Abstract
Cyanate is utilized by many microbes as an organic nitrogen source. The key enzyme for cyanate metabolism is cyanase, converting cyanate to ammonium and carbon dioxide. Although the cyanase gene cynS has been identified in many species, the diversity, prevalence, and expression of cynS in marine microbial communities remains poorly understood. Here, based on the full-length cDNA sequence of a dinoflagellate cynS and 260 homologs across the tree of life, we extend the conserved nature of cyanases by the identification of additional ultra-conserved residues as part of the modeled holoenzyme structure. Our phylogenetic analysis showed that horizontal gene transfer of cynS appears to be more prominent than previously reported for bacteria, archaea, chlorophytes, and metazoans. Quantitative analyses of marine planktonic metagenomes revealed that cynS is as prevalent as ureC (urease subunit alpha), suggesting that cyanate plays an important role in nitrogen metabolism of marine microbes. Highly abundant cynS transcripts from phytoplankton and nitrite-oxidizing bacteria identified in global ocean metatranscriptomes indicate that cyanases potentially occupy a key position in the marine nitrogen cycle by facilitating photosynthetic assimilation of organic N and its remineralisation to NO3 by the activity of nitrifying bacteria.
Similar content being viewed by others
Cyanate (OCN−) is an oxidation product of cyanide and a decomposition product of urea [1, 2]. It is considered as an organic nitrogen source for diverse prokaryotic and eukaryotic microbes in terrestrial and aquatic ecosystems [2,3,4,5,6,7] with concentrations in the nanomolar range [4, 7,8,9]. Cyanate is also formed intracellularly from urea and carbamoyl phosphate, making it part of the central nitrogen metabolism [10,11,12]. In spite of the central metabolic role of cyanate, it has received much less attention than other organic nitrogen compounds, particularly for marine environments. However, it has been found that cyanate is likely an essential N source for cyanobacteria in oligotrophic oceans [2, 6, 13, 14] and an alternate N substrate for marine nitrification and anammox [15,16,17].
Cyanate metabolism relies on the well-characterized enzyme cyanase, which catalyzes the reaction of cyanate with bicarbonate to produce ammonium and carbon dioxide [18]. The cyanase gene cynS has been identified in many terrestrial and aquatic species and was reported to play a significant role in the assimilation of exogenous cyanate and detoxification of internally generated cyanate [6, 13, 16, 19,20,21,22,23,24,25,26,27,28]. However, knowledge on the diversity and evolution of cynS in marine microbes is rather limited, including its prevalence in the oceanic system.
To address this knowledge gap, we retrieved 260 cynS homologs across the tree of life (Table S1) based on the full-length cDNA from the marine dinoflagellate Alexandrium pacificum (APcynS, 653 bp, GenBank accession number: MZ666876) (Fig. S1; Table S2) and its deduced amino acid sequence (APcyanase). This reference dataset was used to query marine metagenomes and metatranscriptomes using the Ocean Gene Atlas (OGA) [29] to explore the biogeography and in situ expression pattern of cynS (full methods were described in supplements).
An amino-acid alignment composed of 260 homologs revealed the presence of nine ultra-conserved residues (Fig. 1a), potentially responsible for the catalytic activity and structural stability. Six of them have not been documented before [25, 30,31,32]. Modeling of the 3D enzyme structure indicated the following subunit organization: a decameric holoenzyme with a core formed by five dimers [30, 31] (Fig. 1b, f; Fig. S2). Five active sites are located between dimers forming an inner ring (Fig. 1c, g), non-covalently bound with five oxalate di-anions (Fig. 1d, e, h, i; Table S3), indicating the possible binding sites of cyanate. In E. coli, there are four types of residue-oxalate interactions known for binding cyanate (Fig. 1j) [30]. However, only type 1 is present in modeled APcyanase (Fig. 1j), suggesting reduced plasticity in binding cyanate. Whether this structural variation translates into different binding affinities and therefore potentially physiological roles of the APcyanase remains to be seen.
a Alignment of the catalytic domain in cyanases from representative species. Numbers in parentheses refer to the sequence ID from the full list (Table S1). b The front view of the decameric cyanase from the dinoflagellate Alexandrium pacificum. Alpha-helix and beta-hairpin is shown in purple and green, respectively. Ten monomers are labeled as chains A-J. c Overall location of the five active sites. d, e Enlarged views of the chain interactions and residues-oxalate anion interactions. Ball-and-stick colored in red and gray denotes oxalate anions. f Structures flipped 90° clockwise around the Y-axis shows the view of two dimer pairs. g Side view of one active site. h, i Enlarged views of proposed catalytic residues. j Four types of residue-oxalate anion interactions in cyanases. k Unrooted maximum likelihood tree of cyanases. Only bootstrap values ≥60% were shown. Lineages are color-coded and marine species are labeled with triangles.
Four major clades of cynases were identified based on their phylogenic relationships (Fig. 1k, Fig. S3). Interestingly, horizontal gene transfer (HGT) of cynS contributed to the evolution of bacteria, archaea, and eukaryotes including microalgae (Bathycoccus, Micromonas) and metazoans, which provides evidence that HGT of cynS is more common than previously documented [3, 6, 13, 16, 27, 33,34,35].
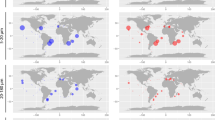
To contextualize cyanate metabolism in the upper ocean from the surface (epipelagic) down to the intermediate depths of ca. 1000 m (mesopelagic (MES)), we analyzed the prevalence and expression of cynS in comparison to ureC, the gene encoding the urease subunit alpha. The urea cycle, unlike cyanate metabolism, is well studied in many marine microbes including the acquisition of urea as an organic nitrogen source. Homologs of both genes and their corresponding transcripts could be retrieved from almost all sampling stations of the OGA (Table S4), which suggests their overall prevalence in many marine microbes. However, the normalized gene activity of cynS and ureC differed depending on the size class, taxonomic group and the water depth (Fig. 2; Fig. S4). Interestingly, the transcriptional activity of ureC appears to be much lower compared to cynS in the larger size fraction (0.8–2000 μm) mostly representing eukaryotic microbes and for both, surface (SRF) and deep chlorophyll maximum (DCM) (Fig. 2a, b, f, g). Pelagophytes, dinophytes, bacillariophytes, and fungi contributed the most cynS transcripts in the epipelagic ocean with pelagophyte transcripts dominating the surface layer (Fig. 2a, b). Transcripts for both genes in the smaller size fraction (0.22–3 μm) were mostly derived from prokaryotes (Figs. 2c–e, 2h-j). In the surface ocean, Synechococcus contributed most of the cynS transcripts in non-polar oceans whereas picochlorophyte cynS transcripts were most dominant in the coastal Arctic (Fig. 2c). In contrast, proteobacteria together with unclassified microbes contributed most of the ureC transcripts in surface ocean metatranscriptomes regardless of geography (Fig. 2h). The taxonomic contributions of the ureC transcripts did not change much for the DCM and not even the MES zone although Gammaproteobacteria appear to have contributed more ureC transcripts in the MES compared to the epipelagic (Fig. 2i, j). By comparison, unclassified microbes together with Prochlorococcus contributed more cynS transcripts in the DCM (Fig. 2d). For the MES, most of the cynS transcripts were contributed by unclassified microbes, Nitrospinae and Proteobacteria (Fig. 2e).
The abundance of cynS transcripts from most prokaryotic and eukaryotic phytoplankton was negatively correlated (p < 0.05) with dissolved inorganic nitrogen concentrations (Fig. S5a, b; Table S5a, b). The latitudinal differences in the taxonomic contributions of cynS correlated negatively with temperature (p < 0.05), suggesting that cyanases are induced in picochlorophytes by low temperature. As no Arctic samples were available for the larger size fraction (0.8–2000 μm) representing mainly eukaryotes, it remains elusive if this group of organisms does have similar cynS expression patterns under polar conditions.
Although much less cynS transcripts were detected in the MES, the contribution of Nitrospinae was more significant compared to the epipelagic (Fig. 2e). Members of the phylum Nitrospinae are known to be the most abundant nitrite-oxidizing bacteria (NOB) in the oceans with an important role in dark-ocean carbon fixation [3, 16]. Cyanate metabolism of NOBs is common and essential for the global nitrogen cycle, supplying ammonia oxidizers with ammonium, which is nitrified by this nitrifying consortium including NOBs [3]. In our study, cynS transcripts from Nitrospinae in the epipelagic layers were limited to only few stations in the Eastern Pacific and Arctic Ocean (Fig. 2c, d). However, more prevalent and abundant were these transcripts in the MES (Fig. 2e). The abundance of cynS transcripts from Nitrospinae was positively correlated with nitrate and nitrite (p < 0.05, Fig. S5b; Table S5b), suggesting that cyanate metabolism in Nitrospinae may facilitate marine nitrification. In contrast, cynS was not detected in marine ammonia-oxidizing archaea of the phylum Thaumarchaeota. However, the ureC transcript from this taxon was detected mainly in MES zone (Fig. 2j) and positively correlated with depth and the concentration of nitrate (p < 0.05, Fig. S5b, Table S6b). This corroborates previous findings as marine Thaumarchaeota genomes lack the canonical cynS gene but the organisms can utilize cyanate and urea to fuel nitrification [15]. The contents of all the retrieved unigenes from OGA have been summarized in Supplementary Tables S7–S14.
Taken together, cynS is a conserved gene ubiquitous across the tree of life, transferred frequently via HGT. Comparative analyses based on the prevalence and expression of cynS and ureC representing intertwined processes of organic N metabolism in marine microbes suggest that cyanate is at least as important as urea in the oceans. Cyanate likely supports the assimilation of organic N in photoautotrophs when inorganic N is scarce and it appears to contribute to remineralisation by the activity of nitrifying bacteria which produce nitrate in deeper layers of the oceans.
References
Nowakowska M, Sterzel M, Szczubiałka K. Photosensitized oxidation of cyanide in aqueous solutions of photoactive modified hydroxyethylcellulose. J Polym Environ. 2006;14:59–64.
Kamennaya NA, Chernihovsky M, Post AF. The cyanate utilization capacity of marine unicellular Cyanobacteria. Limnol Oceanogr. 2008;53:2485–94.
Palatinszky M, Herbold C, Jehmlich N, Pogoda M, Han P, von Bergen M, et al. Cyanate as an energy source for nitrifiers. Nature. 2015;524:105–8.
Mooshammer M, Wanek W, Jones SH, Richter A, Wagner M. Cyanate–a low abundant but actively cycled nitrogen compound in soil. https://www.biorxiv.org/content/10.1101/2020.07.12.199737v1.full. 2020.
Linder T. Cyanase-independent utilization of cyanate as a nitrogen source in ascomycete yeasts. World J Micro Biot. 2019;35:1–7.
Widner B, Fuchsman CA, Chang BX, Rocap G, Mulholland MR. Utilization of urea and cyanate in waters overlying and within the eastern tropical north Pacific oxygen deficient zone. FEMS Microbiol Ecol. 2018;94:fiy138.
Widner B, Mulholland MR, Mopper K. Distribution, sources, and sinks of cyanate in the coastal North Atlantic Ocean. Environ Sci Tech Let. 2016;3:297–302.
Widner B, Mulholland MR, Mopper K. Chromatographic determination of nanomolar cyanate concentrations in estuarine and sea waters by precolumn fluorescence derivatization. Anal Chem. 2013;85:6661–6.
Widner B, Mordy CW, Mulholland MR. Cyanate distribution and uptake above and within the Eastern Tropical South Pacific oxygen deficient zone. Limnol Oceanogr. 2018;63:S177–S192.
Kuypers MMM, Marchant HK, Kartal B. The microbial nitrogen-cycling network. Nat Rev Microbiol. 2018;16:263.
Smith SR, Dupont CL, McCarthy JK, Broddrick JT, Oborník M, Horák A, et al. Evolution and regulation of nitrogen flux through compartmentalized metabolic networks in a marine diatom. Nat Commun. 2019;10:1–14.
Allen JrCM, Jones ME. Decomposition of carbamylphosphate in aqueous solutions. Biochemistry. 1964;3:1238–47.
Kamenaya NA, Post AF. Characterization of cyanate metabolism in marine Synechococcus and Prochlorococcus spp. Appl Enviro Micro. 2011;77:291–301.
Kamennaya NA, Post AF. Distribution and expression of the cyanate acquisition potential among cyanobacterial populations in oligotrophic marine waters. Limnol Oceanogr. 2013;58:1959–71.
Kitzinger K, Padilla CC, Marchant HK, Hach PF, Herbold CW, Kidane AT, et al. Cyanate and urea are substrates for nitrification by Thaumarchaeota in the marine environment. Nat Microbiol. 2019;4:234–43.
Pachiadaki MG, Sintes E, Bergauer K, Brown JM, Record NR, Swan BK, et al. Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science. 2017;358:1046–51.
Ganesh S, Bertagnolli AD, Bristow LA, Padilla CC, Blackwood N, Aldunate M, et al. Single cell genomic and transcriptomic evidence for the use of alternative nitrogen substrates by anammox bacteria. ISME J. 2018;12:2706–22.
Johnson WV, Anderson PM. Bicarbonate is a recycling substrate for cyanase. J Biol Chem. 1987;262:9021–5.
Miller AG, Espie GS. Photosynthetic metabolism of cyanate by the cyanobacterium Synechococcus UTEX 625. Arch Microbiol. 1994;162:151–7.
Harano Y, Suzuki I, Maeda S, Kaneko T, Tabata S, Omata T. Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. J Bacteriol. 1997;179:5744.
Sung YC, Anderson PM, Fuchs JA. Characterization of high-level expression and sequencing of the Escherichia coli K-12 cynS gene encoding cyanase. J Bacteriol. 1987;169:5224.
Sáez LP, Cabello P, Ibáñez MI, Luque-Almagro VM, Roldán MD, Moreno-Vivián C. Cyanate assimilation by the alkaliphilic cyanide-degrading bacterium Pseudomonas pseudoalcaligenes CECT5344: mutational analysis of the cyn gene cluster. Int J Mol Sci. 2019;20:3008.
Wood AP, Kelly DP, McDonald IR, Jordan SL, Morgan TD, Khan S, et al. A novel pink-pigmented facultative methylotroph, Methylobacterium thiocyanatum sp. nov., capable of growth on thiocyanate or cyanate as sole nitrogen sources. Arch Microbiol. 1998;169:148–58.
Elleuche S, Pöggeler S. A cyanase is transcriptionally regulated by arginine and involved in cyanate decomposition in Sordaria macrospora. Fungal Genet Biol. 2008;45:1458–69.
Schlachter CR, Klapper V, Wybouw N, Radford T, Van Leeuwen T, Grbic M, et al. Structural characterization of a eukaryotic cyanase from Tetranychus urticae. J Agr Food Chem. 2017;65:5453–62.
Qian D, Jiang L, Lu L, Wei C, Li Y. Biochemical and structural properties of cyanases from Arabidopsis thaliana and Oryza sativa. PLoS One. 2011;6:e18300.
Zarlenga DS, Mitreva M, Thompson P, Tyagi R, Tuo W, Hoberg EP. A tale of three kingdoms: members of the Phylum Nematoda independently acquired the detoxifying enzyme cyanase through horizontal gene transfer from plants and bacteria. Parasitology. 2019;146:445–52.
Ranjan B, Choi PH, Pillai S, Permaul K, Tong L, Singh S. Crystal structure of a thermophilic fungal cyanase and its implications on the catalytic mechanism for bioremediation. Sci Rep. 2021;11:1–10.
Villar E, Vannier T, Vernette C, Lescot M, Cuenca M, Alexandre A, et al. The Ocean Gene Atlas: exploring the biogeography of plankton genes online. Nucleic Acids Res. 2018;46:W289–W295.
Walsh MA, Otwinowski Z, Perrakis A, Anderson PM, Joachimiak A. Structure of cyanase reveals that a novel dimeric and decameric arrangement of subunits is required for formation of the enzyme active site. Structure. 2000;8:505–14.
Butryn A, Stoehr G, Linke-Winnebeck C, Hopfner KP. Serendipitous crystallization and structure determination of cyanase (CynS) from Serratia proteamaculans. Acta Crystallogr F. 2015;71:471–6.
Pederzoli R, Tarantino D, Gourlay LJ, Chaves-Sanjuan A, Bolognesi M. Detecting the nature and solving the crystal structure of a contaminant protein from an opportunistic pathogen. Acta Crystallogr F. 2020;76:392–7.
Wybouw N, Balabanidou V, Ballhorn DJ, Dermauw W, Grbić M, Vontas J, et al. A horizontally transferred cyanase gene in the spider mite Tetranychus urticae is involved in cyanate metabolism and is differentially expressed upon host plant change. Insect Biochem Molec. 2012;42:881–9.
Spang A, Poehlein A, Offre P, Zumbrägel S, Haider S, Rychlik N, et al. The genome of the ammonia‐oxidizing candidatus nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012;14:3122–45.
Palomo A, Pedersen AG, Fowler SJ, Dechesne A, Sicheritz-Pontén T, Smets BF. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISMEJ. 2018;12:1779–93.
Acknowledgements
This study was supported by the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (2018SDKJ0406-3), National Natural Science Foundation of China (41876156), and Fundamental Research Funds for the Central Universities (201812019) to YZ and the China Scholarship Council (201806330141) to XM. TM and CvO acknowledge the School of Environmental Sciences at the University of East Anglia, Norwich, UK for partial support.
Author information
Authors and Affiliations
Contributions
YZ and TM conceived and coordinated the project with help from CvO. XM did most of the laboratory work with help from YZ, HZ, and GL. XM, JC, and YZ conducted bioinformatic analysis. The manuscript was co-written by XM, TM, YZ, and CvO. All authors edited the final version before they approved submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mao, X., Chen, J., van Oosterhout, C. et al. Diversity, prevalence, and expression of cyanase genes (cynS) in planktonic marine microorganisms. ISME J 16, 602–605 (2022). https://doi.org/10.1038/s41396-021-01081-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-01081-y
This article is cited by
-
Effects of Ocean Acidification on Nitrogen Metabolism of Skeletonema costatum
Journal of Ocean University of China (2024)
-
Effects of cyanate enrichment on growth of natural phytoplankton populations in the subtropical Pacific
Journal of Oceanography (2023)
-
Genomic adaptation of the picoeukaryote Pelagomonas calceolata to iron-poor oceans revealed by a chromosome-scale genome sequence
Communications Biology (2022)