Abstract
The eco-evolutionary interactions among members of the vertebrate gut microbiota that ultimately result in host-specific communities are poorly understood. Here we show that Lactobacillus reuteri coexists with species that belong to the Lactobacillus johnsonii cluster (L. johnsonii, L. gasseri, and L taiwanensis) in a taxonomically wide range of rodents, suggesting cohabitation over evolutionary times. The two dominant Lactobacillus species found in wild mice establish a commensalistic relationship in gastric biofilms when introduced together into germ-free mice in which L. reuteri facilitates colonization of L. taiwanensis. Genomic analysis revealed allopatric diversification in strains of both species that originated from geographically separated locations (Scotland and France). Allopatry of the strains resulted in reduced formation of mixed biofilms in vitro, indicating that interspecies interactions in gastric Lactobacillus-biofilms are the result of an adaptive evolutionary process that occurred in a biogeographical context. In summary, these findings suggest that members within the vertebrate gut microbiota can evolve inter-dependencies through ecological facilitation, which could represent one mechanism by which host-specific bacterial communities assemble across vertebrate species and an explanation for their spatial and biogeographic patterns.
Similar content being viewed by others
Introduction
The gut microbiota makes important contributions to the fitness of vertebrate animals (including humans) [1], but in contrast to host-microbial symbioses in invertebrates, little is known about the basic mechanisms by which symbiotic interactions are maintained and how they evolve [2]. Mammals are colonized by specific bacterial communities that cluster according to host species [3, 4], and recent findings suggest cospeciation of specific microbiota members with their hosts [5]. However, the evolutionary relationships of gut microbes with vertebrates can vary dramatically, with some clades remaining highly host-specific over evolutionary time spans [5, 6], while others evolving generalist lifestyles without host restriction [5, 7, 8]. Little is known about how these lifestyles evolve and how eco-evolutionary interactions within the microbiota shape species associations within the communities.
We have established the species Lactobacillus reuteri as a model to study the evolution of a gut symbiont and the consequences of its symbiosis with the vertebrate host [9]. This species can be divided into several phylogenetic clades that correlate with host origin [10] and whose genome content reflects niche characteristics in respective hosts [11]. Capitalizing on gnotobiotic mice and chickens, we have experimentally demonstrated host specificity [11, 12], determined the ecological role of rodent-specific genes in vivo [11], and identified selective biofilm formation in the mouse stomach as key mechanisms by which host specificity is facilitated [13]. Although our research has provided clear evidence for a persistent and specific evolution of L. reuteri with rodent hosts and the mechanisms by which host specificity is established, nothing is yet known about how this species evolves in the context of the bacterial communities in which it exists in nature.
In rodents, L. reuteri is a component of bacterial biofilms that adhere directly to the stratified squamous epithelium of the forestomach [14]. Recent molecular analysis of the gastric bacterial populations in mice has demonstrated that they are dominated by a small number of Lactobacillus species [15]. The two taxa that consistently dominate Lactobacillus population in the murine gut in both molecular and culture-based studies are L. reuteri and closely related species of the L. johnsonii cluster (e.g., L. johnsonii and L. taiwanensis) [16,17,18,19,20]. Both L. reuteri and L. johnsonii have been shown to be composed of host-specific subpopulations [10, 21] and partition resources during gut colonization [22]. These findings suggest a long-term tripartite evolutionary relationship between these two lineages and their rodent host. The simple composition of gastric biofilms and the fact that they can be studied in gnotobiotic mice under realistic ecological conditions [13] make them an excellent model system to determine the mechanisms by which bacterial members of the vertebrate gut microbiota interact with each other, and how these interactions evolve.
The goal of this study was to characterize the evolutionary relationship between L. reuteri and L. johnsonii-related lineages that dominate natural Lactobacillus populations in rodents. We first characterized the Lactobacillus population from a taxonomically diverse range of wild rodents, and then evaluated the ecological interactions between the two most dominant species in biofilm formation assays using gnotobiotic mice. We next hypothesized that the interactions between Lactobacillus lineages result from an adaptive process over evolutionary time. To test this hypothesis, we characterized the genome variation of isolates obtained from two geographically separated populations of mice and then compared interactions of allopatric strains with sympatric strains during biofilm formation.
Results
L. reuteri and species related to L. johnsonii coexist in the gut of a phylogenetically wide range of rodent hosts
To characterize the Lactobacillus population in rodents, gut samples were collected from zoo animals, as well as animals caught in the wild. Animals belonging to eight rodent species from three main clades within the order Rodentia were included (Table 1). Lactobacilli were isolated by selective culture from gut samples and isolates were taxonomically identified by sequence analysis of the 16S rRNA gene (Table S1).
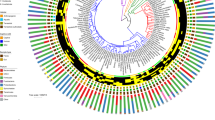
The cultivable Lactobacillus population was dominated by L. reuteri and species related to L. johnsonii (L. johnsonii-like), such as L. rodentium [23] and species that cannot be differentiated from L. johnsonii (the L. johnsonii cluster) based on 16S rRNA sequences, e.g., L. taiwanensis and L. gasseri [24]. Figure 1a depicts the distribution of L. reuteri and species related to L. johnsonii among rodent hosts in the context of the phylogeny established by Fabre and coworkers [25]. L. reuteri was isolated from all rodent species in this study, indicating an ancient history of association dating back to the root of the Rodentia phylogeny. L. johnsonii-like species were isolated from all hosts from the Myodonta (mouse related) clade, but not from hosts in the squirrel-related and Ctenohystrica (guinea-pig related) clades, suggesting a later association of L. johnsonii-like species with rodents. Interestingly, neither L. johnsonii (and related species) nor L. reuteri was isolated from shrews (Fig. 1a and Table 1), which are small mammals belonging to the order Eulipotyphla, suggesting the important role of host phylogeny in these symbiont relationships. Most importantly, the L. johnsonii cluster and L. reuteri were co-isolated from all species in the Myodonta clade, indicating that they may have coexisted with each other in their shared hosts over evolutionary times.
Isolation of L. johnsonii-like species and L. reuteri from rodent hosts. a Schematic clade-level phylogenetic tree of rodents (adapted from [25]) showing detection of L. reuteri and L. johnsonii-like species by culture. b Composition of Lactobacillus community in wild mice originating from France and Scotland (n = 4 each). 200 isolates were cultured from the ceca of mice (25 per mouse), and identities were determined by 16S rRNA sequence analysis
Composition of the Lactobacillus population in the gut of wild house mice
To gain detailed insight into the Lactobacillus community of wild house mice (Mus musculus domesticus) at two separate geographic locations, we selectively cultured lactobacilli from the ceca of four mice obtained from each France and Scotland and identified randomly selected isolates (25 isolates per mouse) by 16S rRNA sequence analysis. In agreement with previous studies [15,16,17,18,19,20], the cultivable Lactobacillus population of wild mice was composed of only a few species and dominated by L. reuteri and the L. johnsonii cluster (Fig. 1b). L. reuteri and the L. johnsonii cluster were also the only two taxonomic groups consistently isolated from all eight animals, and they accounted for more than half of the isolates, confirming their dominance in the Lactobacillus community of wild mice.
For identification of the species within the L. johnsonii cluster, whole-genome sequencing was performed for 31 isolates (16 from Scotland and 15 from France). Sequences of selected housekeeping genes were used to compare isolates with publicly available sequences of L. johnsonii, L. gasseri, and L. taiwanensis using a Multi-locus Sequence Typing (MLST) approach developed for L. johnsonii [21]. As expected, these three species formed distinct phylogenetic clusters (Fig. S1). With the exception of isolates from one Scottish mouse (Mouse 117) identified as L. johnsonii, all other isolates were identified as L. taiwanensis. This implies that in addition to L. reuteri, L. taiwanensis is the most prevalent Lactobacillus species in the gut of wild house mice. L. taiwanensis isolates were therefore used to study the functional interactions with L. reuteri.
C haracterization of species interactions in gastric biofilms
To study the ecological interaction between L. reuteri and L. taiwanensis in their natural niche, we compared biofilm formation in single and dual-species biofilms in the forestomachs of ex-germ-free mice. For this, we selected two pairs of L. reuteri and L. taiwanensis strains from each of two French and two Scottish mice (Table 2). Germ-free mice were inoculated with single strains or mixtures, and biofilm formation on the forestomach epithelium was quantified 48 h after inoculation with confocal microscopy as previously described [13]. Cell numbers for each species were further quantified via culture on mMRS agar.
Although in vivo biofilm quantification was inherently variable, mixed-species biofilms showed on average higher cell density when compared to biofilms in mono-associated mice (Fig. 2a). Quantification of the total forestomach area covered by bacterial cells revealed significant higher biofilm density for two of four strain combinations when compared to L. taiwanensis alone, but clear synergism was only detected for one strain combination (T1R1)(Fig. 2b). Similar findings were obtained when quantifying total bacteria adherent to the forestomach epithelium by culture, with higher cell numbers in mixed-species biofilm in all four pairings when compared to L. taiwanensis mono-associated mice and synergism for the T1R1 combination. L. reuteri biofilms contained a higher number of cells when compared to L. taiwanensis in two of the experiments (Fig. 2c). In two combinations (T1R1 and T2R2), total cell numbers in mixed biofilms were more than double those obtained with either of the single strains, suggesting synergism. Together, these results demonstrate that L. reuteri biofilms were denser than those of L. taiwanensis and that increased biofilm formation was seen in mixed-species biofilms, but synergism was not consistent in all strain pairings.
Biofilm formation in the forestomach of mice (n = 5–7) (See Table 2 and Table S1 for letter designation of the isolates). a Confocal micrographs of representative areas of forestomach showing epithelium autofluorescence (green) and propidium iodide-stained bacteria (red). b Quantification of biofilm density on forestomach epithelia as % red pixel area, as calculated relative to L. tainwanensis mono-associated mice by confocal microscopy. Each point represents one mouse, with 3 z-stacks per mouse. c Total cell count on forestomach epithelia as determined by quantitative culture. Statistical analyses for each group were performed separately. Different letters denote significant differences between groups (one-way ANOVA followed by Tukey’s test; α = 0.05, p < 0.05). CFU of L. taiwanensis (d) and L. reuteri (e) on forestomach epithelium of mice colonized by single strains or mixtures. Strains were differentiated on plates by colony morphology. Different letters denote significant differences between groups (Student’s t-test; α = 0.05, p < 0.05)
To further examine interactions between L. taiwanensis and L. reuteri within dual-species biofilms, we quantified each species by standard plating based on clear differences in colony morphology. L. reuteri formed single-species biofilms of around 5 × 108 CFU per gram of forestomach tissue, while numbers of L. taiwanensis in single-species biofilms were 5–10 times lower. For three out of the four strain pairings, cell numbers of L. taiwanensis were significantly elevated in dual-species biofilms (Fig. 2d), reaching cell number comparable to that of L. reuteri. In contrast, the cell numbers of L. reuteri were not different in mono- and dual-species biofilms (Fig. 2e). Overall, these findings indicate a commensalistic relationship in which L. reuteri facilitates the colonization of L. taiwanensis in gastric biofilms without being affected itself.
Coaggregation with L. reuteri is required for L. taiwanensis to form aggregates
To determine whether the species interactions described above could be attributed to the ability of L. reuteri and L. taiwanensis to auto- and coaggregate, aggregation assays were performed in vitro. Isolates of L. reuteri, but not L. taiwanensis, formed autoaggregates, while aggregation of L. taiwanensis depended on the presence of on L. reuteri (Fig S2A). Treatment with Proteinase K essentially abolished the ability of the lactobacilli to auto- and coaggregate (Fig S2B), showing that aggregation was protein-based, which concurred with our previous finding that coaggregation between L. reuteri and L. taiwanensis (formerly classified as L. johnsonii) was not mediated by exopolysaccharides [26]. Overall, these findings showed that L. taiwanensis could only form cell aggregates when L. reuteri cells were present, while the latter species can autoaggregate, providing a potential mechanism for the facilitation of L. taiwanensis through L. reuteri observed in mice (although additional mechanisms might still apply).
Structural characterization of single- and dual-species biofilms in the mouse forestomach and the spatial distribution of strains
To gain additional insight into the interactions between L. taiwanensis and L. reuteri in gastric biofilms, we developed a Fluorescence in situ Hybridization (FISH) protocol with species-specific probes (Table S2) to elucidate the spatial organization of the two species. Species-specific fluorescent probes were verified in vitro to exclude cross reactivity (Fig. 3a–c). The technique was then used to visualize bacterial cells on the forestomach epithelium after 48 h of colonization. The analysis revealed that L. taiwanensis colonized the epithelium only in small clusters that remained confined within crevices of the forestomach tissue (Fig. 3d, g, Video S1). In contrast, L. reuteri formed biofilms that covered larger areas of the forestomach luminal surface (Fig. 3e, h, Video S2), an observation consistent with previous studies [13, 27]. On forestomach epithelia colonized by both species (Fig. 3f, i, Video S3), L. reuteri and L. taiwanensis cells were both evenly distributed throughout the biofilm in small clusters without obvious distinction in spatial organization or cell number. Altogether, these observations suggest that while both species could colonize the forestomach independently, the biofilm-forming ability of L. reuteri was superior to that of L. taiwanensis and further facilitated the integration of the latter within a dual-species biofilm. This spatial and structural characterization provides an explanation for the beneficial role of L. reuteri in the forestomach colonization of L. taiwanensis (Fig. 2d).
Characterization of structural distribution of L. reuteri 111 v and L. taiwanensis 111z in forestomach biofilms by Fluorescence in situ hybridization (FISH) with species-specific probes. Validation of L. taiwanensis/L. johnsonii-specific probe (red) and L. reuteri-specific probe (yellow) using cultures of L. taiwanensis (a), L. reuteri (b) and dual-species mixtures (c). 2D micrographs of forestomach tissues from mice colonized with L. taiwanensis (d), L. reuteri (e), and dual-species mixtures (f). 3D micrographs of forestomach tissues (stained with DAPI in blue) from mice colonized with L. taiwanensis (g), L. reuteri (h), and dual-species mixtures (i). Mouse experiments were performed in six mice per treatment and representative results are shown
Geographic separation of the host animals resulted in allopatric genome diversification in both L. reuteri and L. taiwanensis
We postulated that if facilitation of L. taiwanensis through L. reuteri has resulted from an adaptive process during a common evolutionary history of coexistence, then reciprocal interactions should enhance traits of L. taiwanesis that support its performance when paired with L. reuteri strains from the same location [28, 29]. Consequently, the pairing of strains from geographically separated locations (allopatric) should result in a reduction of the interactive functions in dual-species biofilms when compared to strains from the same location (sympatric). Before testing the phenotypic interactions of allopatric and sympatric strain combinations in biofilms, we first determined the genetic divergence among L. reuteri and L. taiwanensis strains from France and Scotland, which are likely to have become geographically isolated over timescales sufficient for evolutionary change (i.e., up to several thousand years, [30]).
We analyzed the draft genomes of 25 L. taiwanensis isolates (10 from Scotland and 15 from France) and 32 L. reuteri isolates (19 from Scotland and 13 from France). For each species, core genes without signs of recombination were identified. Phylogenetic trees were constructed using concatenated sequences of the core genes as well as by coalescence of core gene trees. These phylogenetic analyses revealed that isolates from each location clustered separately (Fig. 4a, b, and Fig S3), consistent with allopatric diversification in both species. Average Nucleotide Identity (ANI), Average Amino acid Identity (AAI), and the number of Single Nucleotide Polymorphisms (SNPs) in the core genomes also showed that variation within geographic regions was lower than variation between regions (Fig. 4c). In addition, genetic variation within mice was significantly lower than variation across host animals within the same region, but isolates from the same mouse still showed sufficient diversity to conclude that distinct strains coexisted that did not primarily emerged through clonal expansion of an original colonizer (Fig. 4c).
Genomic analyses of L. taiwanensis and L. reuteri strains obtained from wild mice. Phylogeny of L. taiwanensis (a) and L. reuteri (b) isolates based on concatenated core gene alignment. Color block by the tree represent different BAPS cluster. All isolates of L. reuteri belong to Rodent Lineage III in the species phylogeny [10]. c Intra-host, inter-host and intra-region, and inter-region pairwise ANI, AAI, and the number of SNPs. Different letters denote significant differences between groups (one-way ANOVA; α = 0.05, p < 0.05)
Genetic diversification could result from genetic drift and/or natural selection. To gain insight into potential contributing factors to the observed diversification, a clade model [31] was used to detect the differences in selective constraint between the isolates from Scotland and France. Genes subject to putative changes in selective forces between Scottish and French clades were detected for both L. taiwanensis and L. reuteri (Table S3 and S4), providing candidates for involvement in allopatric diversification. Altogether, these genomic comparisons suggest that geographic isolation resulted in allopatric divergence in the genomes of both L. reuteri and L. taiwanensis.
Reduced formation of dual-species biofilms of allopatric strains
We next sought to determine whether the L. reuteri–L.taiwanensis interrelationship is the result of an evolutionary process resulting in co-adaptation in a biologically relevant phenotype. To that end, we used in vivo and in vitro biofilm assays to compare dual-species biofilm formation by sympatric strains, isolated from the same mouse populations, and allopatric strains, isolated from different mouse populations. We hypothesized that allopatric diversification of each species would have resulted in local adaptation, such that L. taiwanensis benefits more from co-colonization with sympatric L. reuteri than allopatric L. reuteri during biofilm formation. No difference in L. reuteri facilitation of L. taiwanensis according to geographic sympatry was observed in vivo for total biofilm formation or for individual species (Fig S4). However, in vitro, total biofilm biomass was higher in dual-species biofilms composed of sympatric versus allopatric pairings in almost all combinations tested (Fig. 5a, b). Moreover, cell numbers of both L. reuteri and L. taiwanensis were higher in biofilms formed by sympatric combinations of strains (Fig. 5c, d). The enhanced biofilms of sympatric strain pairs suggest that L. taiwanensis and L. reuteri have jointly evolved at the two geographic locations, resulting in local adaptations that improved traits that facilitate interactions between the two species.
The effect of allopatry and sympatry of strains on biofilm formation in dual-species in vitro biofilms. a Photographs of in vitro dual-species biofilms formed by L. taiwanensis and L. reuteri strains that originated from the same (sympatric) or different (allopatric) locations (See Table 2). b OD600 of biomass of dual-species biofilms formed by sympatric and allopatric strain combinations (eight biological replicates). Different letters denotes significant differences between groups (one-way ANOVA; α = 0.05, p < 0.05). CFU of L. taiwanensis (c) and L. reuteri (d) in dual-species biofilms formed by sympatric and allopatric strains (eight biological replicates). Different letters denote significant differences between groups (Student’s t-test; α = 0.05, p < 0.05)
Discussion
Given the complexity of the vertebrate gut microbiota, it is challenging to elucidate the ecological and evolutionary interrelationships that shape communities. The aim of this study was therefore to specifically characterize ecological interactions among bacterial members that were the result of their joint evolution in concert with the host. To achieve this, we characterized, in a biogeographic context, two bacterial lineages that not only closely co-associate in gastric biofilms but are also host-specific, suggesting a stable cohabitation over evolutionary timescales. Indeed, L. reuteri and species related to L. johnsonii were previously isolated together from gut samples of prairie voles and white-throated woodrats [16, 32]. In this study, we isolated the same species combination from gut samples of wild rodents that span three major clades of the order Rodentia (Fig. 1). The phylogenetically wider host distribution of L. reuteri suggests an evolutionarily earlier association of this species with rodents that was later followed by an association with the common ancestor of L. johnsonii, L gasseri, L. taiwanensis, and L. rodentium, and a long history of cohabitation of the two thereafter. As demonstrated by our phenotypic analysis of mono- and mixed-species biofilms in the forestomach of mice, this common evolutionary history of coexistence resulted in a commensalistic relationship between L. reuteri and L. taiwanensis, in which the former species facilitates colonization of the latter.
Although mutualism is common in multi-species biofilms found in soil [33, 34], alga surfaces [35], oral cavity [36], water pipes [37], respiratory tract [38], biliary stents [39], and in food facilities [40], and cooperation has recently been demonstrated within members of the gut microbiota [41], our experiments clearly demonstrated that the interrelationship between L. reuteri and L. taiwanensis in mouse forestomach biofilms is based on commensalism. Although less frequently reported, commensalism has been found in biofilms formed by sewage [42] and soil microbes [43] and also occurs in planktonic cells in liquid culture [44, 45]. Commensalistic interactions between microbes are often based on metabolic exploitation, whereby the metabolite from one organism is utilized by the other [46]. However, L. johnsonii 100-33 and L. reuteri 100-23 partition resources (maltose and glucose) in the mouse stomach [22], and the distinct homofermentative and heterofermentative metabolisms of these two species are likely to be complementary [9]. Our experiments indicate that the commensalism between L. taiwanensis and L. reuteri is mediated through coaggregation, a key mechanism for the formation of biofilms [47,48,49,50]. Coaggregation allows L. taiwanensis to become integrated in the mixed-species biofilm (Fig. 3), thereby increasing its cell numbers in the forestomach (Fig. 2d). This unilateral beneficial interaction is in agreement with a later arrival of L. taiwanensis as a symbiont of the Myodonta, exploiting the presence of pre-established L. reuteri to successfully colonize the host.
Interestingly, our experiments with strain combinations from geographically isolated locations showed a reduction in biofilm density among dual-species biofilms formed by allopatric isolates of L. reuteri and L. taiwanensis compared to sympatric pairings, which supports allopatric diversification resulting in local co-adaptation. Allopatric diversification in bacteria has so far only been inferred by sequence data without any functional assessment of its consequences [51, 52]. However, our findings clearly establish both genomic evidence for allopatric diversification and its phenotypic consequences. Considering that genetic drift alone is limited in its power to generate distinct phylogenetic clusters [53], and that we detected genes subject to differential selection between Scotland and France in both L. reuteri and L. taiwanensis, our findings suggest that region-specific phylogenetic clustering was likely the outcome of local adaption [54]. Most importantly, the increased ability of sympatric strain pairings to form dual-species biofilms suggests that the interspecies interactions of the two gut symbionts were the result of an adaptive process that occurred during a joint evolutionary history impacted by host-microbe biogeography.
Apart from providing insight into the evolutionary interactions between of two bacterial members of gut microbiota, the genomic analysis elucidated spatial factors that shape the population diversity of bacterial gut symbionts, which are likely to ultimately influence their evolution within communities. Genetic variation within populations can arise from dispersal limitation and local adaptation [55]. Accordingly, genomic variation in both L. reuteri and L. taiwanensis was lowest in strains isolated from the same host animal (intra-host), followed by isolates from the same geographic region (inter-host), with the highest diversity found when strains were isolated from the two geographic regions (inter-region) were compared (Fig. 4c). These findings indicate that both host and geographic separation pose dispersal barriers to gut microbes that shape their evolution, with geographic isolation generating the highest genetic variation. Although inter-host dispersal likely allows mixing of genotypes in the same region, we observed reduced diversity within hosts when compared to different hosts for both species. Intra-host diversity among strains was nevertheless too high to be driven solely by a founder effect (i.e., where an early colonizer or superior competitor gains dominance within the population of a particular host animal). Instead, the pattern suggests that subpopulations of L. reuteri and L. taiwanensis are retained within host lineages through familial transmission. Maternally transmitted microbes that colonize early in life could gain a fitness advantage over later arrivers acquired by horizontal transmission due to priority effects (e.g., niche preemption and monopolization) [56, 57]. This could allow specific populations to dominate host lineages over generations, ultimately causing the reduced genetic diversity in individual mice (Fig. 4).
In summary, by establishing that cohabitation of biofilms can result in facilitation between two vertebrate gut symbionts, this study provides important basic information on the eco-evolutionary interactions among gut microbes. Facilitation between organisms is increasingly recognized to underlie both evolutionary and ecological processes within communities [58], but it has not been specifically considered in gut ecosystems although it might be, together with cooperation [41], a cause of the positive associations detected in co-occurrence networks [59, 60]. Since interactions among members do ultimately determine the structure and function of entire communities, the findings obtained here provide a possible mechanism that contributes to the spatial and biogeographic patterns and characteristics described for the gut microbiota in mice. Interspecies interactions that arise during long-term joint evolution of host-confined lineages such as L. reuteri and L. taiwanensis could ultimately drive community profiles that correspond to host phylogeny [3, 4, 7, 61]. It is interesting in this respect that the factors that shape genomic diversity among L. reuteri and L. taiwanensis (e.g., geographic distance and familiar transmission) have also been shown to be among the dominant factors that shape whole gut communities at different scales [62, 63]. Our findings also have implications for the development of strategies to modulate the microbiota, especially considering that probiotic strains rarely engraft into gut microbiomes [64, 65]. Since facilitation can create niches [58], engraftment may be greater in individuals who possess gut microbiota members that facilitate colonization of the incoming probiotic strain, or that probiotics should consist of strain mixtures that maintain commensalistic or mutualistic relationships that potentially enhance colonization ability [65]. Ultimately, only an understanding of the basic ecological principles, which are determined by interactions among community members, will facilitate systematic modulation of the gut microbiome to improve health.
Materials and Methods
Growth of bacteria
Bacteria were cultured anaerobically on modified MRS (mMRS) medium (MRS supplemented with 10 g/L maltose and 5 g/L fructose) at 37 °C unless otherwise noted.
Collection of gut samples from wild small mammals in Lithuania
Between 14 and 27 August 2014, small mammals were snap-trapped at five sites with diverse habitat in Lithuania, all within 30 km of the capital city of Vilnius: a pasture/young planted forest site, the edge of a bog, a regrowing clear-cutting, a natural meadow, and a wheat field/meadow boundary (Table S1). Traps were baited with bread soaked in oil, set at dusk, and retrieved by 10:30 am the following day. All trapped animals were placed in sterile bags and kept in cool boxes filled with ice for transport to the lab. In the lab, the animals were dissected and an ~1-cm section of the jejunum removed. These were stored in RNALater and placed in the fridge at the end of each day. Within two weeks, the samples were spun down, the RNALater decanted, and tissue sections stored at −80 °C before culturing.
Digesta from zoo animals
The Edmonton Valley zoo, the Calgary zoo, The Greater Vancouver Zoo, and the Assiniboine Park Zoo in Winnipeg were contacted via email where institutional information and a research proposal were exchanged. When required, a requisition form for biological sample collection was completed. Once approved, fecal samples were collected during zoo operations (Table S1). Containers were provided to the zoo to collect 1–10 g of feces that was less than 24 h old. The samples were only collected from an enclosure so long as they could be traced back to a single species. Once collected, the samples were frozen at −20 °C until overnight shipment back to the lab for processing. Shipments of samples were kept cool with either dry ice or shipping ice blocks before culturing.
Digesta from natural populations of house mice from France and Scotland
Cecal samples from mice (Mus musculus domesticus) previously described [66] were obtained immediately after the mice were euthanized, placed in liquid nitrogen, stored at −80 °C and shipped frozen to the University of Nebraska for processing. The French mice originated from a wild mouse population sampled in the Massif Central (MC) region of France in 2009 [62] that were subsequently bred at the animal facility of the Max Plank Institute for Evolutionary Biology. The sampled individuals came from the third generation of breeding and were derived from separate families/cages housed in the same room. Mice sampled in Scotland were wild-caught [67].
Isolation of Lactobacillus spp. from gut samples
Lactobacillus spp. were isolated via plating on mMRS agar and incubation at 37 °C anaerobically. Colonies were subcultured to purity and identified by 16S rRNA gene analysis. All isolates from the zoo and wild animals were sequenced. From the French and Scottish mice, 200 isolates (25 from each mouse) were randomly selected. The sequences were classified using the Ribosomal Database Project [68] Seqmatch tool to determine the species identity (Table S1).
Genome sequencing and assembly
Sixty-three isolates from eight mice (four each from Scotland and France) were selected for whole-genome sequencing. We chose 3–5 isolates per mouse (dependent on availability) and per bacterial species (L. reuteri and L. taiwanensis) to allow a determination of intra-host, inter-host and intra-region, and inter-region genomic diversity. DNA was extracted using Qiagen Blood and Tissue kit from overnight cultures, and sequencing was performed at the Applied Genomics Core at the University of Alberta. Libraries were prepared using the NexteraXT Index kit v2 and sequenced on an Illumina Miseq instrument. Pair-end 2 × 300 bp sequences were generated. Paired-end reads from each isolate were trimmed and error-corrected using the BayesHammer program [69] and assembled using SPAdes version 3.1.1 [70]. The SPAdes assembly option “careful” was used with the k-mer lengths 21, 33, 55, 77, 99, and 127, as recommended in the SPAdes documentation. The resulting contigs were assessed for size and gene content using QUAST [71]. BBMap [72] was used to map raw reads back to contigs, to determine percent coverage and average fold coverage. A complete collection of RefSeq prokaryotic sequences was downloaded from NCBI using the Entrez query 'nucleotide genome[Filter] AND (bacteria[Filter] OR archaea[Filter]) AND refseq[Filter] NOT wgs[Filter]' and each contig was compared to the collection using megablast from the blast-2.2.26 package [73] and an e-value threshold of 1e-12. Genome sequences were deposited in NCBI under BioProject PRJNA384810.
Multi-locus sequence typing (MLST)
MLST was performed using three shared hypothetical genes described previously [21]. Published gene sequences from the type strains of L. johnsonii, L. taiwanensis, and L. gasseri were downloaded from NCBI. Gene sequences were extracted from the genomes of our isolates which were identified as L. johnsonii by 16S rRNA gene sequence analysis. Sequences from all strains were aligned with MAFFT, and phylogenetic trees were generated using MrBayes 3.2.6 [74] under GTR + I + G model. The analysis was repeated 10,000,000 times until average standard deviation was below 0.01. The tree was visualized using Interactive tree of life (iTOL) v3.
Genomic analyses
The genomes were annotated using Prokka 1.11 [75] and the core genome alignment were extracted using Roary 3.6.1 [76]. SNPs were extracted from core gene alignment using SNP-sites [77]. Genes with evidence for recombination were detected using GENECONV 1.81a [78] and removed from the core genome. Phylogenetic trees of concatenated core genes were generated using MrBayes 3.2.6 [74] under GTR + I + G model, with 10,000,000 generations, sampling every 5,000th generation with 4 heated chains and a burn percentage of 25%. The analysis was repeated 10,000,000 times until average standard deviation was below 0.01. Coalescent trees were inferred with ASTRAL [79].
Genes with divergent selection was detected with clade model C in PAML 4.9a [80]. The model pairs for each gene were statistically evaluated for significance by likelihood ratio tests (LRT) with a χ2 distribution. The resulting tree was visualized using FigTree 1.4.3 [81]. Pairwise ANI and AAI was calculated using ANI/AAI-Matrix calculator from the enveomics toolbox collection [82]. Statistical clustering was performed using Bayesian analysis of population structure (BAPS) [83].
Mouse experiments
Mouse experiments to test for synergy during in vivo biofilm formation (Fig. 2) were performed with approval of the Institutional Animal Care and Use Committee of the University of Nebraska-Lincoln (Project ID 731). Cultures used for oral gavage were prepared by growing L. reuteri strains for 16 h. The optical density of all strains was adjusted to three. Cells were recovered by centrifugation at 4000 rpm for 5 min and resuspended in phosphate-buffered saline. Cultures for dual-species association were prepared by mixing equal volume of culture of each species. Germ-free Swiss Webster mice (6–10 weeks of age, male and female) were bred and maintained under gnotobiotic conditions at the University of Nebraska-Lincoln Gnotobiotic Mouse Facility. Immediately prior to each experiment, mice were moved from flexible-film isolators into sterile, individually ventilated, positive-pressure biocontainment cages (Allentown Inc., Allentown, NJ, USA). Mice were housed together by treatment (n = 5–7 per treatment), and each mouse was gavaged with 100 μl of a cell suspension containing 108 viable cells. After 48 h of colonization, mice were sacrificed by CO2 asphyxiation. The forestomach was removed, and the tissue was separated from the digesta, washed with PBS, and divided into two sections. One section is stored in PBS for plating and the second section was stored in 3% formalin/phosphate-buffered saline at 4 °C until usage for confocal microscopy.
Mouse experiments for FISH analysis (Fig. 3) were performed with approval of the Institutional Animal Care and Use Committee of the University of Alberta (Project ID 2099), using the same mouse strain and procedures as described above with six mice per treatment. Strains L. taiwanensis 111z and L. reuteri 111 v (Table S1) were used for single- and dual-species inoculation. Forestomach tissues were harvested after two days and fixed in Carnoy’s solution for 6 h before embedding in paraffin.
Visualization and quantification of biofilms in the mouse forestomach by confocal microscopy
Confocal microscopy was performed as described previously [13] at University of Nebraska Lincoln. Briefly, fixed forestomach tissues were transferred to fresh PBS and immersed for 20 minutes for three times to remove residual methanol. Tissues were stained in 5 mg/mL propidium iodide in PBS for 10 min and washed in PBS to remove excess dye and mounted on glass coverslips in Fluorogel (Electron Microscopy Sciences, Hatfield, PA USA) suspended by a CultureWell chambered cover glass (Grace Biolabs Bend, OR USA). Samples were imaged with an Olympus FV500 Confocal Laser Scanning Microscope using an Olympus Ix81 inverted microscope (Olympus, Center Valley, PA, USA). Series of z-axis confocal optical images were collected by a technician with no knowledge of sample identities from three random sites of the forestomach tissue with a 606 oil lens using the dual excitation and emission mode (excitation laser lines: 488 nm and 543 nm, emission filters: 525 nm and 600 nm, respectively). Biofilm area was quantified by determining the area occupied by bacteria stained with propidium iodide using 600 nm red fluorescence while background fluorescence was measured at 525 nm. For each sample (three per mouse), z-stacks were compressed into 2-dimensional images analyzed using a method described previously [13]. ImageJ [84] was used to quantify biofilm formation by determining the red-channel pixel area in images captured from three separate fields of view per individual sample (which results in a total area of 0.144 mm2 per mouse). The autofluorescence of the mouse forestomach tissue at 525 nm was captured as background and removed.
Characterization of mixed-species biofilm structure by Fluorescence in situ hybridization (FISH)
FISH was performed as described previously [85] with modifications. Briefly, for in vitro validation of probes, bacteria were grown in MRS broth at 37 °C to reach OD600 0.6. Cells were washed with PBS, fixed with PFA for 4 h, and stored in 50% ethanol for further use. Cells were fixed on super-frosted slides, permeabilized by treating with 100 mg/mL lysozyme (buffer: 10 mM Tris-HCl + 1 mM EDTA, pH = 7.36) at 37 °C for one hour [86]. The samples were then washed in distilled water and dehydrated in increasing percentage of ethanol. Embedded forestomach tissues were sectioned into 10-μm slices and transferred to super-frosted slides. The samples were washed three times in xylene to remove paraffin and rehydrated by washing in a succession of 100%, 80%, and 60% ethanol. Subsequent permeablization and washing steps were identical as described above for bacterial cells.
The hybridization was carried out with species-specific probes (Table S2) at 50 °C for 2 h and washed in washing buffer at 52 °C, for 1 h. Subsequently, the slides were submerged in quenching solution (10 mM Cupric Sulfate, 50 mM Ammonium Acetate Buffer, pH 5.0) for 60–90 minutes [87]. After quenching, the slides were rinsed with distilled water and air-dried. Samples were stained with DAPI, and a drop of FluorSaveTM Reagent (Millipore, Canada) was added before covering with coverslips. The slides were visualized with Olympus IX-81 (Olympus, Center Valley, PA, USA) and the images were processed with Volocity 6.3 (PerkinElmer Inc., USA).
Biofilm formation in microplates
Cultures at log phase (OD600 = 0.5) were inoculated into 1.5 mL of MRS at 1% individually or in combination with a second strain in Corning® Costar® TC-Treated Multiple Well Plates (Corning Inc., Canada) in eight independent biological replicates. The plates were incubated for 24 h at 37 °C with shaking at 200 rpm. The supernatant was removed and the remaining attached biofilm was photographed before being resuspended with 2 mL of phosphate-buffered saline. The suspended cultures were used for OD600 measurement and quantitative culturing on MRS agar. Strains in mixed biofilms were differentiated by colony morphology.
Aggregation assay
Cell aggregation of bacterial strains was determined as described previously [88, 89] with modifications in three independent biological replicates. Bacterial strains were grown for 18 h at 37 °C in mMRS broth. The concentration of the cells were normalized to an OD600 of three, harvested by centrifugation at 5000× g for 5 min, washed once with PBS, and resuspended in proteinase digestion buffered (50 mM Tris Cl, 10 mM EDTA, pH 8). If used, Proteinase K solution (Qiagen, Canada) was added at 10 μL/mL and the cell suspensions were incubated at 37 °C for 1 h. The cells with and without proteinase digestion were harvested by centrifugation, washed once with PBS, and resuspended in PBS to the original volume. Equal volume of each strain was used to prepare mixtures. A volume of 1.5 mL of each cell suspension was mixed by vortexing for 10 seconds. Auto- and coaggregation were determined after 1 h of incubation at room temperature. At the beginning and end of the incubation, 50 μL of the upper suspension was used to measure OD600 in 96-well plate with spectrophotometer. The aggregation percentage is expressed as: 1 − (At/A0)*100, where At represents the absorbance at 1 h and A0 the absorbance at 0 h [89].
Statistics
Statistical analyses were carried out using Graph Pad Prism 5 (GraphPad Software, Inc., California). Comparisons between single- and dual-species treatments were performed by unpaired Student’s t-test. Comparisons among more than three groups were performed with one-way ANOVA and Tukey’s multiple comparison test.
References
Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904.
Walter J, Britton RA, Roos S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci USA. 2011;108:4645–52.
Groussin M, Mazel F, Sanders JG, Smillie CS, Lavergne S, Thuiller W, et al. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat Commun. 2017;8:14319.
Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR, Kim J. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 2016;14:e2000225.
Moeller AH, Caro-quintero A, Mjungu D, Georgiev AV, Lonsdorf EV, Muller MN, et al. Cospeciation of gut microbiota. Science. 2016;353:380–2.
Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–5.
Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51.
Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207–17.
Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, et al. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev. 2017b;41:S27–S48.
Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, et al. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 2010;4:377–87.
Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, Zhang M, et al. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 2011;7:e1001314.
Duar RM, Frese SA, Lin XB, Fernando SC, Burkey TE, Tasseva G, et al. Experimental evaluation of host adaptation of Lactobacillus reuteri to different vertebrate species Dudley EG (ed). Appl Environ Microbiol. 2017a;83:e00132–17.
Frese SA, Mackenzie DA, Peterson DA, Schmaltz R, Fangman T, Zhou Y, et al. Molecular characterization of host-specific biofilm formation in a vertebrate gut symbiont. PLoS Genet. 2013;9:e1004057.
Savage DC, Dubos R, Schaedler RW. The gastrointestinal epithelium and its autochthonous bacterial flora. J Exp Med. 1968;127:67–76.
Schwab C, Tveit AT, Schleper C, Urich T. Gene expression of lactobacilli in murine forestomach biofilms. Microb Biotechnol. 2014;7:347–59.
Assefa S, Ahles K, Bigelow S, Curtis JT, Köhler GA. Lactobacilli with probiotic potential in the prairie vole (Microtus ochrogaster). Gut Pathog. 2015;7:35.
Leonard MT, Valladares RB, Ardissone A, Gonzalez CF, Lorca GL, Triplett EW. Complete genome sequences of Lactobacillus johnsonii strain N6.2 and Lactobacillus reuteri strain TD1. Genome Announc. 2014;2:e00397–14.
Li D, Chen H, Mao B, Yang Q, Zhao J, Gu Z, et al. Microbial biogeography and core microbiota of the rat digestive tract. Sci Rep. 2017;8:45840.
Marin IA, Goertz JE, Ren T, Rich SS, Onengut-Gumuscu S, Farber E, et al. Microbiota alteration is associated with the development of stress-induced despair behavior. Sci Rep. 2017;7:43859.
Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165:1762–75.
Buhnik-Rosenblau K, Matsko-Efimov V, Jung M, Shin H, Danin-Poleg Y, Kashi Y. Indication for co-evolution of Lactobacillus johnsonii with its hosts. BMC Microbiol. 2012;12:149.
Tannock GW, Wilson CM, Loach D, Cook GM, Eason J, O’Toole PW, et al. Resource partitioning in relation to cohabitation of Lactobacillus species in the mouse forestomach. ISME J. 2012;6:927–38.
Killer J, Havlík J, Vlková E, Rada V, Pechar R, Benada O, et al. Lactobacillus rodentium sp. nov., from the digestive tract of wild rodents. Int J Syst Evol Microbiol. 2014;64:1526–33.
Wang L.-T., Kuo H.-P., Wu Y.-C., Tai C.-J., Lee F.-L.. Lactobacillus taiwanensis sp. nov., isolated from silage. Int J Syst Evol Microbiol. 2009;59:2064–8.
Fabre PH, Hautier L, Dimitrov D, Douzery EJP. A glimpse on the pattern of rodent diversification: A phylogenetic approach. BMC Evol Biol. 2012;12:88.
Walter J, Schwab C, Loach DM, Gänzle MG, Tannock GW. Glucosyltransferase A (GtfA) and inulosucrase (Inu) of Lactobacillus reuteri TMW1.106 contribute to cell aggregation, in vitro biofilm formation, and colonization of the mouse gastrointestinal tract. Microbiol. 2008;154:72–80.
Walter J, Loach DM, Alqumber M, Rockel C, Hermann C, Pfitzenmaier M, et al. D-Alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100-23 results in impaired colonization of the mouse gastrointestinal tract. Environ Microbiol. 2007;9:1750–60.
Bronstein JL. The evolution of facilitation and mutualism. J Ecol. 2009;97:1160–70.
Wellborn GA, Langerhans RB. Ecological opportunity and the adaptive diversification of lineages. Ecol Evol. 2015;5:176–95.
Cucchi T,Vigne JD,Auffray JC, First occurrence of the house mouse (Mus musculus domesticus Schwarz & Schwarz, 1943) in the Western Mediterranean: a zooarchaeological revision of subfossil occurrences. Biol J Linn Soc. 2005;84(429):45
Weadick CJ, Chang BSW. An improved likelihood ratio test for detecting site-specific functional divergence among clades of protein-coding genes. Mol Biol Evol. 2012;29:1297–300.
Miller AW, Kohl KD, Dearing MD. The gastrointestinal tract of the white-throated Woodrat (Neotoma albigula) harbors distinct consortia of oxalate-degrading bacteria. Appl Environ Microbiol. 2014;80:1595–601.
Hansen LBS, Ren D, Burmølle M, Sørensen SJ. Distinct gene expression profile of Xanthomonas retroflexus engaged in synergistic multispecies biofilm formation. ISME J. 2017;11:300–3.
Ren D, Madsen JS, Sørensen SJ, Burmølle M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 2015;9:81–9.
Burmølle M, Webb JS, Rao D, Hansen LH, Sørensen SJ, Kjelleberg S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol. 2006;72:3916–23.
Zhu Y, Dashper SG, Chen YY, Crawford S, Slakeski N, Reynolds EC. Porphyromonas gingivalis and Treponema denticola synergistic polymicrobial biofilm development. PLoS ONE. 2013;8:e71727.
Jones K, Bradshaw SB. Synergism in biofilm formation between Salmonella enteritidis and a nitrogen-fixing strain of Klebsiella pneumoniae. J Appl Microbiol. 1997;82:663–8.
Varposhti M, Entezari F, Feizabadi MM, Varposhti M, Entezari F, Feizabadi MM. Synergistic interactions in mixed-species biofilms of pathogenic bacteria from the respiratory tract. Rev Soc Bras Med Trop. 2014;47:649–52.
Leung JW, Liu YL, Desta T, Libby E, Inciardi JF, Lam K, et al. Is there a synergistic effect between mixed bacterial infection in biofilm formation on biliary stents? Gastrointest Endosc. 1998;48:250–7.
Røder HL, Raghupathi PK, Herschend J, Brejnrod A, Knøchel S, Sørensen SJ, et al. Interspecies interactions result in enhanced biofilm formation by co-cultures of bacteria isolated from a food processing environment. Food Microbiol. 2015;51:18–24.
Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533:255–59.
Cowan SE, Gilbert E, Liepmann D, Keasling JD. Commensal interactions in a dual-species biofilm exposed to mixed organic compounds. Appl Environ Microbiol. 2000;66:4481–5.
Hansen SK, Rainey PB, Haagensen JAJ, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007b;445:533–6.
Christensen BB, Haagensen JAJ, Heydorn A, Molin S. Metabolic commensalism and competition in a two-species microbial consortium. Appl Environ Microbiol. 2002;68:2495–502.
Peterson SB, Dunn AK, Klimowicz AK, Handelsman J. Peptidoglycan from Bacillus cereus mediates commensalism with rhizosphere bacteria from the Cytophaga-Flavobacterium group. Appl Environ Microbiol. 2006;72:5421–7.
Nielsen AT, Tolker-Nielsen T, Barken KB, Molin S. Role of commensal relationships on the spatial structure of a surface- attached microbial consortium. Env Microbiol. 2000;2:59–68.
Hansen SK, Haagensen JAJ, Gjermansen M, Jørgensen TM, Tolker-Nielsen T, Molin S. Characterization of a Pseudomonas putida rough variant evolved in a mixed-species biofilm with Acinetobacter sp. strain C6. J Bacteriol. 2007a;189:4932–43.
Palmer J, Kazmerzak K, Hansen MC, Kolenbrander PE. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect Immun. 2001;69:5794–804.
Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100.
Yamada M, Ikegami A, Kuramitsu HK. Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol Lett. 2005;250:271–7.
Papke RT, Ramsing NB, Bateson MM, Ward DM. Geographical isolation in hot spring cyanobacteria. Environ Microbiol. 2003;5:650–9.
Wollenberg MS, Ruby EG. Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J. 2011;6:352–62.
Vos M. A species concept for bacteria based on adaptive divergence. Trends Microbiol. 2011;19:1–7.
Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7:1225–41.
Orsini L, Vanoverbeke J, Swillen I, Mergeay J, De Meester L. Drivers of population genetic differentiation in the wild: Isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol Ecol. 2013;22:5983–99.
Fukami T. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu Rev Ecol Evol Syst. 2015;46:1–23.
De Meester L, Vanoverbeke J, Kilsdonk LJ, Urban MC. Evolving perspectives on monopolization and priority effects. Trends Ecol Evol. 2016;31:136–46.
Stachowicz JJ. Mutualism, facilitation, and the structure of ecological communities: Postitive interactions play a critical, but underapprciated, role in ecological communities by reducing physical or biotic stresses in existing habitats and by reating new habitats on which many species depend. BioScience. 2001;51:235–46.
Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8: e1002606.
Leamy LJ, Kelly SA, Nietfeldt J, Legge RM, Ma F, Hua K et al. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol. 2014;15:552.
Ochman H, Worobey M, Kuo CH, Ndjango JBN, Peeters M, Hahn BH, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities Achtman M (ed). PLoS Biol. 2010;8:e1000546.
Linnenbrink M, Wang J, Hardouin EA, Künzel S, Metzler D, Baines JF. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol Ecol. 2013;22:1904–16.
Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209:1445–56.
Maldonado-Gómez MX, Martínez I, Bottacini F, O’Callaghan A, Ventura M, van Sinderen D, et al. Stable engraftment of Bifidobacterium longum AH1206 in the human Gut depends on individualized features of the resident microbiome. Cell Host Microbe. 2016;20:515–26.
Walter J, Maldonado-Gómez MX, Martínez I. To engraft or not to engraft: an ecological framework for gut microbiome modulation with live microbes. Curr Opin Biotechnol. 2018;49:129–39.
Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N, et al. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol. 2016;1:16131.
Weldon L, Abolins S, Lenzi L, Bourne C, Riley EM, Viney M. The gut microbiota of wild mice. PLoS ONE. 2015;10:e0134643.
Maidak BL, Larsen N, McCaughey MJ, Overbeek R, Olsen GJ, Fogel K, et al. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–7.
Nikolenko SI, Korobeynikov AI, Alekseyev MA. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genom. 2013;14:S7.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–5.
Joint Genome Institute. BBMap download | SourceForge.net. https://sourceforge.net/projects/bbmap/ Accessed 1 May 2017.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9.
Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3.
Keane JA, Page AJ, Delaney AJ, Taylor B, Seemann T, Harris SR, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016;2:e000056.
Sawyer S. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6:526–38.
Mirarab S, Warnow T. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics. 2015;31:i44–i52.
Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91.
Rambaut A (2007). FigTree. http://tree.bio.ed.ac.uk/software/figtree/ Accessed 1 May 2017.
Rodriguez-R LM, Konstantinidis KT. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016;4:e1900v1.
Corander J, Marttinen P, Sirén J, Tang J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinforma. 2008;9:539.
Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev. 2015;82:518–29.
Bridger JM, Volpi EV, (eds). Fluorescence in situ Hybridization (FISH).. Totowa, NJ: Humana Press; 2010.
de Vries MC, Vaughan EE, Kleerebezem M, de Vos WM. Optimising single cell activity assessment of Lactobacillus plantarum by fluorescent in situ hybridisation as affected by growth. J Microbiol Methods. 2004;59:109–15.
Schnell SA, Staines WA, Wessendorf MW. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J Histochem Cytochem. 1999;47:719–30.
Frece J, Kos B, Svetec IK, Zgaga Z, Mrsa V, Suskovic J. Importance of S-layer proteins in probiotic activity of Lactobacillus acidophilus M92. J Appl Microbiol. 2005;98:285–92.
Golowczyc MA, Mobili P, Garrote GL, Abraham AG, De Antoni GL, Li N, et al. Protective action of Lactobacillus kefir carrying S-layer protein against Salmonella enterica serovar Enteritidis. Int J Food Microbiol. 2007;118:264–73.
Acknowledgements
JW acknowledges support from the Campus Alberta Innovation Program (CAIP). This work was further supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) awarded to JW. TW acknowledges the financial support from China Scholarship Council, and JFB acknowledges support from the German Science Foundation (DFG) CRC 1182. We are grateful to Dr Stephen Ogg and Terri Fangman for assistance with confocal microscopy, and Misaph Alipour for assistance with FISH. Finally, we thank the staff in the animal facilities at both University of Nebraska Lincoln and University of Alberta for their support with animal husbandry.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Lin, X.B., Wang, T., Stothard, P. et al. The evolution of ecological facilitation within mixed-species biofilms in the mouse gastrointestinal tract. ISME J 12, 2770–2784 (2018). https://doi.org/10.1038/s41396-018-0211-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-018-0211-0
This article is cited by
-
A phylogenomic analysis of Limosilactobacillus reuteri reveals ancient and stable evolutionary relationships with rodents and birds and zoonotic transmission to humans
BMC Biology (2023)
-
A citizen-science-enabled catalogue of the vaginal microbiome and associated factors
Nature Microbiology (2023)
-
Complete genome sequencing of Peyer’s patches-derived Lactobacillus taiwanensis CLG01, a potential probiotic with antibacterial and immunomodulatory activity
BMC Microbiology (2021)