Abstract
The cosmopolitan coccolithophore Emiliania huxleyi is a unicellular eukaryotic alga that forms vast blooms in the oceans impacting large biogeochemical cycles. These blooms are often terminated due to infection by the large dsDNA virus, E. huxleyi virus (EhV). It was recently established that EhV-induced modulation of E. huxleyi metabolism is a key factor for optimal viral infection cycle. Despite the huge ecological importance of this host–virus interaction, the ability to assess its spatial and temporal dynamics and its possible impact on nutrient fluxes is limited by current approaches that focus on quantification of viral abundance and biodiversity. Here, we applied a host and virus gene expression analysis as a sensitive tool to quantify the dynamics of this interaction during a natural E. huxleyi bloom in the North Atlantic. We used viral gene expression profiling as an index for the level of active infection and showed that the latter correlated with water column depth. Intriguingly, this suggests a possible sinking mechanism for removing infected cells as aggregates from the E. huxleyi population in the surface layer into deeper waters. Viral infection was also highly correlated with induction of host metabolic genes involved in host life cycle, sphingolipid, and antioxidant metabolism, providing evidence for modulation of host metabolism under natural conditions. The ability to track and quantify defined phases of infection by monitoring co-expression of viral and host genes, coupled with advance omics approaches, will enable a deeper understanding of the impact that viruses have on the environment.
Similar content being viewed by others
Introduction
Marine viruses are considered to be major ecological, evolutionary, and biogeochemical drivers in the marine environment, responsible for nutrient recycling and determining species composition within microbial food webs [1,2,3,4]. Recent investigations into the molecular mechanisms that mediate host–virus interactions revealed that viruses rewire their host metabolic network during infection, generating a unique metabolic state (the “virocell” [5]) that supports their high metabolic requirements during active infection [6]. This phase of the viral infection cycle drives host–virus co-evolution and is considered to have a large effect on biogeochemical cycles [7,8,9].
The cosmopolitan coccolithophore Emiliania huxleyi is a unicellular eukaryotic alga that forms vast oceanic blooms [10,11,12,13]. Its intricate calcite exoskeleton accounts for ~1/3 of the total marine CaCO3 production [14,15,16]. Furthermore, E. huxleyi is also a key producer of dimethyl sulfide, a bioactive gas with a significant climate-regulating role that enhances cloud formation [17, 18]. Therefore, biotic interactions that regulate the fate of these blooms play a profound role in determining carbon and sulfur flow in the ocean.
Infection by E. huxleyi viruses (EhV) has been established as an important mortality factor and a major cause of E. huxleyi bloom demise [19,20,21,22]. High-throughput gene expression profiling during infection, using controlled laboratory cultures of E. huxleyi infected by EhV strains, enabled to associate expression of viral genes to different phases of infection [23,24,25]. In addition, viral infection led to rapid remodeling of the host transcriptome and resulted in rewiring of metabolic pathways encoding antioxidant networks, de novo fatty acid and sphingolipid biosynthesis, and membrane recycling via autophagy machinery [24, 26,27,28,29]. Quantification of such metabolic products can serve as novel biomarkers to detect viral infection in natural E. huxleyi populations. Recent studies successfully utilized a unique virus-derived glycosphingolipid to detect E. huxleyi–EhV interactions in the lab, mesocosm and natural communities in the North Atlantic [22, 30, 31].
The current understanding on the impact of viruses on the marine environment is mainly derived from “virus-centric” approaches which are based on assessment of abundance and diversity of viral particles (i.e., virions). These approaches are focus on quantification of virions or~viral DNA using microscopy, flow cytometry, qPCR [20, 32,33,34,35], or by applying advanced ecogenomics approaches [36,37,38]. While these approaches allow quantification of viral DNA during the infection process, they cannot directly detect active viral infection in the form of virocells. Consequently we still lack quantitative tools to measure the direct impact of viral infection on its micro and macro-environment. Here we show that assessment of host and virus co-expressed genes can increase our resolution of in situ detection and tracking of the virocell dynamics during viral infection of E. huxleyi blooms in the ocean.
Materials and methods
Satellite images
Surface chlorophyll (Chl) and particulate inorganic carbon (PIC) data are derived products from radiances measured by the Moderate resolution imaging spectroradiometer (MODIS) aboard the Aqua satellite. The spatial and temporal resolutions are 1 km and 1 day. Level 2 MODIS data were downloaded from Ocean Color Web (http://oceancolor.gsfc.nasa.gov/), and mapped using the SeaDAS software. Station locations were clustered as core or periphery (blue and red dots in Fig. 1c, d) according to the clustering of water temperature profiles (Supplementary Fig. S1a).
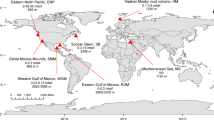
a Sampling stations in the North Atlantic (black dots) obtained during NAVICE cruise (June–July 2012). b Sampling stations within an eddy (black dots). Stations that were included in the data analysis are within the red rectangle. A cross section through the eddy is represented by a red dashed line. Satellite imagery (MODIS aqua) of c Particulate inorganic carbon (PIC, mg m−3) and d chlorophyll (Chl, mg m−3) used as proxies for a coccolithophore bloom. Dots in c and d represent sampling locations in the core (blue) or periphery (red) of the eddy, determined by the clustering of water temperature profiles at the stations (Supplementary Fig. S1a)
Environmental sampling and biomass collection
In situ measurements were taken during the North Atlantic Virus Infection of Coccolithophore Expedition (NAVICE; KN207-03, http://www.bco-dmo.org/dataset-deployment/455468), aboard the R/V Knorr (www.bco-dmo.org/project/2136). Water samples collection within the eddy (61.5–61.87°N/33.5–34.1°W) began on 30 June and continued for 6 days while drifting over the same water mass. The area was sampled again between 7 and 10 July. Samples were obtained from 5-6 depths using a Sea-Bird SBE 911plus CTD carrying 10 L Niskin bottles. Biomass from 1 to 2 L of seawater was collected on 0.8 μm polycarbonate filters (Millipore), flash-frozen in liquid nitrogen, and stored at −80 °C until further processing. In addition, biomass from 250 mL of laboratory samples of uninfected E. huxleyi cultures and 48 h post infection were collected on 1.0 μm polycarbonate filters (Millipore) flash-frozen in liquid nitrogen, and stored at −80 °C until further processing.
Enumeration of phytoplankton populations abundances by flow cytometry
Flow cytometry analyses were performed on an Eclipse iCyt flowcytometer (Sony Biotechnology Inc., Champaign, IL, USA) equipped with 405 and 488 nm solid-state air cooled lasers, and with standard optic filter set-up. Calcifying cells were identified by plotting the chlorophyll fluorescence (663–737 nm, Chl) against light side scattering (SS) and were quantified by counting the high-chlorophyll high SS events in addition to nano-eukaryotes (Neuks), pico-eukaryotes and Cyanobacteria (Peuks + Cyano, gated together) and phycoerythrin-rich Synechococcus (PE-rich Synechococcus; Supplementary Fig. S2).
Quantification of E. huxleyi and biomass-associated EhV by quantitative PCR (qPCR)
We quantify viral DNA copy number and estimated relative abundance of E. huxleyi DNA derived from biomass collected on 0.8 μm polycarbonate filters (Millipore) as indicated above. Genomic DNA was extracted using an adapted phenol–chloroform method previously described by Schroeder et al. [39]. Filters were cut into small, easily dissolved pieces and placed in a 2 mL tube. Following addition of 800 μL of GTE buffer (50 mM glucose, 25 mM Tris-HCl (pH 8.0), and 10 mM EDTA), 10 μg mL−1 proteinase K, and 100 μL of 0.5 M filter-sterilized EDTA, samples were incubated at 65 °C for 1–2 h. An aliquot of 200 μL of a 10% stock solution of SDS was then added and DNA was then purified by phenol extraction and ethanol precipitation. Sample were then diluted 1:10 with DDW and cleaned using AMPure XP beads (Beckman Coulter) as described by the manufacturer using 1:1.8 sample to beads volumetric ratio. For all reactions Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen) was used as described by the manufacturer. Reactions were performed on StepOnePlus real-time PCR System (Applied Biosystems) as follows: 50 °C for 2 min, 95 °C for 2 min, 40 cycles of 95 °C for 15 s, 60 °C for 30 s.
E. huxleyi DNA relative abundance was estimated using qPCR with primers designed to target E. huxleyi cytochrome c oxidase subunit 3 (cox3) gene: Eh-COX3 F1: 5′-tcctacacttggatatttag-3′, Eh-COX3 R1: 5′-tcgcatttttggtttggaagacc-3′ [40]. Biomass-associated EhV DNA was quantified using primers against the Major Capsid Protein (mcp) gene, mcp1F: 5′-acgcaccctcaatgtatggaagg-3′ and mcp90Rv: 5′-agccaactcagcagtcgttc-3′. All reactions were carried out in triplicates. Results were calibrated against serial dilutions of EhV201 DNA at known concentrations.
RNA isolation, RT-PCR, and high-throughput qPCR gene expression evaluation of host and viral genes
RNA from biomass of environmental and laboratory samples was isolated with the RNeasy Plant Mini kit (Qiagen) according to manufacturer’s instructions, followed by DNAse treatment with Turbo DNAse (Ambion). RNA and DNA concentrations were measured using QUBIT Fluorometer with high sensitivity RNA and dsDNA detection kits (Life Technologies) in order to quantify residual DNA level in RNA extractions after DNAse treatment (Supplementary Table S1). RNA was reverse transcribed to cDNA with the ThermoScript RT-PCR system (Invitrogen). The same procedure was conducted, for specific samples with high DNA concentration, without RT enzyme to control for DNA contamination. To assess the efficiency of RT reactions, an External RNA Controls Consortium (ERCC) RNA Spike In-Mix (Ambion) was introduced into the reaction buffer and compared for its level of amplification between samples (e.g., Et level; Supplementary Fig. S3 and Supplementary Table S1) by means of qPCR. StepOnePlus real-time PCR System (Applied Biosystems) was used as follows: 50 °C for 2 min, 95 °C for 2 min, 40 cycles of 95 °C for 15 s, 60 °C for 30 s. qPCR reaction was targeted for the most abundant RNA molecule in the spike mix (ERCC-00074), using the primer pair: F: 5′- CTTCCCATCTTCTTTGAGAGTTGTT-3′ R: 5′-GCCTTATGTGATAGATGCCTCTTTAA-3′. For all reactions Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen) was used as described by the manufacturer. Data was analyzed using StepOnePlus Software (Applied Biosystems).
The BioMark HD system was used for high-throughput qPCR using standard fast cycling conditions and melt-curve analysis, generating an amplification curve for each gene of interest (Supplementary Table S3) in each sample, following the manufacturer's instructions (Fluidigm). Primers for qPCR amplification were either used as described in previous work or designed using Primer Express software (Applied Biosystems, Supplementary Table S3). Reactions were performed in duplicates for each sample and analyzed using Real-time PCR Analysis software (Fluidugm). Primer efficiency was calculated according to reaction dynamics using the Real-time PCR Miner algorithm (http://ewindup.info/miner/, [41], Supplementary Table S3). Cycle threshold (Ct) values for each reaction which correspond to the logarithm of the fold change in gene expression was used for analysis. Minimal threshold level of detection was set as 30 Ct and the expression threshold values (Et) was calculated by linearly transforming the data so that minimal Et is zero (30-Ct). Mean Et value was calculated between technical duplicates. Any average value with Et difference greater than 2 between duplicates was removed from the analysis. In order to calibrate the system and validate our primers, we used samples of the calcifying E. huxleyi RCC 1216 strain, this host cells were either uninfected or infected with EhV201 or EhV86 for 48 h. These RNA samples were used to validate the robustness of the Biomark HD technology, and to examine the efficiency of the primers as well as positive controls for the samples from natural populations. Accordingly, Et values for any primer sets (host genes: Supplementary Table S4, or viral genes: Supplementary Table S5) that did not yield positive amplification in laboratory samples were removed from the analysis. Et values were normalized by the volume of water sampled during the cruise.
Statistical analysis
For hierarchical clustering with heat-map visualization, normalized Et values were first standardized using z-score function. The data was then analyzed and displayed using the function clustrogram in MATLAB (MathWorks). Principle component analysis was conducted on normalized Et values using the principle component analysis (PCA) function in MATLAB (MathWorks). Pearson’s linear correlation, and Student's t test were performed, using MATLAB (MathWorks). Multiple linear regression was done using R, v. 3.4.1., with infection index or phase index as the dependent variable, and with depth, RNA concentration, and ERCC spike Et value as the independent variables. Box plots were produced using BoxPlotR web tool (http://shiny.chemgrid.org/boxplotr/) [68].
Results and discussion
We aimed to follow active E. huxleyi–EhV interaction during a natural algal bloom in the North Atlantic Ocean. In situ data was obtained aboard the R/V Knorr as part of the “North Atlantic Virus Infection of Coccolithophore Expedition” (NAVICE; KN207-03, http://www.bco-dmo.org/project/2136; Fig. 1a, b). We followed a patch of an E. huxleyi bloom occupying an eddy by satellite imagery of PIC (Fig. 1c) and chlorophyll (Fig. 1d), features that are typically used as specific proxies for detection of coccolithophore blooms by remote sensing [11, 42,43,44,45]. These large scale signatures were coupled to micro-scale measurement of abundance of calcifying coccolithophore cells based on flow cytometry analysis (Supplementary Fig. S2). In order to plot the spatial distribution of in situ parameters we aligned all sample sites within the eddy to a section through its center (Fig. 1b; red dashed line). To define the eddy’s physical boundaries we examined the water temperature profiles that showed a difference between the core and periphery of the eddy. Namely, cooler water was detected below the surface water layer in the core of the eddy (Supplementary Fig. S1a, c). This physical property (Supplementary Fig. S1a–d), combined with the clockwise rotation of the eddy [46] suggested the occurrence of an anticyclonic mode-water eddy. This physical condition was previously shown to provide a flux of nutrients that are essential for the stimulation of algal blooms [47, 48].
The abundance of calcifying cells in the eddy reached a maximal density of ~3300 cells mL−1 which is similar to previous studies of E. huxleyi blooms in the North Atlantic [19, 49,50,51]. Maximal calcifying cells abundance differed in its depth between the core and periphery of the eddy and was situated at ~15 and ~30 m, respectively (Fig. 2a). This pattern coincided with the distribution of physical parameters of the water within the eddy (Supplementary Fig. S1) and corresponds to a known feature of E. huxleyi blooms whereby algal cells occupy the surface mixed layer, above the pycnocline (the depth of maximum gradient in water density), usually not deeper than 30 m [45, 49, 50, 52, 53]. As a complementary approach, we quantified the relative abundance of E. huxleyi using qPCR using E. huxleyi specific primers for COX3 gene (Fig. 2b). This analysis corresponded with the abundance pattern of calcifying cells determined by flow cytometry and confirmed that E. huxleyi was present in the bloom. The qPCR analysis also revealed high abundance of E. huxleyi DNA deeper in the water column than was detected by the flow cytometry analysis (Fig. 2a, b).
Section distance of the eddy (Fig. 1b, dashed red line) and depth distributions of (a) E. huxleyi cell abundance in the water measured by flow cytometry, b E. huxleyi relative abundance measured by qPCR for COX3 gene, and c EhV abundance measured by qPCR for the MCP gene (presented in log10 values) in biomass samples collected during NAVICE. Black dots indicate sampling points. Figure panels were prepared using Ocean Data View (ODV) version 4.7.7, [67]
In order to quantify EhV abundance we first applied an established methods for quantifying abundance of free virions in the water by flow cytometry [54] and qPCR [24]. Nevertheless, both methods were unable to detect viral abundance in most of the samples throughout the NAVICE cruise, even when water samples were concentrated between 100–500 times. To overcome this, we quantified viral DNA copy number derived from bloom-biomass collected from several liters of water. Quantification of biomass-associated viral DNA showed that EhV was most abundant in the eddy periphery, reaching concentrations of ~1.6 × 105 viral DNA copies mL−1 (Fig. 2c), similar to the viral concentrations found in the Western English Channel [19]. The maximal abundance of EhV in the core of the eddy was two orders of magnitude lower than in the periphery, reaching 1.3 × 103 viral DNA copies mL−1 (Fig. 2c). Using biomass-associated viral DNA we could increase detection sensitivity, but lacked the ability to discriminate between free virions, viral DNA within infected cells or viruses attached to cells. In order to detect active viral infection within infected cells (virocell) we used host and virus gene expression profiling. We quantified the level of expression of 47 E. huxleyi and 25 EhV genes in biomass collected throughout the bloom by high-throughput qPCR (Materials and methods). Specific genes were chosen based on previous culture-based transcriptomic studies that examined E. huxleyi–EhV interactions during different phases [23, 24]. We examined the expression levels of specific host genes involved in diverse metabolic pathways that were essential for optimal EhV infection, such as reactive oxygen species (ROS), sphingolipids, triacylglycerols, and autophagy [24, 26,27,28,29]. Viral genes were chosen according to their temporal expression pattern during different phases of infection (early, mid, and late. Supplementary Table S3, [23, 24]).
Hierarchical clustering of the 37 environmental samples (Supplementary Table S1 for sample description) according to their expression profiles of viral genes divided the samples into two main clusters. A cluster of 14 samples in which most of the viral genes were expressed, and a second cluster of 23 samples where only few were expressed (Fig. 3a). In order to reduce the high complexity and multi-dimensionality of the gene expression data we performed a PCA (Fig. 3b). The coefficient values of the first component (PC1, Fig. 3c), namely, the correlation between each gene expression profile and the PC1 score values, indicated that all viral gene expression profiles contributed positively to PC1. PC1 was also highly correlated to the mean viral gene expression in all samples (Supplementary Fig. S4, r = 0.92, p = 4.37×10−11 by Pearson linear correlation). Accordingly, we used the score value of PC1 as a novel index for the level of expression of viral genes in each sample, and termed it “infection index”. This new proxy for viral infection retained the separation of the samples into two clusters as seen in the hierarchical clustering (Fig. 3a, b, red “x” symbol and blue “+” symbol).
a Heat-map representation of viral gene expression levels (Et value, color scale bar) of 25 viral genes (x axis, infection phase and gene number) at 37 environmental samples, specifying station number and water depth (y axis), arranged by hierarchical clustering. b Principle component analysis (PCA) of the viral expression values for each environmental sample plotted over the axes of 1st component (PC1) vs. 2nd component (PC2). Samples are marked in a blue “+” or a red “x” according to the hierarchical clustering in the heat-map (a). c Coefficient values of viral genes for PC1 (x axis) and PC2 (y axis). The labels “Early” in blue, “Mid” in green, and “Late” in red represent the association of each viral gene to different phases of infection based on transcriptomic analysis [23, 24]. The gene number is depicted according to Supplementary Table S3
The coefficient values of the second component (PC2) for each gene expression profile (Fig. 3c) correlated with the phase of viral infection as determined based on the expression profiles of early, mid and late viral genes [23, 24] (Supplementary Fig. S5, difference between early and late tested using Student's t test, p = 0.05). Accordingly, genes with higher coefficient values of PC2 are more likely to be expressed at late stages of infection. We used the score values of PC2 to categorize between samples that exhibited expression signatures typical to either early, mid or late stages of infection, and termed it “phase index”. Interestingly, the phase index was more variable within samples with low infection index (red “x” symbols in Fig. 3b), while in samples with high infection index (blue “+” symbols in Fig. 3b) the distribution of the phase index (PC2) was narrower. This suggests that it may be possible to resolve the distinct phases of viral infection with more sensitivity in samples with low infection index (e.g., low extent of infection in the cells population).
We further applied our newly developed viral infection indices for detection of active infection and examined its spatial distribution in our study site. We detected high levels of active infection in the periphery of the eddy (Fig. 4a). In the core of the eddy this index exposed two hotspots of active infection which were not apparent by using quantification of viral DNA (Fig. 2c). Interestingly, high E. huxleyi abundance, (determined by flow cytometry, Fig. 2a), did not coincide with high active viral infection (infection index, Fig. 4a). Active viral infection was more prominent in deeper depths, where E. huxleyi abundance seems to be low. Since flow cytometry can discriminate only calcifying single cells, it excludes naked (non-calcifying) cells and cell’s aggregates from the analysis. Examination of the E. huxleyi DNA relative abundance (Fig. 2b) and the expression levels of E. huxleyi genes (Supplementary Fig. S6) revealed significant signals below the pycnocline, where the concentration of free calcifying single cells was low (Fig. 2a). Intriguingly, both the infection index (Fig. 4a) and the phase index (Fig. 4b) correlated with depth (r = 0.35, p = 0.032 and r = 0.38, p = 0.02, respectively by Pearson linear correlations, Fig. 4c, d). Taken together, these observed patterns suggest that infected cells from surface areas may be exported to the depths by an active virus-induced aggregation and sinking mechanism in infected population.
Section distance of the eddy (Fig. 1b, dashed red line) and depth distributions of a infection index, b phase index in biomass samples collected during NAVICE. Black dots indicate sampling points. Figure panels were prepared using Ocean Data View (ODV) version 4.7.7, [67]. Scatter plots indicating the correlation between depth and c infection index or d phase index (r = 0.35, p = 0.032 and r = 0.38, p = 0.02, respectively, by Pearson linear correlation)
In order to validate that the correlation between depth and infection indices was not derived from variability in other parameters that could influence gene expression analysis (e.g., RNA concentrations), we conducted a multiple linear regression model. Infection index or phase index were used as the dependent variables and depth, RNA concentration, and ERCC RNA spike Et value (Material and methods) as the independent variables. RNA concentration and ERCC spike effects were not significant and were therefore removed from the models, leaving only depth as the significant factor (F1,28 = 4.97, p = 0.034 for infection index and F1,28 = 5.26, p = 0.030 for phase index).
To detect in situ host response to viral infection, we calculated the correlations between the infection index and the respective expression of E. huxleyi genes in all samples. Ten E. huxleyi metabolic genes showed significant correlation to the viral infection index (r > 0.3, p < 0.05 by Pearson’s linear correlation, Table 1). Most genes were related to life cycle, metabolism of ROS and sphingolipids which are strongly remodeled during viral takeover of the virocell [24, 26, 28, 29]. This analysis demonstrates a virus-induced modulation of host metabolism under natural bloom conditions.
By investigating host–virus micro-scale interactions within the bloom, we aimed to shed light on the biogeochemical impact of viral infection. Viral infection of E. huxleyi was recently reported to induce production of transparent exopolymer particles (TEP) which may potentially modulate the stickiness of cells and hence their ability to aggregate and, consequently, affect carbon export [22, 55]. With estimated density lower than 0.85 g cm−3 TEP is considered to contribute to particle ascending velocity, and not to sinking [56]. Nevertheless, TEP was suggested to be important factor promoting the high carbon export rate measured during E. huxleyi blooms [57, 58]. E. huxleyi cells are decorated by biogenic minerals (CaCO3 in the coccoliths) that could contribute the missing ballast for the initiation of sinking [59,60,61]. Recent evidence from the Tara-Ocean global survey has directly linked enhanced carbon flux with specific plankton communities and cyanophages groups [62]. Additionally, increased sinking rates were reported for the alga Heterosigma akashiwo undergoing viral infection [63]. Moreover, 7000 years old EhV DNA was retrieved from the sediments in the black sea, indicating an effective export mechanism of viruses from the surface bloom [33]. Accordingly, we propose that aggregation and sinking of infected cells may be the mechanism explaining the high active infection below the pycnocline as presented here (Fig. 4). Co-expression of host and virus genes below the surface layer (Fig. 4 and Table 1) corroborated the finding that intact coccolith-bearing cells, rather than free coccoliths, are the main contributors to PIC export bellow the mixed layer [64]. Therefore, sensitive detection of early stages of active viral infection could potentially predict the occurrence of high carbon flux. Indeed, carbon flux at 50 m, from the same E. huxleyi patch, which we examined in this this study, reached above 350 mg −2 per day (249 and 109 mg C m−2 per day for POC and PIC, respectively). The highest along the entire NAVICE cruise track [65]. This carbon flux measurement was within the range of carbon export measured previously for E. huxleyi blooms in the North Atlantic [61].
Despite the acknowledgment that marine viruses have a huge ecological importance in shaping metabolic fluxes in the marine environment, the ability to accurately assess their ecological impact is still challenging. Current approaches in aquatic virology are mainly virocentric, focusing on quantification of viral abundance [20, 32,33,34,35], diversity [19, 21] and global distribution using ecogenomics approaches [36,37,38, 66]. However, it is necessary to develop sensitive tools to assess the activity and dynamics of infected cells (virocells), in order to achieve insights into the impact of viruses on marine microbial food webs. We propose that quantification of host and virus transcriptomic signatures allows sensitive tracking of the state of viral infection and serves as a complementary approach to the classical methods. As we demonstrated here, this approach allows us to detect active infection below the pycnocline which is typically undetectable due to low virion concentrations. This approach can specifically identify the state of the virocell including detection of early phases of infection. Other approaches, such as quantification of specific viral derived metabolites (e.g., viral glycosphingolipids-vGSL for detection of EhV) provide detection of the later phases of infection, in which intracellular concentrations reach detectable levels. These specific metabolites can act as biomarkers for in situ monitoring of active viral infection [22, 31]. vGSL is also a major component of the virion membranes [26, 31], its quantification will also correspond to the level of virion abundance and not only to infected cells.
To conclude, the ability to track and quantify defined phases of infection by host and virus gene co-expression, coupled with enumeration of free virions, greatly facilitates the quantification of active viral infection in the ocean. This approach opens new avenues to assess the dynamics of infection and its impact on the surrounding micro-environment and macro-environment. Together with recent efforts to map viral diversity on a global scale, future studies will enable better assessment of the impact of viral infection on microbial food webs and, consequently, the flux of carbon in the marine environment.
References
Breitbart M. Marine viruses: truth or dare. Ann Rev Mar Sci. 2012;4:425–48.
Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–12.
Suttle Ca. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12.
Wilhelm SW, Suttle CA. Viruses and nutrient cycles in the sea. Bioscience. 1999;49:781.
Forterre P. The virocell concept and environmental microbiology. ISME J. 2013;7:233–6.
Rosenwasser S, Ziv C, Creveld SG, van, Vardi A. Virocell metabolism: metabolic innovations during host–virus interactions in the ocean. Trends Microbiol. 2016;24:821–32.
Mojica KDA, Huisman J, Wilhelm SW, Brussaard CPD. Latitudinal variation in virus-induced mortality of phytoplankton across the North Atlantic Ocean. ISME J. (2015) 10; 500–513.
Sheik AR, Brussaard CPD, Lavik G, Lam P, Musat N, Krupke A, et al. Responses of the coastal bacterial community to viral infection of the algae Phaeocystis globosa. ISME J. 2013;8:1–14.
Bidle KD, Vardi A. A chemical arms race at sea mediates algal host-virus interactions. Curr Opin Microbiol. 2011;14:449–57.
Brown CW, Yoder JA. Coccolithophorid blooms in the global ocean. J Geophys Res Ocean. 1994;99:7467–82.
Holligan PM, Viollier M, Harbour DS, Camus P, Champagne-Philippe M. Satellite and ship studies of coccolithophore production along a continental shelf edge. Nature. 1983;304:339–42.
Balch WM, Holligan PM, Kilpatrick Ka. Calcification, photosynthesis and growth of the bloom-forming coccolithophore, Emiliania huxleyi. Cont Shelf Res. 1992;12:1353–74.
Winter A, Henderiks J, Beaufort L, Rickaby REM, Brown CW. Poleward expansion of the coccolithophore Emiliania huxleyi. J Plankton Res. 2014;36:316–25.
Bolton CT, Hernández-Sánchez MT, Fuertes M-Á, González-Lemos S, Abrevaya L, Mendez-Vicente A, et al. Decrease in coccolithophore calcification and CO2 since the middle Miocene. Nat Commun. 2016;7:10284.
Gal A, Wirth R, Kopka J, Fratzl P, Faivre D, Scheffel A. Macromolecular recognition directs calcium ions to coccolith mineralization sites. Science. 2016;353:590–3.
Rost B, Riebesell U. Coccolithophores and the biological pump: responses to environmental changes. In: Thierstein HR, Young JR, editors. Coccolithophores. Berlin, Heidelberg: Springer; 2004. p. 99–125.
Alcolombri U, Ben-Dor S, Feldmesser E, Levin Y, Tawfik DS, Vardi A. Identification of the algal dimethyl sulfide-releasing enzyme: A missing link in the marine sulfur cycle. Science. 2015;348:1466–9.
Simó R. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol Evol. 2001;16:287–94.
Highfield A, Evans C, Walne A, Miller PI, Schroeder DC. How many Coccolithovirus genotypes does it take to terminate an Emiliania huxleyi bloom? Virology. 2014;466-467:138–45.
Lehahn Y, Koren I, Schatz D, Frada M, Sheyn U, Boss E, et al. Decoupling physical from biological processes to assess the impact of viruses on a mesoscale algal bloom. Curr Biol. 2014b;24:2041–6.
Martínez JM, Schroeder DC, Wilson WH. Dynamics and genotypic composition of Emiliania huxleyi and their co-occurring viruses during a coccolithophore bloom in the North Sea. FEMS Microbiol Ecol. 2012;81:315–23.
Vardi A, Haramaty L, Van Mooy BaS, Fredricks HF, Kimmance Sa, Larsen A, et al. Host-virus dynamics and subcellular controls of cell fate in a natural coccolithophore population. Proc Natl Acad Sci USA. 2012;109:19327–32.
Allen MJ, Forster T, Schroeder DC, Hall M, Roy D, Ghazal P, et al. Locus-specific gene expression pattern suggests a unique propagation strategy for a giant algal virus. J Virol. 2006;80:7699–705.
Rosenwasser S, Mausz MAMA, Schatz D, Sheyn U, Malitsky S, Aharoni A, et al. Rewiring host lipid metabolism by large viruses determines the fate of emiliania huxleyi, a bloom-forming alga in the ocean. Plant Cell. 2014;26:2689–707.
Wilson WH, Schroeder DC, Allen MJ, Holden MTG, Parkhill J, Barrell BG, et al. Complete genome sequence and lytic phase transcription profile of a coccolithovirus. Science. 2005;309:1090–2.
Malitsky S, Ziv C, Rosenwasser S, Zheng S, Schatz D, Porat Z, et al. Viral infection of the marine alga Emiliania huxleyi triggers lipidome remodeling and induces the production of highly saturated triacylglycerol. New Phytol. 2016;210:88–96.
Schatz D, Shemi A, Rosenwasser S, Sabanay H, Wolf SG, Ben-Dor S, et al. Hijacking of an autophagy-like process is critical for the life cycle of a DNA virus infecting oceanic algal blooms. New Phytol. 2014;204:854–63.
Sheyn U, Rosenwasser S, Ben-Dor S, Porat Z, Vardi A. Modulation of host ROS metabolism is essential for viral infection of a bloom-forming coccolithophore in the ocean. ISME J. 2016;10:1742–54.
Ziv C, Malitsky S, Othman A, Ben-Dor S, Wei Y, Zheng S, et al. Viral serine palmitoyltransferase induces metabolic switch in sphingolipid biosynthesis and is required for infection of a marine alga. Proc Natl Acad Sci USA. 2016;113:E1907–E1916.
Fulton JM, Fredricks HF, Bidle KD, Vardi A, Kendrick BJ, Ditullio GR, et al. Novel molecular determinants of viral susceptibility and resistance in the lipidome of Emiliania huxleyi. Environ Microbiol. 2013;16: 1137–49.
Vardi A, Van Mooy BAS, Fredricks HF, Popendorf KJ, Ossolinski JE, Haramaty L, et al. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science. 2009;326:861–5.
Brussaard CPD. Optimization of procedures for counting viruses by flow cytometry. Appl Environ Microbiol. 2004;70:1506–13.
Coolen MJL. 7000 years of Emiliania huxleyi viruses in the Black Sea. Science. 2011;333:451–2.
Frada MJ, Schatz D, Farstey V, Ossolinski JE, Sabanay H, Ben-Dor S, et al. Zooplankton may serve as transmission vectors for viruses infecting algal blooms in the ocean. Curr Biol. 2014;24:2592–7.
Schroeder DC, Oke J, Hall M, Malin G, Wilson WH. Virus succession observed during an Emiliania huxleyi bloom. Appl Environ Microbiol. 2003;69:2484–90.
Brum JR, Ignacio-Espinoza JC, Roux S, Doulcier G, Acinas SG, Alberti A, et al. Ocean plankton. Patterns and ecological drivers of ocean viral communities. Science. 2015;348:1261498.
Culley AI, Lang AS, Suttle CA. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–8.
Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368.
Schroeder DC, Oke J, Malin G, Wilson WH. Coccolithovirus (Phycodnaviridae): characterisation of a new large dsDNA algal virus that infects Emiliana huxleyi. Arch Virol. 2002;147:1685–98.
Bendif EM, Probert I, Carmichael M, Romac S, Hagino K, de Vargas C. Genetic delineation between and within the widespread coccolithophore morpho-species Emiliania huxleyi and Gephyrocapsa oceanica (Haptophyta). J Phycol. 2014;50:140–8.
Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–64.
Brown C. Global distribution of coccolithophore blooms. Oceanography. 1995;8:59–60.
Sharoni S, Trainic M, Schatz D, Lehahn Y, Flores MJ, Bidle KD, et al. Infection of phytoplankton by aerosolized marine viruses. Proc Natl Acad Sci USA. 2015;112:6643–7.
Siegel H, Ohde T, Gerth M, Lavik G, Leipe T. Identification of coccolithophore blooms in the SE Atlantic Ocean off Namibia by satellites and in-situ methods. Cont Shelf Res. 2007;27:258–74.
Tyrrell T, Merico A. Emiliania huxleyi: bloom observations and the conditions that induce them. In: Thierstein HR, Young JR, editors. Coccolithophores. Berlin, Heidelberg: Springer; 2004. p. 75–97.
Lehahn Y, Koren I, Rudich Y, Bidle KD, Trainic M, Flores JM, et al. Decoupling atmospheric and oceanic factors affecting aerosol loading over a cluster of mesoscale North Atlantic eddies. Geophys Res Lett. 2014;41:4075–81.
Ledwell JR, McGillicuddy DJ, Anderson LA. Nutrient flux into an intense deep chlorophyll layer in a mode-water eddy. Deep Sea Res Part II Top Stud Oceanogr. 2008;55:1139–60.
McGillicuddy DJ, Anderson LA, Bates NR, Bibby T, Buesseler KO, Carlson CA, et al. Eddy/wind interactions stimulate extraordinary mid-ocean plankton blooms. Science. 2007;316:1021–6.
Holligan PM, Fernández E, Aiken J, Balch WM, Boyd P, Burkill PH, et al. A biogeochemical study of the coccolithophore, Emiliania huxleyi, in the North Atlantic. Glob Biogeochem Cycles. 1993;7:879–900.
Townsend DW, Keller MD, Holligan PM, Ackleson SG, Balch WM. Blooms of the coccolithophore Emiliania huxleyi with respect to hydrography in the Gulf of Maine. Cont Shelf Res. 1994;14:979–1000.
Wilson WH, Tarran GA, Schroeder D, Cox M, Oke J, Malin G. Isolation of viruses responsible for the demise of an Emiliania huxleyi bloom in the English Channel. J Mar Biol Assoc UK. 2002;82:369–77.
Cokacar T, Kubilay N, Oguz T. Structure of Emiliania huxleyi blooms in the Black Sea surface waters as detected by SeaWIFS imagery. Geophys Res Lett. 2001;28:4607–10.
Berge G. Discoloration of the sea due to Coccolithus huxleyi ‘bloom’. Sarsia. 1962;6:27–40.
Brussaard CPD, Payet JP, Winter C, Weinbauer MG. Quantification of aquatic viruses by flow cytometry Wilhelm S, Weinbauer M, Suttle C (eds). Man Aquat Viral Ecol. 2010;11:102–9.
Mari X, Kerros M-E, Weinbauer MG. Virus attachment to transparent exopolymeric particles along trophic gradients in the southwestern lagoon of New Caledonia. Appl Environ Microbiol. 2007;73:5245–52.
Azetsu-Scott K, Passow U. Ascending marine particles: significance of transparent exopolymer particles (TEP) in the upper ocean. Limnol Oceanogr 2004;49:741–8.
Harlay J, De Bodt C, Engel A, Jansen S, D’Hoop Q, Piontek J, et al. Abundance and size distribution of transparent exopolymer particles (TEP) in a coccolithophorid bloom in the northern Bay of Biscay. Deep Sea Res Part I Oceanogr Res Pap. 2009;56:1251–65.
Schmidt S, Harlay J, Borges AV, Groom S, Delille B, Roevros N, et al. Particle export during a bloaom of Emiliania huxleyi in the North-West European continental margin. J Mar Syst. 2013;109-110:S182–S190.
Francois R, Honjo S, Krishfield R, Manganini S. Factors controlling the flux of organic carbon to the bathypelagic zone of the ocean. Glob Biogeochem Cycles. 2002;16:34-1–34–20.
Iversen MH, Ploug H. Ballast minerals and the sinking carbon flux in the ocean: Carbon-specific respiration rates and sinking velocity of marine snow aggregates. Biogeosciences. 2010;7:2613–24.
Klaas C, Archer DE. Association of sinking organic matter with various types of mineral ballast in the deep sea: Implications for the rain ratio. Glob Biogeochem Cycles. 2002;16:63–1-63–14.
Guidi L, Chaffron S, Bittner L, Eveillard D, Larhlimi A, Roux S, et al. Plankton networks driving carbon export in the oligotrophic ocean. Nature. 2016;532:465–70.
Lawrence JJE, Suttle CA. Effect of viral infection on sinking rates of Heterosigma akashiwo and its implications for bloom termination. Aquat Microb Ecol. 2004;37:1–7.
Balch WM, Kilpatrick KA, Trees CC. The 1991 coccolithophore bloom in the central North Atlantic. 1. Optical properties and factors affecting their distribution. Limnol Oceanogr. 1996;41:1669–83.
Collins JR, Edwards BR, Thamatrakoln K, Ossolinski JE, DiTullio GR, Bidle KD, et al. The multiple fates of sinking particles in the North Atlantic Ocean. Glob Biogeochem Cycles. 2015;29:1471–94.
Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537:689–93.
Schlitzer R. Interactive analysis and visualization of geoscience data with Ocean Data View. Comput Geosci. 2002;28:1211–8.
Spitzer M, Wildenhain J, Rappsilber J, Tyers M. BoxPlotR: a web tool for generation of box plots. Nat Methods. 2014;11:121–2.
Acknowledgments
We thank the captain and crew of the R/V Knorr during the NAVICE cruise as well as the Marine Facilities and Operations at the Woods Hole Oceanographic Institution for assistance and cooperation at sea. We thank Shifra Ben-Dor for her contribution to the bioinformatics analysis. This research was supported by a European Research Council (ERC) StG (INFOTROPHIC grant # 280991) and CoG (VIROCELLSPHERE grant # 681715) as well as by the National Science Foundation (NSF) grant OCE-1061883 (to AV and KDB).
Author Contributions
US, SR, and AV conceived and design the experiments and analyses. US and SR analyzed the expression data. DS and NBG preformed and analyzed E. huxleyi and viral DNA qPCR enumeration. US analyzed all data including the cruise data. YL and IK analyzed satellite imagery. KDB was the chief scientist on the NAVICE cruise and implemented corresponding ship-based sampling strategies. RR preformed statistical analysis. US, AV, NBG, SR, and DS contributed to the writing of the manuscript. AV supervised the project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Sheyn, U., Rosenwasser, S., Lehahn, Y. et al. Expression profiling of host and virus during a coccolithophore bloom provides insights into the role of viral infection in promoting carbon export. ISME J 12, 704–713 (2018). https://doi.org/10.1038/s41396-017-0004-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-017-0004-x
This article is cited by
-
The consequences of viral infection on protists
Communications Biology (2024)
-
Disease resistance and infectivity of virus susceptible and resistant common carp strains
Scientific Reports (2024)
-
Characterization of the sphingolipid profiling of Emiliania huxleyi against virus infection
Journal of Oceanology and Limnology (2023)
-
Viral infection switches the balance between bacterial and eukaryotic recyclers of organic matter during coccolithophore blooms
Nature Communications (2023)
-
Analyses of key gene networks controlling carotenoid metabolism in Xiangfen 1 banana
BMC Plant Biology (2022)