantification of volatile fatty acids (VFAs) and 16S rRNA gene abundance (sampling times = 0, 115 and 325 h). The top panel shows the amounts of N2 produced (mmol N2 L−1 digestate, log scale) and 16S rRNA gene copy numbers. The mid panel shows the concentration of N2O in the digestate (log scale), which was replenished by repeated injections (20 mL N2O, resulting in 5 mM N2O in the liquid) from t = 140 h and onwards (indicated by black arrows). The bottom panel shows the rate of methane production. Standard deviations (n = 3) are shown as vertical lines in all panels. Panels B and C show the results of a repeated enrichment experiment where N2O-depletion (as seen at t = 100–140 h in panel A) was avoided, to allow more precise assessment and modeling of growth kinetics. Panel B: N2O concentration in the digestate (mM N2O), rate of N2-production (VN2) and N2 produced (mmol N2 mL−1 digestate), all log scaled. The curved black line shows the modeled VN2 assuming two populations, one growing exponentially (µ = 0.1 h−1), and one whose activity was dying out gradually (rate = −0.03 h−1). The dotted black line is the activity of the exponentially growing population extrapolated to time = 0. Panel C shows the modeled density (cells mL−1) of cells growing by N2O respiration, extrapolated back to t = 0 h (dashed line), and the respiratory electron flow rate per cell (Ve-, fmol electrons cell−1 h−1), which declined gradually after 110 h. Standard deviations (n = 3) are shown as vertical lines. Supplementary Fig. S1 provides additional data for the experiment depicted in Panel A, as well as a detailed description of the modeling procedures and their results.
The rates of N2-production (VN2) declined during the first 50 h, followed by exponential increase (Fig. 2B). This was modeled as the activity of two groups of NRB, one growing exponentially from low initial abundance, and one which was more abundant initially, but whose activity declined gradually (further explained in Supplementary Fig. S1). The modeling indicated that the cell density of the growing NRB increased exponentially (specific growth rate, µ = 0.1 h−1) from a very low initial density (~2.5·103 cells mL−1) to 1.6·108 cells mL−1 after 110 h, and continued to increase at a gradually declining rate to reach ~3·109 cells mL−1 at the end of the incubation period (215 h). The modeled electron flow rate per cell for the growing NRB (cell specific e-flow, Ve-, Fig. 2C) was sustained at around 5 fmol e− cell−1 h−1 during the exponential growth, and declined gradually thereafter (note that the model assumes constant cell yield per mole electrons), as the number of cells continued to increase, while the overall rate of N2O-respiration remained more or less constant (VN2, Fig. 2B). Enrichment culturing was repeated several times, demonstrating that the characteristic N2 production kinetics was highly reproducible (Supplementary Figs. S2 and S14).
The provision of substrate for the N2O-respiring bacteria can be understood by considering the enrichment culture as a continuation of the metabolism of the anaerobic digester (AD), albeit slowed down by the lower temperature (20 °C versus 37 °C in the digester). In AD, organic polymers are degraded and converted to CO2 and CH4 through several steps, conducted by separate guilds of the methanogenic microbial consortium: 1) hydrolysis of polysaccharides to monomers by organisms with carbohydrate-active enzymes, 2) primary fermentation of the resulting monomers to volatile fatty acids (VFAs), 3) secondary fermentation of VFAs to acetate, H2 and CO2, and 4) methane production from acetate, CO2, H2, and methylated compounds. By providing N2O to this (anaerobic) system, organisms that respire N2O can tap into the existing flow of carbon, competing with the methanogenic consortium for intermediates, such as monomeric sugars, VFAs (such as acetate) and hydrogen [26]. Thus, the respiration and growth of the N2O-respiring bacteria is plausibly sustained by a flow of carbon for which the primary source is the depolymerization of organic polymers. It is possible that the retardation of growth after ~100 h of enrichment was due to carbon becoming limiting. Thus, at this point, the population of N2O-respiring organisms may have reached high enough cell densities to reap most of the intermediates produced by the consortium.
Parallel incubations of digestates without N2O confirmed the presence of an active methanogenic consortium, sustaining a methane production rate of ~0.2 µmol CH4 mL−1 h−1 throughout (Supplementary Fig. S3). Methane production was inhibited by N2O, and partly restored in periods when N2O was depleted (Fig. 2A, Supplementary Figs. S3 and S4). We also conducted parallel incubations with O2 and NO3− as electron acceptors. These incubations showed that methanogenesis was completely inhibited by NO3−, and partly inhibited by O2 (concentration in the liquid ranged from 20 to 90 µM O2) (Supplementary Fig. S3). The rates of O2 and NO3- reduction indicated that the digestate contained a much higher number of cells able to respire O2 and NO3− than cells able to respire N2O (Supplementary Fig. S5A–C). During the enrichment culturing with NO3− almost all reduced NO3−N appeared in the form of N2O-N during the first 50 h (Supplementary Fig. S5E), another piece of evidence that in the digestate, prior to enrichment culturing, the organisms reducing NO3− to N2O outnumbered those able to reduce N2O to N2. The measured production of CH4 and electron flows to electron acceptors deduced from measured gases (N2, O2 and CO2) were used to assess the effect of the three electron acceptors (N2O, NO3− and O2) on C-mineralization. While oxygen appeared to have a marginal effect, NO3− and N2O caused severe retardation of C-mineralization during the first 50 and 100 h, respectively (Supplementary Fig. S5A–D). This retarded mineralization is plausibly due to the inhibition of methanogenesis, causing a transient accumulation of H2 and VFAs until the N2O-reducing bacteria reach a cell density that allowed them to effectively reap these compounds. This was corroborated by measurements of H2 and VFAs (Supplementary Fig. S13).
To track the origin of the enriched N2O-respiring bacteria in the digestate, we considered the possibility that these are indigenous wastewater-sludge bacteria that survive the passage through the anaerobic digester, which had a retention time of 20–24 days. We assessed survival of N2O-respiring bacteria by comparing the N2O reduction potential of wastewater sludge and the digestate. The results indicated that ≤1/3 of the N2O-respiring bacteria in the sludge survived the passage (Supplementary Fig. S6). We also did enrichment culturing with a digestate from a thermophilic digester (52 °C) operated in parallel with the mesophilic digester (the same feed), and found that it contained N2O reducers that could be enriched, although the estimated initial numbers were orders of magnitude lower than in the mesophilic digestate (Supplementary Fig. S7).
MAG-centric metaproteomic analysis of the enrichment cultures
We analyzed the metagenome and metaproteome at three timepoints (0, 115 and 325 h) during the first enrichment culturing (gas kinetics and sampling times shown in Fig. 2A), to explore the effect of the anaerobic incubation with N2O on the entire microbial consortium, and to identify the organisms growing by N2O reduction. Metagenomic sequences were assembled and resultant contigs assigned to 278 metagenome-assembled genomes (MAGs), of which 149 were deemed to be of sufficient quality (completeness > 50% and contamination < 20%, Supplementary Data S1) for downstream analysis. The phylogenetic relationship and the relative abundance of the MAGs throughout the enrichment are summarized in Fig. 3, which also shows selected features revealed by the combined metagenomic and metaproteomic analyses, including information about genes and detected proteins involved in N2O reduction, other denitrification steps, methanogenesis, syntrophic acetate oxidation and methane oxidation.
Maximum likelihood tree indicating the phylogenetic placement of MAGs from the anaerobic enrichment. The tree was constructed from a concatenated set of protein sequences of single-copy genes. Taxonomic classification of the MAGs was inferred using the Genome Taxonomy Database (GTDB) and is displayed at the phylum level by label and branch coloring. Branch label decorations indicate the presence of genes involved in selected metabolic traits in the MAGs. The relative abundance of the MAG in the community as calculated from sequence coverage is indicated by bubbles at branch tips and bar charts indicate the number of detected proteins affiliated with each MAG at the three time points during incubation. Four of the 149 MAGs that met the completeness and contamination threshold for construction of the metaproteome database were lacking the universal single-copy marker genes and were omitted from the tree. Total protein counts per MAG were calculated by aggregating both secretome and cell-associated proteomes.
Closer inspections of the abundance of individual MAGs, based on their coverage in the metagenome and metaproteome, showed that the majority of the MAGs had a near constant population density throughout the incubation, while two MAGs (260 and 268) increased substantially (Fig. 4; further analyses in Supplementary Section B, Supplementary Figs. S8–S11). The stable abundance of the majority indicates that the methanogenic consortium remained intact despite the downshift in temperature (20 °C versus 37 °C) and the inhibition of methanogenesis by N2O. Only 9 MAGs showed a consistent decline in abundance throughout the enrichment (Supplementary Table S1). These MAGs could theoretically correspond to microbes whose metabolism is dependent on efficient H2 scavenging by methanogens [27], but we found no genomic evidence for this, and surmise that organisms circumscribed by the declining MAGs were unable to adapt to the temperature downshift from 37 °C to 20 °C.
Panel A shows the quality (completeness, strain heterogeneity and contamination), taxonomic classification based on GTDB and NCBI, presence of denitrifying genes and proteins, and the detected levels of Nos (N2O reductase, encoded by nosZ) throughout the enrichment culturing for the six MAGs containing the nosZ gene (Supplementary Data S1 and S2). Nos was detected in the proteome of five MAGs, but the detection level increased significantly throughout for only MAG260 and 248, respectively. None of the MAGs produced detectable amounts of the other denitrification reductases. (a) Lable Free Quantification (LFQ) values for one of the two detected predicted Nos proteins for MAG268 is shown. Panel B shows the apparent growth rates of the MAGs, based on their coverage in the metagenome and metaproteome (regression of ln(N) against time; see Supplementary Fig. S11 for more details). Panel C shows the taxonomic classification (16S rDNA), working names (abbreviations) and denitrification genotypes of three isolates from the enrichment culturing. The genes coding catalytic subunits of denitrification reductases are shown in bold, above the accessory genes [55] that were also identified. More information about accessory genes is presented in Supplementary Fig. S15. The isolate AN has 98.2% ANI to MAG260.
Six MAGs, including the two that were clearly growing (MAG260 & MAG268) contained the nosZ gene and thus had the genetic potential to produce N2O-reductase (Nos) (Fig. 4). Nos proteins originating from five of these MAGs were detected in the metaproteome. Importantly, while all but one of these MAGs contained genes encoding the other denitrification reductases, none of these were detected in the metaproteome, suggesting that the organisms can regulate the expression of their denitrification machinery to suit available electron acceptors, in this case N2O. Three of the MAGs with detectable Nos in the proteome (MAG004, MAG059, MAG248) appeared to be non-growing during the enrichment. The detected levels of their Nos proteins remained more or less constant, and their estimated abundance in the metagenome and -proteome did not increase (Fig. 4B). It is conceivable that these three MAGs belong to the initial population of N2O reducers whose N2O-reduction activity was present initially but gradually decreased during the early phase of the enrichment (Fig. 2A). The two growing MAGs (MAG260 and MAG268) showed increasing Nos levels and increasing abundance both in terms of coverage and metaproteomic detection (Fig. 4B), in proportion with the N2 produced (Supplementary Fig. S11). MAG260 reached the highest abundance of the two and accounted for 92% of the total detectable Nos pool at the final time point. MAG260 is taxonomically most closely affiliated with the genus Dechloromonas (GTDB, 97.9% amino acid similarity). Interestingly, Nap rather than Nar takes the role of nitrate reductase in MAG260 (Fig. 4), which makes it a promising organism for N2O mitigation since organisms with Nap only (lacking Nar) preferentially channel electrons to N2O rather than to NO3− [18]. MAG260, MAG004 and MAG088 contain a clade II nosZ, characterized by a sec-dependent signal peptide, in contrast to the more common tat-dependent clade I nosZ [16] The physiological implications of clade I versus clade II nosZ remains unclear. Organisms with nosZ Clade II have high growth yield and high affinity (low ks) for N2O, compared to those with nosZ Clade I [28], suggesting a key role of nosZ Clade II organisms for N2O reduction in soil, but this was contested by Conthe et al. [29], who found that Clade I organisms had higher “catalytic efficiency” (Vmax/ks) than those with Clade II.
The apparent inhibition of methanogenesis by N2O seen in the present study has been observed frequently [30] and is probably due to inhibition of coenzyme M methyltransferase [31], which is a membrane bound enzyme essential for methanogenesis and common to all methanogenic archaea [32]. The gas kinetics demonstrate that the inhibition was reversible, being partly restored whenever N2O was depleted (Fig. 2). In the enrichment culture where metagenomics and metaproteomics was monitored, several such incidents of N2O depletion occurred (Fig. 2A) and during these periods CH4 accumulated to levels amounting to 10% of levels in control vials without N2O (Supplementary Fig. S4B). These observations suggest that methanogens would be able to grow, albeit sporadically, during the enrichment, which is corroborated by the sustained detection of the complete methanogenesis pathway, including the crucial coenzyme M methyl-transferase, of Methanothrix (MAG025), Methanoregulaceae (MAG014) and Methanobacterium (MAG124) at high levels in the metaproteome. In fact, both MAG coverage data and 16S rRNA gene copy numbers assessed by ddPCR suggested that the majority of the original methanogenic consortium continued to grow (Supplementary Section B). A tentative map of the metabolic flow of the methanogenic consortium, including the reaping of intermediates (monosaccharides, fatty acids, acetate and H2) by N2O-respiring bacteria is shown in Supplementary Fig. S12. Since methane production was inhibited from the very beginning of the incubation, while it took ~100 h for the N2O-respiring bacteria to reach high enough numbers to become a significant sink for intermediates (Fig. 2), one would expect transient accumulation of volatile fatty acids and H2, which was corroborated by measurements of these metabolites (Supplementary Fig. S13).
Of note, we detected methane monooxygenase and methanol dehydrogenase proteins from MAG087 and MAG059, respectively, in the metaproteome. This opens up the tantalizing hypothesis of N2O-driven methane oxidation, a process only recently suggested to occur [33, 34]. However, a close inspection of the N2O- and CH4-kinetics indicated that N2O-driven methane oxidation played a minor role (Supplementary Fig. S4CD).
In a follow-up experiment, 7 parallel enrichment cultures were analyzed by 16S rRNA gene amplicon sequencing, demonstrating reproducibility of the selective enrichment of organisms circumscribed by MAG 260 (Fig. S14).
Isolation of N2O-respiring bacteria and their geno- and phenotyping
Whilst this enrichment culture could be used directly as a soil amendment, this approach is likely to have several disadvantages. First, it would require the use of large volumes of N2O for enrichment, a process which would be costly and require significant infrastructure. An alternative approach would be to introduce an axenic or mixed culture of digestate-derived, and likely digestate-adapted, N2O-respiring bacteria to sterilized/sanitized digestates. This approach has multiple benefits: (1) it would remove the need for N2O enrichment on site as isolates could be grown aerobically in the digestate material, (2) one could chose organisms with favorable denitrification genotypes and regulatory phenotypes, (3) the sanitation would eliminate the methanogenic consortium hence reducing the risk of methane emissions from anoxic micro-niches in the amended soil, and (4) sanitation of digestates aligns with current practices that require such a pretreatment prior to use for fertilization. For these reasons an isolation effort was undertaken to obtain suitable digestate-adapted N2O-respiring microorganisms from the N2O-enrichment cultures (Supplementary Material and Methods, Chapter 6). These efforts resulted in the recovery of three axenic N2O-respiring bacterial cultures, which were subjected to subsequent genomic and phenotypic characterization.
The isolates were phylogenetically assigned to Pseudomonas sp. (PS), Azospira sp. (AS) and Azonexus sp. (AN) (working names in bold) based on full length 16 S rRNA genes obtained from the sequenced genomes (accessions ERR4842639 − 40, Supplementary Table S2, phylogenetic trees shown in Supplementary Fig. S15). All were equipped with genes for a complete denitrification pathway (Fig. 4C). AN and AS carried napAB, encoding the periplasmic nitrate reductase (Nap) and nosZ clade II, whilst PS carried genes for the membrane bound nitrate reductase (Nar), encoded by narG, and nosZ clade I. All had nirS and norBC, coding for nitrite reductase (NirS) and nitric oxide reductase (Nor), respectively. Pairwise comparison of average nucleotide identities (ANI) with MAGs from the enrichment metagenomes showed that the isolate AN matched the Dechloromonas-affiliated MAG260 with 98.2% ANI, suggesting the isolate is circumscribed by MAG260 [35]. Given the GTDB phylogeny of AN and MAG260 and the 16S rRNA gene homology of AN (95.2% sequence identity to Azonexus hydrophilus DSM23864, Supplementary Fig. S15C), we conclude that AN likely represents a novel species within the Azonexus lineage. Unfortunately, the 16S rRNA gene was not recovered in MAG260, preventing direct comparison with related populations. No significant ANI matches in our MAG inventory were identified for the genomes of PS and AS indicating they were not captured via our metagenomic approaches, which highlights the complementarity of applying culture-dependent methods in parallel.
The carbon catabolism profiles of the isolates were assayed using Biolog PM1 and PM2 microplates, to screen the range of carbon sources utilized (Supplementary Section E: Supplementary Table S3). PS utilized a wide spectrum of carbon sources (amino acids, nucleic acids, volatile fatty acids (VFA), alcohols, sugar alcohols, monosaccharides and amino sugars), but only one polymer (laminarin). AN and AS could only utilize small VFAs (e.g. acetate, butyrate), intermediates in the TCA cycle and/or the β-oxidation/methyl malonyl-CoA pathways of fatty acid degradation (e.g. malate, fumarate, succinate), and a single amino acid (glutamate). Thus, all three would be able to grow in a live digestate by reaping the VFA’s produced by the methanogenic consortium. While the utilization of VFAs as C-substrates is one of several options for PS, AN and AS appear to depend on the provision of VFAs. This was confirmed by attempts to grow the three isolates in an autoclaved digestate: while PS grew well and reached high cell densities without any provision of extra carbon sources, AN and AS showed early retardation of growth unless provided with an extra dose of suitable carbon source (glutamate, acetate, pyruvate or ethanol) (Supplementary Figs. S26 and S27). A high degree of specialization and metabolic streamlining may thus explain the observed dominance of AN (MAG260) during enrichment culturing.
To evaluate the potentials of these isolates to act as sinks for N2O, we characterized their denitrification phenotypes, by monitoring kinetics of oxygen depletion, subsequent denitrification and transient accumulation of denitrification intermediates (NO2−, NO, N2O). The experiments were designed to assess properties associated with strong N2O reduction such as 1) bet hedging, i.e. that all cells express N2O reductase while only a fraction of the cells express nitrite- and/or nitrate-reductase, as demonstrated for Paracoccus denitrificans [17]; 2) strong metabolic preference for N2O-reduction over NO3− -reduction, as demonstrated for organisms with periplasmic nitrate reductase [18]. Supplementary section F (Supplementary Figs. S16–S25) provides the results of all the experiments and a synopsis of the findings. In short: Azonexus sp. (AN) had a clear preference for N2O over NO3− reduction, but not over NO2− reduction, ascribed to bet hedging with respect to the expression of nitrate reductase (a few cells express Nap, while all cells express Nos), which was corroborated by proteomics: the Nos/Nap abundance ratio was ~25 during the initial phase of denitrification (Supplementary Fig. S18). Azospira sp. (AS) had a similar preference for N2O over NO3− reduction, albeit less pronounced than in AN, and no preference for N2O over NO2−. Pseudomonas sp. (PS) showed a phenotype resembling that of Paracoccus denitrificans [17], with denitrification kinetics indicating that Nir is expressed in a minority of cells in response to O2 depletion, while all cells appeared to express N2O reductase. This regulation makes PS a more robust sink for N2O than the two other isolates, since it kept N2O extremely low even when provided with NO2−.
In summary, PS appeared to be the most robust candidate as a sink for N2O in soil for two reasons; 1) it can utilize a wide range of carbon substrates, and 2) its N2O sink strength is independent of the type of nitrogen oxyanion present (NO2− or NO3−). In contrast, AN and AS appear to be streamlined for harvesting intermediates produced by anaerobic consortia, hence their metabolic activity in soil could be limited. In addition, they could be sources rather than sinks for N2O if provided with NO2−, which is likely to happen in soils, at least in soils of neutral pH, during hypoxic/anoxic spells [36].
Effects on N2O emissions
To assess if fertilization with digestates containing N2O-reducing bacteria could reduce N2O emissions from denitrification in soil, we conducted a series of incubation experiments with soils fertilized with digestates with and without N2O-reducing bacteria. The fertilized soils were incubated in closed culture vials containing He + 0.5 vol % O2, and O2, NO, N2O and N2 were monitored during oxygen depletion and subsequent denitrification. The experiments included soils amended with digestates in which indigenous N2O-reducing bacteria had been enriched by anaerobic incubation with N2O (Fig. 2), as well as autoclaved digestates in which the isolates from the current study had been grown by aerobic cultivation (see Supplementary Figs. S26 and S27 for cultivation details). The experiments included three types of control digestates: digestate (directly from the digester), digestate heated to 70 °C for 2 h (to eliminate most of the indigenous consortium), and autoclaved digestate in which the strain PS had been grown aerobically and then heated to 70 °C for 2 h, to kill PS. In all cases, 3 mL of digestate was added to 10 g of soil. Since soil acidity has a pervasive effect on the synthesis of functional N2O reductase [24], we tested the digestates with two soils from a liming experiment [37] with different pH (pHCaCl2 = 5.5 and 6.6).
The transient N2O accumulation during denitrification was generally higher in the acid than in the near-neutral soil (Fig. 5), which was expected since the synthesis of functional Nos is hampered by low pH [23, 24]. Based on the kinetics of both N2 and N2O (see Supplementary Figs. S28 and S29), we calculated the N2O-index (IN2O) which is a measure of the molar amounts of N2O relative to N2 + N2O in the headspace for a specific period (0-T), see equation at top of Fig. 5). Low values of IN2O indicate efficient N2O-reduction. In this case, we calculated IN2O for the incubation period until 40% of the available NO3− had been recovered as N2 + N2O (=IN2O 40) and for the incubation period until 100% was recovered (IN2O 100).Statistical analyses showed significant effects of digestate treatments and soil pH, and the interaction between the two (p < 0.001).
N2O kinetics during incubation of soils amended with seven different digestates and a control (soil only); panel A: pH 5.5 soil, panel B: pH 6.6 soil. The digestates treatments are: Soil = soil without any amendment; Digestate = digestate directly from the anaerobic digester; Dig_70 °C = digestate heat treated to 70 °C for 2 h; PS, AN and AS = autoclaved digestate in which isolates PS, AN and AS (respectively) had been grown aerobically (see Supplementary Figs. S26 and S27); PS_70 °C = digestate in which PS had been grown (as for the PS treatment), subsequently heated to 70 °C for 2 h, N2O enr. = digestate in which indigenous N2O-respiring bacteria had been enriched (as shown in Fig. 2). The left panels show measured N2O throughout soil incubations; the insets with altered scaling show N2O levels for treatments that accumulated very little N2O. The bar graphs to the right show the N2O indexes (IN2O) expressed as % (equation shown in the panel), with confidential intervals (ANOVA + Tukey’s range test). IN2O is a proxy for potential N2O emission from denitrification in soil [56]. Two IN2O values are shown: one for the timespan until 40% of the NO3− -N was recovered as N2 + N2O + NO (IN2O 40), and one for 100% recovery (IN2O 100). More details (including N2 and NO kinetics) are shown in Supplementary Figs. S28 and S29.
Extremely low IN2O values were recorded for the treatments with digestate in which N2O-reducing bacteria were enriched by anaerobic incubation with N2O, even in the acid soil. This is in line with the current understanding of how pH affects N2O-reduction: low pH slows down the synthesis of functional Nos, but once synthesized, it remains functional even at low pH [23]. Functional Nos had already been expressed during the enrichment and was evidently activeafter amendment to the soils.
The presence of the isolates in the digestates had clear but variable effects on IN2O. The most relevant control-treatment for evaluating the effect of the isolates would be PS_70 °C, because this digestate had been treated exactly the same way as that with metabolically active isolates present (autoclaved, aerated and aerobic cultivation). These contrasts (IN2O for PS_70 °C versus PS, AS and AN) were all statistically significant (confidential intervals did not overlap), thus all strains reduced IN2O compared to PS_70 °C. AN and AS resulted in much higher IN2O in the acid than in the neutral, suggesting that the expression of functional N2O reductase in these strains was hampered by low pH. In contrast to AN and AS, PS resulted in very low IN2O values in both soils, suggesting that this organism has an exceptional capacity to synthesize functional Nos at low pH.
These results show that the emission of N2O from soil fertilized with digestates can be manipulated by tailoring the digestate microbiome. Interestingly, measurements of methane in these soil incubations showed that the methanogenic consortia in digestates that had not been heat treated (i.e. the live digestate and the N2O enrichment) remained metabolically intact in the soil, and started producing methane as soon as N2O and nitrogen oxyanions had been depleted, while no methane was produced in the soils amended with autoclaved digestate, and that heated to 70 °C (Supplementary Fig. S30).
In an effort to determine the survival of the N2O-scavenging capacity of a digestate enriched with N2O reducers, we also tested its effect on soil N2O emissions after a 70-h aerobic storage period (in soil or as enrichment culture, at 20 °C). These experiments demonstrated a sustained beneficial effect on IN2O after 70 h of aerobic storage (Supplementary Fig. S31). This result indicates that the enrichment strategies discussed here are robust, although long-lasting storage experiments as well as field trials are needed.
Concluding remarks
This feasibility study identifies an avenue for large scale cultivation of N2O reducers for soil application, which could be low cost if implemented as an add-on to biogas production systems. Further efforts should be directed towards selecting organisms that are both strong sinks for N2O and able to survive and compete in soil, to secure long-lasting effects on N2O emissions. A tantalizing added value would be provided by selecting organisms (or consortia of organisms) that are not only strong N2O-sinks, but also promote plant growth and disease resistance [38, 39].
Gas kinetics, metagenomics and metaproteomics revealed that the methanogenic consortium of the digestate remains active during anaerobic incubation with N2O, and that bacteria with an anaerobic respiratory metabolism grew by harvesting fermentation intermediates. The inhibition of methanogenesis by N2O implies that the respiring organisms would have immediate access to the electron donors that would otherwise be used by the methanogens, i.e. acetate and H2, while they would have to compete with fermentative organisms for the “earlier” intermediates such as alcohols and VFA. The importance of fermentation intermediates as a carbon source for the N2O-respiring bacteria would predict a selective advantage for organisms with a streamlined (narrow) catabolic capacity, i.e. limited to short fatty acids, and our results lend some support to this: the catabolic capacity of the organism that became dominant (MAG260, isolate AN) was indeed limited, as was also the case for isolate AS. Such organisms are probably not ideal N2O-sinks in soil because their ability to survive in this environment would be limited. Organisms with a wider catabolic capacity, such as the isolated Pseudomonas sp. (PS), are stronger candidates for long term survival and N2O-reducing activity in soil. The ideal organisms are probably yet to be found, however, and refinements of the enrichment culturing process are clearly needed.
The digestate used in this study contained N2O-respiring bacteria, most likely survivors from the raw sludge, which however, were clearly outnumbered by bacteria that are net producers of N2O. We surmise that the relative amounts of N2O-producers and N2O-reducers in digestates may vary, depending on the feeding material and configuration for the anaerobic digestion. This could explain the observed variable effects of digestates on N2O emission from soils [40, 41]. The high abundance of both NO3− - and O2-respiring organisms in digestates has practical implications for the attempts to grow isolated strains in digestates: they could be outnumbered by the indigenous NO3− - and O2-respiring organisms (Supplementary Fig. S5). Hence, we foresee that future implementation of this strategy will require a brief heat treatment or other sanitizing procedure. A bonus of such sanitation is that it eliminates methane production by the digestate in soil.
We failed to enrich organisms lacking all other denitrification genes than nosZ; the only reconstructed genome with nosZ only (MAG004) did not grow at all. Failure to selectively enrich such organisms by anaerobic incubation with N2O was also experienced by Conthe et al. [29]. The organisms that did grow by respiring N2O in our enrichment, were all equipped with genes for the full denitrification pathway, although the only denitrification enzyme expressed/detected during the enrichment was Nos. This agrees with the current understanding of the gene regulatory network of denitrification; nosZ is the only gene whose transcription does not depend on the presence of NO3−, NO2− or NO [42], which were all absent during the enrichment.
Two of the reconstructed MAGs had genes encoding periplasmic nitrate reductase (nap), as was the case for two of the three isolates (AN and AS). This in itself would predict preference for N2O- over NO3− reduction at a metabolic level [43], but otherwise their potential for being N2O sinks cannot be predicted by their genomes. The phenotyping of the isolates revealed conspicuous patterns of bet hedging as demonstrated for Paracoccus denitricans [17]. The bet hedging in P. denitrificans is characterized by expression of Nir (and Nor) in a minority of the cells, while Nos is expressed in all cells, in response to oxygen depletion, hence the population as a whole is a strong sink for N2O. The isolated Pseudomonas sp. (PS) displayed denitrification kinetics that closely resembles that of P. denitrificans. The two other isolates (AN and AS) showed indications of bet hedging as well, but of another sort: Nap appears to be expressed in a minority of the cells. This different regulatory phenotype had clear implications for the ability of organisms to function as N2O-sinks: while all isolates were strong N2O sinks when provided with NO3− only, AN and AS accumulated large amounts of N2O if provided with NO2−.
The N2O sink capacity of the organisms was tested by fertilizing soils with digestates with and without the organisms, and monitoring the gas kinetics in response to oxygen depletion, thus imitating the hot spots/hot moments of hypoxia/anoxia [44]. Since the isolates were raised by aerobic growth in autoclaved digestates, they would have to synthesize all denitrification enzymes in the soil, hence the synthesis of functional Nos was expected to be hampered by low pH [24]. The results for isolates AS and AN lend support to this (high IN2O in the soil with pH = 5.5). AN was also dominating in the digestate enrichment culture, and in this case the organism had a strong and pH-independent effect on N2O emission, plausibly due synthesis of Nos prior to incorporation into the soils.
In summary, we have demonstrated that a digestate from biogas production can be transformed into an effective agent for mitigating N2O emission from soil, simply by allowing the right bacteria to grow to high cell densities in the digestate prior to fertilization. The technique is attractive because it can be integrated in existing biogas production systems, and hence is scalable. If we manage to treat a major part of waste materials in agroecosystems by AD, the resulting digestates would suffice to treat a large share of total farmland, as illustrated by Fig. 1. Estimation of the potential N2O-mitigation effect is premature, but the documented feasibility and the scalability of the approach warrant further refinement as well as rigorous testing under field condition. Our approach suggests one avenue for a much needed valorization of organic wastes [45] via anaerobic digestion. Future developments of this approach could extend beyond the scope of climate change mitigation and include the enrichment of microbes for pesticide- and other organic pollutant degradation [46], plant growth promotion [47] and inoculation of other plant symbiotic bacteria [48].
Data availability
The sequencing data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB41283 (isolates AN, AS and PS) and PRJEB41816 (metagenome) (https://www.ebi.ac.uk/ena/browser/view/PRJEBxxxx). Functionally annotated MAGs and metagenomic assembly are available in FigShare (https://doi.org/10.6084/m9.figshare.13102451 and https://doi.org/10.6084/m9.figshare.13102493). The proteomics data has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [49] with the dataset identifier PXD022030* and PXD023233** for the metaproteome and proteome of Azonexus sp. AN, respectively.
* Reviewer access: Username: reviewer_pxd022030@ebi.ac.uk. Password: GdTR3biE
** Reviewers access: Username: reviewer_pxd023233@ebi.ac.uk Password: nMz62S8O
References
Tian H, Xu R, Canadell JG, Thompson RL, Winiwarter W, Suntharalingam P, et al. A comprehensive quantification of global nitrous oxide sources and sinks. Nature. 2020;586:248–56.
Snyder CS, Davidson EA, Smith P, Venterea RT. Agriculture: sustainable crop and animal production to help mitigate nitrous oxide emissions. Curr Opin Environ Sustain. 2014;9–10:46–54.
Sutton MA, Oenema O, Erisman JW, Leip A, Van Grinsven H, Winiwarter W. Too much of a good thing. Nature. 2011;472:159–61.
Erisman JW, Sutton MA, Galloway J, Klimont Z, Winiwarter W. How a century of ammonia synthesis changed the world. Nat Geosci. 2008;1:636–9.
D’Hondt K, Kostic T, McDowell R, Eudes F, Singh BK, Sarkar S, et al. Microbiome innovations for a sustainable future. Nat Microbiol. 2021;6:138–42.
Bakken LR, Frostegård Å. Emerging options for mitigating N2O emissions from food production by manipulating the soil microbiota. Curr Opin Environ Sustain. 2020;47:89–94.
Butterbach-Bahl K, Baggs EM, Dannenmann M, Kiese R, Zechmeister-Boltenstern S. Nitrous oxide emissions from soils: how well do we understand the processes and their controls? Philos Trans R Soc B Biol Sci. 2013;368:13.
Hu HW, Chen D, He JZ. Microbial regulation of terrestrial nitrous oxide formation: understanding the biological pathways for prediction of emission rates. FEMS Microbiol Rev. 2015;39:729–49.
Prieme A, Christensen S. Natural perturbations, drying–wetting and freezing–thawing cycles, and the emission of nitrous oxide, carbon dioxide and methane from farmed organic soils. Soil Biol Biochem. 2001;33:2083–91.
Song X, Liu M, Ju X, Gao B, Su F, Chen X, et al. Nitrous oxide emissions increase exponentially when optimum nitrogen fertilizer rates are exceeded in the North China plain. Environ Sci Technol. 2018;52:12504–13.
Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616.
Qu Z, Bakken LR, Molstad L, Frostegård Å, Bergaust LL. Transcriptional and metabolic regulation of denitrification in P aracoccus denitrificans allows low but significant activity of nitrous oxide reductase under oxic conditions. Environ Microbiol. 2016;18:2951–63.
Shapleigh JP. Denitrifying prokaryotes. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (editors). Prokaryotes - prokaryotic physiology and biochemistry. Springer (New York): 2013; p 405–25.
Lycus P, Bøthun KL, Bergaust L, Shapleigh JP, Bakken LR, Frostegård Å. Phenotypic and genotypic richness of denitrifiers revealed by a novel isolation strategy. ISME J. 2017;11:2219–32.
Sanford RA, Wagner DD, Wu Q, Chee-Sanford JC, Thomas SH, Cruz-García C, et al. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc Natl Acad Sci USA. 2012;109:19709–14.
Hallin S, Philippot L, Löffler FE, Sanford RA, Jones CM. Genomics and ecology of novel N2O-reducing microorganisms. Trends Microbiol. 2018;26:43–55.
Lycus P, Soriano-Laguna MJ, Kjos M, Richardson DJ, Gates AJ, Milligan DA, et al. A bet-hedging strategy for denitrifying bacteria curtails their release of N2O. Proc Natl Acad Sci USA 2018;115:11820–5.
Mania D, Woliy K, Degefu T. Frostegård åsa. A common mechanism for efficient N2O reduction in diverse isolates of nodule‐forming bradyrhizobia. Environ Microbiol. 2020;22:17–31.
Domeignoz-Horta LA, Putz M, Spor A, Bru D, Breuil MC, Hallin S, et al. Non-denitrifying nitrous oxide-reducing bacteria - an effective N2O sink in soil. Soil Biol Biochem. 2016;103:376–9.
Scarlat N, Dallemand JF, Fahl F. Biogas: developments and perspectives in Europe. Renew Energy 2018;129:457–72.
Molstad L, Dörsch P, Bakken LR. Robotized incubation system for monitoring gases (O2, NO, N2O and N2) in denitrifying cultures. J Microbiol Methods. 2007;71:202–11.
Molstad L, Dörsch P, Bakken LR. Improved robotized incubation system for gas kinetics in batch cultures. Researchgate. 2016. https://doi.org/10.13140/RG.2.2.30688.07680.
Bergaust L, Mao Y, Bakken LR, Frostegård Å. Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal ph on nitrogen oxide reductase in paracoccus denitrificans. Appl Environ Microbiol. 2010;76:6387–96.
Liu B, Frostegård Å, Bakken LR. Impaired reduction of N2O to N2 in acid soils is due to a posttranscriptional interference with the expression of nosZ. MBio. 2014;5:e01383–14.
Lu H, Chandran K, Stensel D. Microbial ecology of denitrification in biological wastewater treatment. Water Res. 2014;64:237–54.
Stams AJ, Oude Elferink SJ, Westermann P. Metabolic interactions between methanogenic consortia and anaerobic respiring bacteria. Adv Biochem Eng Biotechnol. 2003;81:31–56.
Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61:262–80.
Yoon S, Nissen S, Park D, Sanford RA, Löffler FE. Nitrous oxide reduction kinetics distinguish bacteria harboring clade I NosZ from those harboring clade II NosZ. Appl Environ Microbiol. 2016;82:3793–3800.
Conthe M, Wittorf L, Kuenen JG, Kleerebezem R, Van Loosdrecht MCM, Hallin S. Life on N2O: Deciphering the ecophysiology of N2O respiring bacterial communities in a continuous culture. ISME J. 2018;12:1142–53.
Andalib M, Nakhla G, McIntee E, Zhu J. Simultaneous denitrification and methanogenesis (SDM): review of two decades of research. Desalination. 2011;279:1–14.
Kengen SWM, Mosterd JJ, Nelissen RLH, Keltjens JT, van der Drift C, Vogels GD. Reductive activation of the methyl-tetrahydromethanopterin: coenzyme M methyltransferase from Methanobacterium thermoautotrophicum strain ΔH. Arch Microbiol. 1988;150:405–12.
Fischer R, Gärtner P, Yeliseev A, Thauer RK. N5-Methyltetrahydromethanopterin: coenzyme M methyltransferase in methanogenic archaebacteria is a membrane protein. Arch Microbiol. 1992;158:208–17.
Valenzuela EI, Padilla-Loma C, Gómez-Hernández N, López-Lozano NE, Casas-Flores S, Cervantes FJ. Humic substances mediate anaerobic methane oxidation linked to nitrous oxide reduction in wetland sediments. Front Microbiol. 2020;11:587.
Cheng C, Shen X, Xie H, Hu Z, Pavlostathis SG, Zhang J. Coupled methane and nitrous oxide biotransformation in freshwater wetland sediment microcosms. Sci Total Environ. 2019;648:916–22.
Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–31.
Lim NYN, Frostegård Å, Bakken LR. Nitrite kinetics during anoxia: the role of abiotic reactions versus microbial reduction. Soil Biol Biochem. 2018;119:203–9.
Nadeem S, Bakken LR, Frostegård Å, Gaby JC, Dörsch P. Contingent effects of liming on N2O-emissions driven by autotrophic nitrification. Front Env Sci. 2020;8:598513.
Gao N, Shen W, Camargo E, Shiratori Y, Nishizawa T, Isobe K, et al. Nitrous oxide (N2O)-reducing denitrifier-inoculated organic fertilizer mitigates N2O emissions from agricultural soils. Biol Fertil Soils. 2017;53:885–98.
Gao N, Shen W, Kakuta H, Tanaka N, Fujiwara T, Nishizawa T, et al. Inoculation with nitrous oxide (N2O)-reducing denitrifier strains simultaneously mitigates N2O emission from pasture soil and promotes growth of pasture plants. Soil Biol Biochem. 2016;97:83–91.
Baral KR, Labouriau R, Olesen JE, Petersen SO. Nitrous oxide emissions and nitrogen use efficiency of manure and digestates applied to spring barley. Agric Ecosyst Environ. 2017;239:188–98.
Herrero M, Henderson B, Havlík P, Thornton PK, Conant RT, Smith P, et al. Greenhouse gas mitigation potentials in the livestock sector. Nat Clim Chang. 2016;6:452–61.
Spiro S. Regulation of denitrification, (Chapter 13). In Isabel M, José JGM, Sofia RP, Luisa BM (editors). RSC metallobiology series 9: metalloenzymes in denitrification: applications and environmental impacts. Cambridge: The Royal Society of Chemistry; 2017. p. 312–31.
Gao Y, Mania D, Mousavi SA, Lycus P, Arntzen M, Wolly K, et al. Competition for electrons favors N2O reduction in denitrifying Bradyrhizobium isolates. Env Microbiol. 2021;23:223–59.
Kuzyakov Y, Blagodatskaya E. Microbial hotspots and hot moments in soil: concept & review. Soil Biol Biochem. 2015;83:184–99.
Peng W, Pivato A. Sustainable management of digestate from the organic fraction of municipal solid waste and food waste under the concepts of back to earth alternatives and circular economy. Waste Biomass Valoriz. 2019;10:465–81.
Sun S, Sidhu V, Rong Y, Zheng Y. Pesticide pollution in agricultural soils and sustainable remediation. Methods: a Rev Curr Pollut Rep. 2018;4:240–50.
Backer R, Rokem JS, Ilangumaran G, Lamont J, Praslickova D, Ricci E, et al. Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci. 2018;871:1473.
Poole P, Ramachandran V, Terpolilli J. Rhizobia: from saprophytes to endosymbionts. Nat Rev Microbiol. 2018;16:291–303.
Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, et al. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:1063–9.
Foged R, Lyngsø H, Flotats X, August B, Blasi J, Palatsi A, et al. Inventory of manure processing activities in Europe. Technical report No. I concerning “Manure Processing Activities in Europe” to the European Commission, Directorate-General Environment. 2011;138. Publications Office of the European Union, Brussels.
Holm-Nielsen JB, Al Seadi T, Oleskowicz-Popiel P. The future of anaerobic digestion and biogas utilization. Bioresour Technol. 2009;100:5478–84.
Stenmarck. Estimates of European food waste levels, Report of the project FUSIONS (contract number: 311972) granted by the European Commission (FP7). https://doi.org/10.13140/RG.2.1.4658.4721.
Meyer AKP, Ehimen EA, Holm-Nielsen JB. Future European biogas: animal manure, straw and grass potentials for a sustainable European biogas production. Biomass Bioenergy. 2018;111:154–64.
Eurostat (2017) Agri-environmental indicator – greenhouse gas emissions. ISSN 2443-8219, https://ec.europa.eu/eurostat/statistics-explained/pdfscache/16817.pdf.
Vaccaro BJ, Thorgersen MP, Lancaster WA, Price MN, Wetmore KM, Poole FL, et al. Determining roles of accessory genes in denitrification by mutant fitness analyses. Appl Environ Microbiol. 2016;82:51–61.
Russenes AL, Korsaeth A, Bakken LR, Dörsch P. Spatial variation in soil pH controls off-season N2O emission in an agricultural soil. Soil Biol Biochem. 2016;99:36–46.
Funding
The work was funded by The Research Council of Norway through project no. 286888 (New generation biofertilizers for improved nitrogen management, sustainable food production and low greenhouse gas emissions); and project no. 260868 (Optimization of biogas production and stabilization of sludge).
Author information
Authors and Affiliations
Contributions
KRJ conducted the research, analyzed and interpreted the data, and wrote the manuscript, LHH conducted genomic and proteomic analyses of the enrichments, SHWV conducted the research and wrote the manuscript, MA contributed to analysis of the genomics and proteomics, VE and ÅF contributed to the writing of the manuscript, PL did proteomics on the isolate, LM was responsible for gas measurements and kinetics analyses, PBP contributed to the analysis of genomics and proteomics and to the writing of the manuscript, LRB designed the experiment, analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jonassen, K.R., Hagen, L.H., Vick, S.H.W. et al. Nitrous oxide respiring bacteria in biogas digestates for reduced agricultural emissions. ISME J 16, 580–590 (2022). https://doi.org/10.1038/s41396-021-01101-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-01101-x