Abstract
A lasting imbalance between fatty acid synthesis and consumption leads to non-alcoholic fatty liver disease (NAFLD), coupled with hepatitis and insulin resistance. Yet the details of the underlying mechanisms are not fully understood. Here, we unraveled that the expression of the transcription factor Zbtb18 is markedly decreased in the livers of both patients and murine models of NAFLD. Hepatic Zbtb18 knockout promoted NAFLD features like impaired energy expenditure and fatty acid oxidation (FAO), and induced insulin resistance. Conversely, hepatic Zbtb18 overexpression alleviated hepato-steatosis, insulin resistance, and hyperglycemia in mice fed on a high-fat diet (HFD) or in diabetic mice. Notably, in vitro and in vivo mechanistic studies revealed that Zbtb18 transcriptional activation of Farnesoid X receptor (FXR) mediated FAO and Clathrin Heavy Chain (CLTC) protein hinders NLRP3 inflammasome activity. This key mechanism by which hepatocyte’s Zbtb18 expression alleviates NAFLD and consequent liver fibrosis was further verified by FXR’s deletion and forced expression in mice and cultured mouse primary hepatocytes (MPHs). Moreover, CLTC deletion significantly abrogated the hepatic Zbtb18 overexpression-driven inhibition of NLRP3 inflammasome activity in macrophages. Altogether, Zbtb18 transcriptionally activates the FXR-mediated FAO and CLTC expression, which inhibits NLRP3 inflammasome’s activity alleviating inflammatory stress and insulin resistance, representing an attractive remedy for hepatic steatosis and fibrosis.
Similar content being viewed by others
Introduction
Chronic or excessive lipids exposure resulting from prolonged high caloric intake frequently leads to steatohepatitis (steatosis with inflammation), liver fibrosis, cirrhosis, and eventually even hepatocellular carcinoma, representing the most relevant etiological factor of chronic liver diseases.1,2,3,4 Alarmingly, the incidence of lipid disorders in both adults and children is steadily rising due to the ongoing metabolic syndrome epidemic, which also entails obesity and diabetes.5,6
Mounting lines of evidence indicate that imbalances between intrinsic lipogenesis and FAO-derived lipid consumption contributes to lipotoxicity and NAFLD occurrence.7,8,9,10 In mammals, an abundant lipid deposition within the hepatocytes severely impairs the lysosomal-mitochondrial interaction in a vicious ROS-JNK feed-forward loop, eventually causing hepatocytes’ death.11,12 Moreover, lipotoxic injury stimulates liver macrophages accumulation and activates the NLRP3 inflammasome assembly and activation, driving the release of pro-inflammatory cytokines, such as TNF-α, IL-6 and IL-1β, providing a key pathogenetic link to nonalcoholic steatohepatitis (NASH) progression.13,14 Previous studies proved that pharmacological or genetic restoration of an impaired liver FAO alleviated steatohepatitis and hindered liver fibrosis progression by decreasing ROS overproduction or by reducing lipid peroxidation and NLRP3 inflammasome activity.15 However, the detailed molecular mechanisms causing steatohepatitis development remain incompletely understood and no effective or prospective therapeutic approaches to this highly prevalent disease are presently available.
Currently, the worthy and promising but limited therapeutic approaches to hepatic lipid disorders focus mainly on nuclear receptor remodeling.16 Among them, the activation of PPARα (peroxisome proliferator-activated receptor α) and FXR (farnesoid X receptor) have shown discrete effects.17 As a hepatic and intestinal highly expressed nuclear receptor, FXR is closely involved in bile acid metabolism and, as proven by a recent study, has organ-dependent different functions on lipid metabolism by remodeling NLRP3-mediated inflammasome activity.18,19 In NAFLD patients or mice models, the treatment with optocollic acid (OCA), an FXR agonist, exerted a set of relevant hepatic effects, decreasing TAGs (triacylglycerols) and inflammation, improving insulin sensitivity, and mitigating steatohepatitis and liver fibrosis.20 Deletion of hepatic FXR or its target genes predisposes mice to hyperlipemia and insulin resistance, quickly resulting in the acute NASH phenotype when fed on an HFD due to irreversible lipotoxicity, oxidative burden, and steatohepatitis.21 The stability and transcriptional activity of FXR might be regulated by endogenous proteins, such as SIRT6, and KLF16.22 Yet, the details about the involved molecular mechanisms are far from clear.
The Zbtb18 (Zinc Finger and BTB Domain Containing 18) gene encodes a C2H2-type zinc finger protein and shares a subclass of conservative POK (POZ and Krüppel)/BTB domains. Previous reports revealed that Zbtb18 interacts with CtBP2 (C-Terminal Binding Protein 2) gene to promote glioblastoma malignancy.23 Recent studies also showed that Zbtb18 repressed transcriptional programs closely linked to non-neuronal cell identity and glioblastoma subtypes, participating in neurodevelopmental diseases.24 However, it is obscure that whether Zbtb18 participates the hepatic glucolipid metabolism.
In this study, we showed that the hepatic Zbtb18 could activates FXR transcription, and subsequently accelerates the hepatic FAO, thereby preventing the onset and development of NAFLD. Moreover, the Zbtb18/FXR axis-stimulated CLTC protein expression remarkably inhibits NLRP3 inflammasome’s activity and alleviated liver inflammatory infiltrations and liver fibrosis. Therefore, the Zbtb18/FXR axis represents a novel candidate to target for the treatment of NAFLD and NASH.
Results
Hepatic Zbtb18 down-regulation is closely related to the development of NAFLD
To achieve a systematic and comprehensive identification of key signaling molecules involved in the development of steatohepatitis, we first performed a mRNA microarray analysis of liver tissues from NAFLD patients and normal control, and subsequently crossed this clinic transcriptome data with published transcriptome data (GSE213621 from NAFLD patients and GSE35961 from mice model with NAFLD) to identify the differentially expressed genes (DEGs). The results exhibited there were 274, 411, and 792 DEGs from the 3 transcriptomes respectively. Additionally, 33 DEGs were overlapped among the three datasets. Of note, we found that hepatic Zbtb18 expression was downregulated among these transcriptome data (Fig. 1a & Supplementary Fig. 1a–d). Furthermore, we confirmed the significant decrease in Zbtb18 mRNA and protein in liver biopsies from NAFLD patients (Fig. 1b & Supplementary Table 1). Consistently, we also found the reduction of hepatic Zbtb18 expression in several NAFLD mice models, including db/db diabetic mice, ob/ob obese mice, and HFD-induced obese mice (Fig. 1c–e). In addition, hepatic Zbtb18 expression was markedly reduced in oleic acid & palmitic acid (OA & PA) cultured mouse primary hepatocytes (MPHs) (Fig. 1f–g). To preliminarily probe the physiological function of Zbtb18, we generated Ad-Zbtb18-infected MPHs and found that Zbtb18 overexpression effectively increased the expression of genes involved in FAO and oxidative phosphorylation (OXPHOS), while it significantly suppressed the genes involved in gluconeogenesis (Fig. 1h, i). Accordingly, the Zbtb18 overexpression increased fatty acid catabolism while decreasing lipid deposition in the MPHs (Fig. 1j–l). Therefore, our findings suggested that the hepatic Zbtb18 was negatively related to the development and progression of NAFLD.
Hepatic Zbtb18 mRNA and protein down-regulation is closely related to the development of NAFLD. a Venn diagram representing common significantly changed transcripts in liver from mice with NAFLD (GSE35961, P < 0.01), NAFLD patients (GSE213621, P < 0.05) and our clinic transcript data (P < 0.05). b–e Representative quantitative PCR and Western blot analysis of hepatic Zbtb18 in clinical samples (b, normal controls = 7, NAFLD patients = 8); diabetic mice (c, n = 6); obese mice (d, n = 6); HFD-fed mice (e, n = 6). f, g Representative quantitative PCR, Western blot and immunofluorescence (IF) analysis of Zbtb18 in MPHs treated with OA&PA; n = 4. h Representative quantitative PCR and Western blot data show an effective overexpression of Zbtb18 in Ad-Zbtb18 infected MPHs; n ≥ 5. i Representative quantitative PCR data show that Zbtb18 overexpression significantly upregulates the genes involved in FAO and OXPHOS; n ≥ 5. j Zbtb18 overexpression elevates the FAO rates in MPHs; n = 4. k, l Zbtb18 overexpression decreases the lipid accumulation (k) and TGs contents (l) in MPHs; n = 6. Data are shown as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001
Ablation of liver Zbtb18 makes mice more prone to steatohepatitis
To explore the function of hepatic Zbtb18, we first generated hepatocyte-specific Zbtb18 deletion (Zbtb18LKO) mice (Supplementary Fig. 2a). The Zbtb18flox/flox mice were used as the control of Zbtb18LKO mice. We found that Zbtb18 deficiency led to severe lipid accumulation in cultured MPHs isolated from the Zbtb18LKO mice (Supplementary Fig. 2b). Interestingly, hepatic Zbtb18 deletion did not affect the body weight gain and energy expenditure of mice kept on a standard diet (SD) (Supplementary Fig. 2c, d). Yet, the blood glucose and serum insulin levels were increased in Zbtb18LKO mice, even when fed on the SD (Supplementary Fig. 2e). Moreover, these mice displayed an impairment of glucose tolerance and insulin sensitivity, suggesting that hepatic Zbtb18 was required to maintain glucose homeostasis (Supplementary Fig. 2f). Subsequent assays revealed that hepatic Zbtb18LKO mice obviously increased the liver weight/body weight ratio value, and the circulating and hepatic TGs contents, which might lead to slight but visible lipid deposition in the liver (Supplementary Fig. 2g–i). Besides, Zbtb18LKO mice exhibited increased serum ALT and AST levels (Supplementary Fig. 2j). In mechanistic terms, we preliminarily found that Zbtb18 deletion suppressed the expression of genes involved in FAO and OXPHOS, which confirmed that Zbtb18 is an important mediator of lipid catabolism in the liver (Supplementary Fig. 2k). Moreover, increased fatty acid oxidation by Zbtb18 overexpression resulted in an increased serum ketone body, which is closely related to abnormal lipid deposition (Supplementary Fig. 2l).
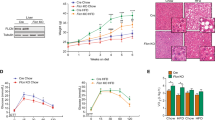
Next, we prolongedly kept these mice on a high-fat diet (HFD) to further explore the physiological role of hepatic Zbtb18 in lipid metabolism. We found that hepatic Zbtb18 deletion easily predisposed the mice to obesity after a lengthy HFD exposure, as illustrated by their higher body weight and fat mass (Fig. 2a, b). Consistently, histological examination (H&E staining) also revealed an increased adipocyte hypertrophy of epididymal white adipose tissue (WAT), inguinal WAT, and interscapular brown adipose tissue (BAT) in Zbtb18LKO mice (Fig. 2c). Correspondingly, these mice displayed a suppressed expression of thermogenic genes in the interscapular BAT, which might have contributed to their gain in body weight (Fig. 2d). Then, we housed the mice in metabolic cages and found that, compared to control mice, the Zbtb18LKO mice showed lower degrees of energy expenditure, respiratory O2 consumption, and CO2 production (Fig. 2e). More importantly, Zbtb18LKO mice showed an increase in fasting blood glucose and insulin levels in mice following HFD feeding (Fig. 2f & Supplementary Fig. 3a). Besides, Zbtb18LKO mice exhibited an impairment of glucose tolerance and insulin sensitivity (Fig. 2g). Subsequent western blotting analyses also demonstrated the decrease in AKT and GSK-3β phosphorylation in livers from the Zbtb18LKO mice, further suggesting the impaired insulin sensitivity in these mice (Fig. 2h). Moreover, hepatic Zbtb18 deletion decreased the genes involved in FAO and OXPHOS, and up-regulated glycogenic genes, collectively increasing lipid deposition in the livers of mice fed on an HFD (Fig. 2i, j & Supplementary Fig. 3b, c). We also observed the decreased serum ketone body levels, increased serum TG and TC, coupled with elevation of serum pro-inflammatory cytokines levels in Zbtb18LKO mice, which might be the result of the development and progression of NAFLD (Supplementary Fig. 3d–f).
Hepatic ablation of Zbtb18 aggravates steatohepatitis in mice fed on HFD. a, b Hepatic Zbtb18 deletion leads to increases in body weight (a) and fat mass percentage (b) in mice fed on HFD; n ≥ 5. c Adipocyte hypertrophy of epididymal WAT, inguinal WAT and interscapular BAT of hepatic Zbtb18 deleted mice fed on HFD. d, e Hepatic Zbtb18 deletion decreases thermogenic genes (d) and energy expenditure (e) in BAT of mice fed on HFD; n ≥ 5. f, g Hepatic Zbtb18 deletion increases the fasting blood glucose (f) and impairs glucose tolerance, and insulin sensitivity (g) in mice fed on HFD; n = 6. h Hepatic Zbtb18 deficiency decreases the phosphorylation of AKT and GSK-3β in the livers of mice fed on HFD. i, j Hepatic Zbtb18 deficiency alters the function of genes related to glucose and lipid metabolism (i) and leads to severe fatty liver phenotype (j) in mice fed on HFD, n = 6. Data are shown as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005
Hepatic Zbtb18 overexpression safeguards from HFD-induced hepato-steatosis
To verify whether an increase in hepatic Zbtb18 could improve the hepatic lipid and glucose homeostasis, we generated liver-specific Zbtb18 overexpressing (Zbtb18LKI) mice via crossing Alb-Cre mice and Rosa26-Zbtb18 mice (Supplementary Fig. 4a, b). The ZTBT18Rosa26 knock-in mice were used as the control of Zbtb18LKI. We first exposed these mice to HFD and found that the Zbtb18 overexpression reduced body weight, which might ascribe to the decreases in fat mass and liver weight (Fig. 3a, b & Supplementary Fig. 4c). Moreover, compared to littermate control mice, Zbtb18LKI exhibited enhanced energy expenditure, respiratory O2 consumption and CO2 production coupled with an increased Ucp1 (Uncoupling protein 1) gene expression in BAT eventually contributing to the decrease in body weight (Fig. 3c, d). Of note, Zbtb18 overexpression effectively alleviated HFD-induced hepatic steatosis, including the improvements in hepatocyte ballooning and lipid deposition (Fig. 3e). Besides, Zbtb18LKI mice showed an increased serum ketone body levels and reduced hepatic and serum TGs levels (Fig. 3f, g). Correspondingly, Zbtb18 overexpression increased the expression of genes involved in lipolysis, FAO, and OXPHOS, while genes related to lipogenesis were slightly changed (Fig. 3h). Furthermore, the improvement of excessive hepatic lipid accumulation effectively mitigated HFD-induced liver lipotoxicity. This alleviated the infiltration of inflammatory cells and the release of inflammatory cytokines, eventually reversing the hepatitis caused by HFD and improving liver function, which was reflected by reductions in the expression of inflammatory genes and serum ALT and AST levels (Fig. 3i–l). Consistently, due to the hepatic Zbtb18 overexpression, these mice displayed lower fasting blood glucose and insulin levels (Fig. 3m, n). Hepatic Zbtb18 overexpression also improved glucose tolerance and insulin sensitivity in the mice fed on HFD, and elevated the phosphorylation of AKT and GSK-3β in livers, suggesting a recovery from the altered insulin signaling due to HFD exposure (Fig. 3o, p). Moreover, the improvement of insulin sensitivity was further confirmed by the increase in phosphorylation of AKT and GSK-3β in MPHs after Zbtb18 overexpression (Supplementary Fig. 5).
Hepatic Zbtb18 overexpression defends against HFD-induced hepatic steatosis. a, b Hepatic Zbtb18 overexpression decreases body weight (a) and fat mass (b) of mice fed on HFD; n ≥ 4. c, d Hepatic Zbtb18 overexpression increases energy expenditure (c) and Ucp1 expression in the BAT (d) of mice fed on HFD; n ≥ 5. e, f Hepatic Zbtb18 overexpression improves the hepatic steatosis (e), increases serum ketone body levels (f) and decreases TGs contents in serum and livers of mice fed on HFD; n = 6. g, h Hepatic Zbtb18 overexpression decreased serum and hepatic TG contents (g) and altered hepatic genes related to glucose and lipid metabolism (h) in mice fed on HFD; n ≥ 5. i Hepatic Zbtb18 overexpression decreases F4/80+ cells in the livers of mice fed on HFD. j, k Hepatic Zbtb18 overexpression decreases the mRNA levels of inflammatory genes (j) and serum proinflammatory cytokines levels (k) of mice fed on HFD; n = 6. l Hepatic Zbtb18 overexpression decreases serum ALT and AST levels in mice fed on HFD; n = 6. m–o Hepatic Zbtb18 overexpression decreases fasting blood glucose (m), and insulin levels (n) and improves glucose intolerance and insulin resistance (o) in mice fed on HFD; n = 6. p Hepatic Zbtb18 overexpression enhances the phosphorylation of AKT and GSK-3β in the livers of mice fed on HFD. Data are shown as means ± SEM. *P < 0.05; **P < 0.01
To determine the physiological function of hepatic Zbtb18 in defending from glucose and lipid disorders, we next injected adeno-associated virus (AAV)-Zbtb18 into hepatic Zbtb18LKO mice to rescue hepatic Zbtb18 expression. Thus, we found that a rescued expression of Zbtb18 slightly decreased the body weight of the hepatic Zbtb18-deficient mice (Supplementary Fig. 6a). It also decreased the abnormal lipid deposition in the liver, as shown by the lowered serum and hepatic TG levels, and by an attenuation of the otherwise massive accumulations of large lipid droplets and of the ballooning degeneration of hepatocytes (Supplementary Fig. 6b, c). Besides, the fasting blood glucose and insulin levels were significantly decreased in the Zbtb18LKO mice after AAV-Zbtb18 injection (Supplementary Fig. 6d). Moreover, the hepatic Zbtb18 rescue improved the glucose intolerance and insulin resistance of Zbtb18LKO mice, also beneficially inducing enhanced phosphorylation of AKT and GSK-3β in Zbtb18-deficient liver (Supplementary Fig. 6e, f). Furthermore, the rescued expression of hepatic Zbtb18 accelerated the liver FAO rate, as shown by the up-regulation of genes involved in FAO and OXPHOS, coupled with the increased serum ketone body levels (Supplementary Fig. 6g, h). Moreover, the rescued expression of hepatic Zbtb18 significantly reduced the serum ALT and AST levels, indicating an improvement of hepatic function in these mice (Supplementary Fig. 6i). Collectively, these data proved that hepatic Zbtb18 played an essential role in keeping the whole body’s glucose and lipid balance.
Forced liver Zbtb18 expression alleviates hepatic steatosis in diabetic mice
Given the predominant role of hepatic Zbtb18 in protecting diet-induced glucose and lipid disorders, we next tested whether its overexpression would improve the fatty liver phenotype in diabetic db/db mice. Thus, the hepatic Zbtb18 expression was increased via tail vein injection with AAV-Zbtb18 in db/db mice (Supplementary Fig. 7). Consistently, AAV-Zbtb18-infected db/db mice displayed reduced body weight and decreased liver weight/body weight ratio values (Fig. 4a, b). AAV-mediated hepatic Zbtb18 overexpression also markedly improved the fatty liver phenotype in db/db mice, as revealed by the gross changes of TGs contents and by histological analysis (Fig. 4c, d). Likewise, the AAV-Zbtb18 infected db/db mice showed decreased fasting blood glucose and insulin levels (Fig. 4e). The latter results implied an improvement of the glucose disorder in db/db mice, which was further confirmed by the ameliorated glucose intolerance and insulin resistance (Fig. 4f). Moreover, elevated serum ketone body levels were also noticed in AAV-Zbtb18 injected db/db mice (Fig. 4g). Correspondingly, hepatic Zbtb18 overexpression significantly increased the expression of genes involved in fatty acid oxidation, while glucogenic genes including Pgc-1α and Pepck were suppressed (Fig. 4h). A concurrently increased phosphorylation of AKT and GSK-3β in the liver, contributed to the recovery from insulin dysfunction in db/db mice (Fig. 4i). At the same time, Zbtb18 overexpressing db/db mice showed significantly decreased gathering of hepatic F4/80 and Cd11b-positive macrophages within the liver, which together with the reduced inflammatory cytokines levels (Fig. 4j, k). All of these changes contributed to the improvement of hepatic dysfunction, as proven by the decreased serum ALT and AST levels in these mice (Fig. 4l). Altogether, these data indicated that targeting hepatic Zbtb18 represents an effective approach to hinder the progression of liver steatosis in db/db mice.
Rescued hepatic Zbtb18 expression alleviates hepato-steatosis in diabetic mice. a–c Rescued hepatic Zbtb18 expression decreases the body weight (a), the ratio values of liver weight to body weight (b) and TGs contents in the serum and livers (c) of db/db mice; n = 6. d AAV-Zbtb18 infected db/db mice show an improved liver steatosis phenotype. e, f Hepatic Zbtb18 overexpression significantly reduces fasting blood glucose and insulin levels (e), and improves glucose tolerance and insulin sensitivity (f) of db/db mice; n = 6. g, h Hepatic Zbtb18 overexpression increases serum ketone body levels (g) and alters the expression of genes related to glucose and lipid metabolism (h) in db/db mice; n = 6. i Rescued Zbtb18 expression in livers increases the phosphorylation of AKT and GSK-3β in db/db mice. j Hepatic Zbtb18 overexpression decreases F4/80+ and Cd11b+ cells in the livers of db/db mice. k, l Hepatic Zbtb18 overexpression reduces serum proinflammatory cytokines levels (k) and ALT, and AST levels (l) in db/db mice, n = 6. Data are shown as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001
Zbtb18 accelerates lipid catabolism via the transcriptional activation of FXR-mediated FAO
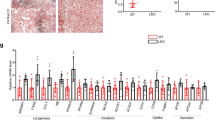
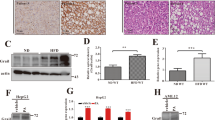
To investigate the details about the effectors or signaling nodes involved in Zbtb18-mediated protective effects of the liver, the RNA-seq was performed on the Zbtb18 overexpressing MPHs. Thus, we found that 7347 genes were differently changed due to the Zbtb18 overexpression, of which 3628 were up-regulated and 3719 were down-regulated (Fig. 5a). Subsequent Gene Ontology (GO) analysis indicated that Zbtb18 overexpression along with an obvious significant altering of the hepatic inflammation and fatty acid metabolism pathway (Fig. 5b). To gain insight into the underlying mechanisms by which Zbtb18 regulated hepatic fatty acid metabolism and inflammation, we analyzed the overlapping DEGs identified from the Zbtb18 overexpressing MPHs and the liver from the NAFLD patients. Notably, the results showed that most overlapping DEGs were involved in the FXR target genes (Fig. 5c, d). Moreover, gene set enrichment analysis (GSEA) of the above DEGs identified in Zbtb18 overexpressing MPHs and the liver from the NAFLD patients showed that “FXR signaling pathway” was positively correlated with Zbtb18 expression, suggesting that the expression of these DEGs may be driven by FXR-dependent mechanisms (Fig. 5e). In addition, up-regulated FXR mRNA and protein levels were noticed in the Zbtb18 overexpressing livers (Fig. 5f–g). Consistently, Zbtb18 overexpression also significantly increased both of the total and nuclear levels of FXR protein and its target genes in cultured MPHs (Fig. 5h).
Zbtb18 transcriptional activation of FXR accelerates lipid catabolism via FAO. a, b RNA-seq data of Zbtb18-overexpressing and control MPHs show that Zbtb18 alters the expression of genes related to FAO and the FXR signaling pathway, as shown by Scatter plot (a) and KEGG analysis (b). c Venn diagram showing the overlapping significantly differentially expressed transcripts identified in MPHs overexpressing Zbtb18 (P < 0.01) and the liver form the NAFLD patients (P < 0.01). d Heatmap of transcriptome data showing the mRNA levels of genes involved in the FAO and FXR signaling pathway. e GSEA analysis of cellular components of DEGs identified in Zbtb18-overexpressing primary hepatocytes and the patients with NAFLD. f, g Representative quantitative PCR and Western blot analysis of FXR and of its target genes in hepatic Zbtb18 overexpressing mice and control mice, n ≥ 5. h Immunofluorescence analysis reveals that Zbtb18 overexpression increases FXR and SHP levels in the cytoplasm and nucleus of cultured MPHs. i Map of the FXR locus revealing Zbtb18 binding in MPHs; aligned reads were visualized by Integrated Genomics Viewer 2. The signal of the IgG or Anti-Zbtb18 is represented with gray or red peaks, respectively. j The predicted Zbtb18-binding motif. k, l Luciferase assays indicate that Zbtb18 protein binds to a unique site (−993 bp to −984 bp) to transcriptionally activate FXR expression, n ≥ 5. m ChIP-qPCR analyses of the Zbtb18 protein occupancy on the FXR promoter; n = 3. n FXR deletion diminished the Zbtb18 protein-driven protective effects on lipid accumulation. o, p FXR deletion reduces the stimulatory effects on FAO induced by Zbtb18 protein overexpression; n = 4 (o) and TGs contents (p) in MPHs; n ≥ 3. q Forced expression of FXR counteracts the Zbtb18-deficiency-induced elevation of TGs contents in MPHs; n ≥ 4. Data are shown as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001
To further explore the underlying mechanisms by which Zbtb18 activates FXR, Zbtb18-specific ChIP-seq was performed in MPHs. The result revealed a significant Zbtb18-binding promoter region in the FXR gene (Fig. 5i). Besides, we further analyzed the ChIP-seq data and confirmed that “AACTCACTCT”, which was located in the promoter region of FXR, was a key binding motif of Zbtb18 (Fig. 5j). Then, the luciferase assays using a series of reported constructs (pFXR-1838, pFXR-1074, and pFXR-677) in HepG2 and MIHA cells. We found that Zbtb18 markedly stimulated the transcriptional activity of pFXR-1838 and pFXR-1074, while pFXR-677 elicited no effects, indicating a potential binding site for the Zbtb18 transcription factor from −1074 bp to −677 bp (Fig. 5k, l, left). More importantly, mutation of the AACTCACTCT in the promoter region of FXR to TTTTTTTTTT diminished the Zbtb18-induced stimulatory effects on FXR transcriptional activity, suggesting that its binding site is located in the region of FXR (Fig. 5k, l, right). Additionally, our ChIP-qPCR analysis revealed that the occupancy of Zbtb18 transcription factor on FXR’s promoter was significantly decreased in the livers of db/db mice and HFD-fed mice, which further details the mechanism underlying FXR dysregulation in these mice (Fig. 5m). To further validate the key role of FXR in mediating the Zbtb18-induced protective effects, we generated FXR-knockout MPHs and found that FXR deletion significantly weakened the Zbtb18-derived protective effects on FAO leading to an unaltered lipid accumulation level in MPHs (Fig. 5n–p). Conversely, FXR overexpression alleviated the Zbtb18 deficiency stimulated lipid deposition in MPHs (Fig. 5q). Collectively, these data implied that a strong correlation existed between Zbtb18-mediated transcriptional activation of FXR and hepatocellular lipid homeostasis.
Hepatic Zbtb18 inhibits NLRP3 inflammasome’s activation in macrophages via an FXR-mediated CLTC protein expression
Emerging evidence suggests that a hepatocellular lipid imbalance always initiates and promotes an inflammation in the liver which in turn aggravates NAFLD progression.25 To comprehensively explore the impact of Zbtb18 on NAFLD development, we also investigated the changes in NLRP3 inflammasome’s assembly and activation, which plays a key role in the liver inflammation in hepatic Zbtb18 gain or loss models. Consistently, hepatic Zbtb18 deletion substantially intensified the HFD-induced liver expression of the inflammasome-related proteins, including NLRP3, ASC, and Caspase-1. These effects were significantly reversed by a hepatic Zbtb18 overexpression (Fig. 6a, b). Our results suggested a suppressing role of hepatic Zbtb18 on NLRP3-mediated inflammasome activation, which was also verified in AAV-Zbtb18 infected db/db mice (Fig. 6c). Subsequently, we performed the proteomic analysis of the supernatants of Ad-Zbtb18 infected hepatocytes to uncover the key transmitter involved in the hepatocellular Zbtb18-driven suppression of inflammation. Thus, we found that Zbtb18 overexpression significantly increased the abundance of proteins related to the inflammatory response, especially the CLTC protein’s amount, which is closely related to macrophage activity and potentially suppresses hepatic inflammation (Supplementary Fig. 8a–c), as previously reported.26,27,28 Subsequent experiments conducted on macrophages treated with the indicated cultured medium, also confirmed this assumption by showing that inflammasome’s activity was suppressed in macrophages cultured in Ad-Zbtb18 infected hepatocytes conditioned medium (Fig. 6d). Interestingly, FXR deletion effectively abrogated the Zbtb18-elicited inhibition of NLRP3 inflammasome related proteins, while FXR overexpression effectively blocked the NLRP3 inflammasome’s abnormal activation in hepatic Zbtb18-deleted mice (Fig. 6e–f). Of note, the Zbtb18-stimulated CLTC protein expression was almost abolished by the FXR deletion in the hepatocytes and mice (Supplementary Fig. 8c, d). And deletion of CLTC in hepatocytes effectively diminished the inhibition of the NLRP3 inflammasome in macrophages induced by Zbtb18 conditioned medium in vitro (Fig. 6g–h). Collectively, these results suggested that hepatic Zbtb18 expression inhibited NLRP3 inflammasome’s activation in macrophages via an FXR-mediated CLTC protein expression, which helped mitigate NAFLD in mice.
Hepatic Zbtb18 protein inhibits NLRP3 inflammasome’s activation in macrophage via an FXR-mediated CLTC protein expression. a Hepatic Zbtb18 deletion increases HFD-induced inflammasome-related proteins NLRP3, ASC, Caspase-1, and NF-кB phosphorylation in livers. b Hepatic Zbtb18 overexpression decreases HFD-induced inflammasome-related proteins NLRP3, ASC, Caspase-1, and NF-кB phosphorylation in livers. c Hepatic Zbtb18 overexpression decreases inflammasome-related proteins NLRP3, ASC, Caspase-1, and NF-кB phosphorylation in the livers of db/db mice. d Representative Western blot analyses indicated that the treatment with the conditioned medium of Ad-Zbtb18 infected MPHs cultures suppressed the OA&PA&LPS-induced expression of inflammasome related proteins, NLRP3, ASC, Caspase-1, and NF-κb phosphorylation in BMDM macrophages. e FXR deletion diminishes the hepatic Zbtb18 overexpression-induced protective effects on inflammasome-related proteins NLRP3, ASC, Caspase-1, and NF-кB phosphorylation in livers. f Hepatic FXR forced expression rescued the Zbtb18 protein deficiency-induced expression of inflammasome-related proteins NLRP3, ASC, Caspase-1, and NF-кB phosphorylation in livers. g, h Representative Western blot and Immunofluorescence data show that the treatment with a conditioned medium of Ad-Zbtb18-infected CLTC knockout MPHs cultures fails to alter the OA&PA&LPS-induced expression of inflammasome related proteins NLRP3, ASC, Caspase-1, and NF-кB phosphorylation in BMDM cells
FXR activity effectively mediated the Zbtb18-induced attenuation of hepato-steatosis
In keeping with the above results, we tested the physiological function of FXR in mediating the Zbtb18-driven beneficial effects in vivo via tail vein injection of AAV-Zbtb18 into FXR-deleted mice. Thus, we found that FXR deletion almost abolished the Zbtb18-induced reduction of body weight and fat mass, with rare changes in the energy expenditure in HFD mice (Fig. 7a, b & Supplementary Fig. 9a). Besides, the serum ketone body level was rarely changed in FXR-deleted mice after AAV-Zbtb18 injection (Fig. 7c). Moreover, hepatic Zbtb18 overexpression failed to alter the expression of hepatic genes involved in FAO (Fig. 7d & Supplementary Fig. 9b). This results in unchanged TGs contents in the serum and the livers of AAV-Zbtb18 infected FXR-knockout mice, coupled with unchanged ballooning and lipid droplet accumulation in the hepatocytes (Fig. 7e, f & Supplementary Fig. 9c). Similarly, FXR deletion abrogated the decline of fasting blood glucose and insulin levels in serum after Zbtb18 overexpression (Fig. 7g). In addition, the AAV-Zbtb18 infected FXR-deficient mice showed no changes in glucose tolerance and insulin resistance (Fig. 7h). These results suggested a critical role of FXR in mediating hepatic Zbtb18-induced protective effects on glucose disorder. Notably, the Zbtb18-induced increase of phosphorylation of AKT and GSK-3β in livers was not observed in FXR-deleted mice (Fig. 7i). Also, no difference in the hepatic expression of glucogenic genes, such as Pgc-1α and Pepck, was detectable between AAV-Zbtb18 infected mice and AAV-Gfp infected mice due to FXR’s deletion (Fig. 7j). Again, Zbtb18 failed to reduce serum inflammatory cytokines, such as TNF-α and IL-1β, as well as ALT and AST levels in FXR-deficient mice (Fig. 7k–l).
FXR ablation diminished hepatic Zbtb18-induced protective effects against steatohepatitis. a, b Hepatic Zbtb18 overexpression fails to reduce the body weight (a) and fat mass (b) of FXR knockout mice; n = 6. c, d FXR deficiency abrogates the Zbtb18 protein-stimulated elevation of serum ketone body (c) and expression of hepatic genes related to FAO (d); n = 6. e, f Hepatic Zbtb18 overexpression fails to alter the TGs contents in the serum and liver (e), and the fatty liver phenotype (f) due to FXR deletion; n = 6. g Hepatic Zbtb18 overexpression fails to change the fasting blood glucose and insulin levels in FXR knockout mice; n = 6. h The Zbtb18-driven improvement of glucose and insulin resistance was diminished in FXR knockout mice; n = 6. i–j Hepatic Zbtb18 overexpression fails to change the phosphorylation of AKT and GSK-3β (i) and of glucogenic genes (j) in FXR knockout mice, n = 6. k, l Hepatic Zbtb18 overexpression does not change serum proinflammatory cytokines (k) and ALT and AST levels (l) following FXR ablation; n = 6. Data are shown as means ± SEM. ns=no significant, *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001
On the contrary, FXR’s forced expression effectively increased the expression of its target genes in the livers of hepatic Zbtb18LKO mice (Fig. 8a). These mice showed a lesser fat mass and enhanced energy expenditure due to FXR expression (Fig. 8b, c). Correspondingly, the changes in liver weight/body weight ratio values, serum ketone body levels, and TGs contents in hepatic Zbtb18LKO mice were reversed due to the rescued activation of FXR, along with an improvement in the expression of hepatic genes involved in FAO and on the hepatic steatosis in these mice, as shown by the decreased ballooning due to lipid droplets accumulation in the hepatocytes (Fig. 8d–f). Moreover, increased FXR expression reduced fasting blood glucose and insulin levels in Zbtb18LKO mice. Also, increased expression of FXR significantly alleviates glucose intolerance and insulin resistance in the liver caused by Zbtb18’s absence (Fig. 8g, h). Similar stimulatory effects on the phosphorylation of AKT and GSK-3β in livers were observed following FXR overexpression, indicating rescue of hepatic insulin signaling activity in Zbtb18LKO mice, coupled with a beneficial regulation of the genes involved in glycogenesis (Fig. 8i, j). Of note, forced hepatic FXR expression effectively mitigated the Zbtb18-deficiency stimulated CD11b+ and F4/80+ macrophages gathering in livers, which attenuated the inflammatory stress and eventually contributed to the improvement of NAFLD in these mice (Fig. 8k–m). Collectively, these data strongly indicated a potential working scheme by which Zbtb18-mediated transcriptional activation of FXR preserved hepatic glucose and lipid homeostasis.
Hepatic FXR forced expression alleviates NAFLD phenotype in hepatic Zbtb18 deleted mice. a, b Hepatic FXR overexpression (a) decreases the fat mass (b) of Zbtb18LKO mice; n = 6. c Hepatic FXR overexpression rescues Zbtb18 deficiency-induced impairment of energy expenditure; n ≥ 4. d Hepatic FXR overexpression decreases Zbtb18 deficiency-induced liver weight gain and TGs accumulation in liver and serum, while recovering normal levels of serum ketone body; n = 6. e, f Hepatic FXR overexpression reduces Zbtb18 deficiency-induced fatty liver phenotype (e), and the dysregulation of genes related to lipid metabolism (f); n = 6. g Hepatic FXR overexpression decreases the fasting blood glucose and insulin levels in Zbtb18LKO mice; n = 6. h Hepatic FXR overexpression improves glucose and insulin resistance in Zbtb18LKOmice; n = 6. i Hepatic FXR overexpression decreases Zbtb18 deficiency-induced glucogenic genes in mice; n = 6. j Hepatic FXR overexpression alleviates the impaired phosphorylation of AKT and GSK-3β in the livers of Zbtb18LKO mice. k Hepatic FXR overexpression reduces the Zbtb18 deficiency-induced gathering of F4/80+ and Cd11b+ cells in livers. l, m Hepatic FXR overexpression decreases the hepatic mRNA expression of inflammatory genes (l) and serum proinflammatory cytokines levels (m) in Zbtb18LKO mice, n = 6. Data are shown as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001
Hepatic Zbtb18 protein protects mice against MCD-induced liver fibrosis by transcriptionally activating FXR
Accumulating evidence suggests that a prolonged abnormal hepatic accumulation of lipids leads to excessive deposition of fibrous connective tissue and an imbalance of extracellular matrix synthesis and degradation in the liver, thereby advancing NASH progression.29 Considering the obvious effects of Zbtb18 in decreasing hepatic lipid deposition, we suspected that it might exert a significant protective effect against liver fibrosis. Consequently, we first found that hepatic Zbtb18 protein level was significantly decreased in (Methionine choline deficient) MCD diet fed mice (Supplementary Fig. 10a). Next, we fed Zbtb18LKO mice with MCD diet to prove Zbtb18’s physiological function in the MCD diet induced development of liver fibrosis. As expected, in contrast to their littermate control mice, after MCD diet exposure Zbtb18LKO mice had elevated ALT and AST levels (Fig. 9a), suggesting that a severe liver injury and hepatocytic death had occurred in these mice. Also, after MCD diet feeding, the TGs levels and hepatic lipid deposition were both aggravated due to Zbtb18 deletion, as illustrated by the lipid droplet accumulation and consequent ballooning of the hepatocytes (Fig. 9b, c). Moreover, our Sirius Red staining analysis indicated that the MCD diet induced development of a fatal intraparenchymal pericellular fibrosis concurring with an acute elevation of α-SMA staining levels was promoted in hepatic Zbtb18 ablated mice (Fig. 9d). Moreover, western blotting results also confirmed that Zbtb18 deficiency accelerated the accumulation of α-SMA and Col1a1 in the liver (Supplementary Fig. 11a). Correspondingly, the MCD diet elicited expression of liver genes related to liver fibrosis development, including α-SMA, Col1a1, and TGF-β, was also stimulated in hepatic Zbtb18LKO mice, along with an impaired expression of FXR and of its target genes (Fig. 9e, f). Moreover, hepatic Zbtb18 deletion exacerbated the MCD diet induced inflammatory stress in the liver, as illustrated by the up-regulation of inflammatory genes, such as F4/80 and TNF-α, and by the elevated levels of serum inflammatory cytokines, along with increased hepatic gathering of F4/80+ and CD11b+ macrophages (Fig. 9e, g, h). All of these events contribute to a fast steatohepatitis development.
Hepatic Zbtb18 deletion aggravates MCD-induced progression of liver fibrosis. a, b Serum ALT, and AST levels (a) and TGs contents in serum and livers (b) of hepatic Zbtb18 deleted mice and control mice fed on MCD diet or on normal diet, n = 6. c Hepatic Zbtb18 deletion aggravates MCD-induced liver injury and lipid deposition in livers. d Sirius and α-SMA staining of liver sections from hepatic Zbtb18 deleted mice and control mice fed on MCD diet or a normal diet. e Hepatic Zbtb18 deletion alters the expression of genes related to liver fibrosis and to an inflammatory response in the liver of mice fed on MCD diet or on a normal diet; n = 6. f Hepatic Zbtb18 deletion aggravates the MCD-induced suppression of FXR and its downstream target genes in the liver. g, h Hepatic Zbtb18 deletion aggravates the MCD-induced elevation of serum proinflammatory cytokines (g) and the hepatic gathering of F4/80+ and CD11b+cells (h); n = 6. Data are shown as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005
By contrast, a hepatic Zbtb18 overexpression protected mice against MCD diet induced liver fibrosis, as illustrated by the normal serum ALT and AST levels as compared to littermate control mice (Fig. 10a). Moreover, MCD diet stimulated TGs elevation and excess hepatic lipid deposition were effectively improved by Zbtb18 overexpression, as shown by the H&E and Oil Red O staining analysis (Fig. 10b, c). Consistently, the Sirius staining and α-SMA immunohistochemistry analysis indicated that the development of intraparenchymal pericellular fibrosis was partly hindered in hepatic Zbtb18 overexpressing mice, even when fed on MCD diet (Fig. 10d). Moreover, increased Zbtb18 repressed the expression of a-SMA and Col1a1 in the liver (Supplementary Fig. 11b). This was coupled with a reduced expression of closely related genes, including α-SMA, Col1a1, and TGF-β (Fig. 10e). Moreover, hepatic Zbtb18 overexpression suppressed the MCD diet induced inflammatory genes, such as F4/80, TNF-α, CXCL1, and CXCL10, in the liver, thereby reducing inflammatory cytokines’ serum levels and hepatic infiltration of inflammatory cells, helping improve the steatohepatitis (Fig. 10e–g). Collectively, these findings suggest a potentially therapeutic role of hepatic Zbtb18 in defending against steatohepatitis partly via the activation of FXR and its downstream target genes (Fig. 11).
Hepatic Zbtb18 overexpression protects mice against MCD-induced liver fibrosis. a, b Serum ALT, and AST levels (a) and TGs contents in serum and liver (b) samples from hepatic Zbtb18 overexpressing mice and control mice fed on MCD diet or normal diet; n = 6. c Hepatic Zbtb18 overexpression attenuates MCD-induced liver injury and hepato-steatosis. d Sirius and α-SMA staining of liver sections from hepatic Zbtb18 transgenic mice and control mice fed on MCD diet or normal diet. e Hepatic Zbtb18 overexpression alters the expression of genes related to liver inflammatory response and fibrosis in mice fed on MCD diet or normal diet; n = 6. f, g Hepatic Zbtb18 overexpression reduces the MCD-induced elevation of serum proinflammatory cytokines (f) and the hepatic accumulation of F4/80+ and CD11b+ cells (g); n = 6. Data are shown as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005
Discussion
In mammals, the imbalance of de novo lipogenesis and FAO leads to a surplus hoarding of TGs in the hepatocytes, which contributes to the development of NAFLD, and even to its progression to NASH.30 As yet, the underlying causative mechanisms are far from clear. In the current study, we identified a close link between the hepatic dysregulation of Zbtb18 and the onset and progression of NAFLD and NASH for the first time. Hepatic Zbtb18 protein could transcriptionally activate FXR and its downstream target genes by directly binding to its promoter region: the upshot would be an accelerated FAO and consumption of excess lipids in the liver, helping improve steatohepatitis.
Accumulating evidence indicated that multiple especially endogenous factors, including environmental or physiological regulators, affect the lipogenesis and FAO in hepatocytes to keep lipid homeostasis.31,32 We and others previously found that cellular transcriptional factors, such as KLFs, which encode a variety of Krüppel-like factor subfamily of zinc finger proteins, are closely involved in the regulation of the hepatic glucose and lipid balance. KLF16 and KLF11 potentially bind to the promoter region of PPARα to activate FAO and improve the fatty liver phenotype.33,34 KLF9, a critical sensor of GRs, participates in fasting-induced hyperglycemia via the transcriptional activation of Pgc-1α.35 In this study, we found that the Zbtb18, another member of the transcriptional factors family (Zbtbs), which shares an alike C2H2-type Zinc finger and BTB Domain, is closely related to the development of NAFLD. As previously reported, ZBTBs play key roles in systemic metabolism homeostasis. Zbtb11 cooperates with NRF-2 to control mitochondrial function.36 Zbtb20 acts as a key regulator of hepatic lipogenesis, influencing systemic lipid homeostasis.37 Likewise, we found that Zbtb18 played effective roles in stimulating FAO and defending against the diet-induced NAFLD phenotype. Hepatic Zbtb18 deletion severely hinders FAO in the liver while dramatically suppressing the genes involved in FAO, resulting in a conspicuous hepato-steatosis. By contrast, hepatic Zbtb18 overexpression or rescue protects mice against HFD-induced fatty liver due to its stimulatory effects on fatty acid catabolism and FAO. Besides, chronic HFD exposure often fires “multiple hits” in the liver, resulting in impaired insulin response cascades and leading to insulin resistance, which has become an important complication and diagnostic index of NAFLD.33 In this regard, after Zbtb18 overexpression we also found a glucose intolerance improvement and a reduced insulin resistance, coupled with an enhanced phosphorylation of AKT and GSK-3β in the liver. Since hepatic Zbtb18 deletion predisposes HFD-fed mice to serious glucose intolerance and insulin resistance, it is easy to take it as an indication that Zbtb18 plays a predominant role in alleviating the HFD-induced dysfunctional glucose and insulin responses.
Several studies have reported a Zbtbs-driven activation of ChREBP-α and PCK1 in mediating the regulation of glucose and lipid metabolism by Zbtb20 or Zbtb16.37,38 Yet, we didn’t find alike changes after altering Zbtb18 expression. In the present work, we subjected hepatocytes overexpressing the Zbtb18 to RNA-seq to systematically explore the potential targets of the Zbtb18-mediated beneficial effects. Thus, we found that Zbtb18 protein could effectively stimulate FXR expression and hence its downstream target genes. As previously reported, FXR protein, a highly expressed hepatic and intestinal nuclear receptor, works as a key regulator of bile acid metabolism in mammals and participates in the upkeeping of whole-body glucose and lipid homeostasis in an organ-dependent working fashion.39 We and others showed that increasing by pharmacological or genetic means the transcriptional activity or stability of the FXR protein exerted remarkably protective effects on inflammatory stress and acute damage in the liver. FXR activation by hepatocellular Cystathionine γ lyase/H2S significantly alleviates the diet-induced NAFLD, while iron-induced FXR suppression leads to fatal hepatotoxicity in humans and mice.40 In this work, we found that Zbtb18 overexpression remarkably increased the FXR mRNA and protein levels and activated its target genes in livers and cultured MPHs by enhancing the nuclear-translocation of the FXR protein. These results suggest a transcriptional modulation of FXR by the Zbtb18. Subsequent luciferase assays and mutation analysis indicate the potential binding site for Zbtb18 transcription factor on the −800bp to −789bp promoter region of FXR, a finding subsequently validated by ChIP and ChIP-qPCR data. The latter also complements and perfects the mechanism underlying FXR dysfunction, which senses and responds to a surplus lipid deposition. To prove FXR’s critical role in mediating the Zbtb18-induced protective effects against lipid and glucose metabolic disorders, we employed FXR gain or loss cultured MPHs and HFD-fed mice combining them with hepatic Zbtb18 alterations. Consistently, we found that FXR deletion almost completely diminished the Zbtb18-induced beneficial effects on lipid deposition and glucose metabolism dysfunction in vivo and in vitro, coupled with a lack of effects on insulin resistance. Since FXR exerts different effects on lipid metabolism, including lipogenesis, FAO, and lipid absorption according to the anatomic location considered, we next injected Ad-FXR into hepatic Zbtb18 deleted mice to monitor its effects on the glucose and lipid metabolism while suppressing FXR’s function in other tissues. Consistent with our hypothesis, the activation of hepatic FXR effectively offset the NAFLD phenotype and insulin resistance induced by Zbtb18 silencing, suggesting a novel yet essential role for FXR as the mediator of hepatic Zbtb18’s key functions in maintaining glucose and lipid balance.
Several works have confirmed that a prolonged lipid overload always stimulates the gathering of macrophages inside the liver, resulting in chronic inflammation, which via the release of inflammatory cytokines, such as TNF-α, IL-6, and IL-1β, promotes NAFLD progression.41,42 As a consequence, the abnormal inflammatory stress destroys the balance between extracellular matrix synthesis and degradation, which leads to a surplus deposition of fibrous connective tissue in the liver, and NASH.43 Previous studies reported effective approaches preventing NASH development by improving the impaired hepatic FAO and the abnormal inflammatory response proper of the advanced stages of NAFLD.44 Functioning as critical transcriptional factors, Zbtbs were proven to be involved in the pathogenesis of many inflammatory disorders by flexibly modulating the development, differentiation, distribution, and effectors activity of immune cells.45 Our present results showed for the first time the critical role of hepatic Zbtb18 in blocking NLRP3 inflammasome activity via an FXR-regulated CLTC protein expression, while hepatic Zbtb18 deletion accelerated the gathering of F4/80+ and CD11b+ macrophages in the liver, leading to an increased expression of inflammatory genes and of circulating proinflammatory cytokines.
On the contrary, hepatic Zbtb18 overexpression exerted opposite effects on HFD-induced hepatic inflammatory response partly via FXR activation, as illustrated by the decreased gathering of F4/80+ and CD11b+ macrophages in the liver, and by the suppression of inflammatory genes and of the release of proinflammatory cytokines in these mice. Subsequent MCD exposure experiments also revealed the vital role of Zbtb18 in alleviating the development of steatohepatitis, as shown by the more intense hepatocytic death and liver injury that occurred in hepatic Zbtb18 deleted mice. Conversely, hepatic Zbtb18 overexpression protected the mice against MCD-induced intraparenchymal pericellular fibrosis by activating FXR and its downstream target genes. Altogether, our findings may explain the complex mechanisms by which FXR was dysregulated during the progress of liver fibrosis.
In summary, we proved that the hepatic Zbtb18 could increase the FXR mRNA and protein expression through direct binding to the AACTCTCT element on the promoter region of FXR. In turn, the FXR could stimulate its target genes to accelerate FAO, thereby preventing the onset and development of NAFLD. Moreover, the Zbtb18/FXR axis-stimulated CLTC protein remarkably alleviates the liver inflammatory infiltrations and fibrosis. Therefore, the Zbtb18/FXR axis represents a novel candidate to target for the treatment of NAFLD and NASH.
Methods
Clinical tissue preparation
Fifteen liver biopsy samples were taken from patients with preoperative testing at the First Affiliated Hospital of Guangzhou University of Chinese Medicine according to the ethical guidelines of the Declaration of Helsinki. A previous approval was obtained from the Ethics Committee of the First Affiliated Hospital of Guangzhou University of Chinese Medicine (No. K [2023] 013). Written informed consent was obtained from each patient included in the study. Liver samples were divided into the normal group and NAFLD group based on a gold-standard histological classification (i.e., NAFLD activity score [NAS]) and liver pathology.
Animals and treatments
Male db/db, ob/ob and their control mice (aged 6–8 weeks) were purchased from GemPharmatech Co., Ltd (Guangdong, China). C57BL/6 J mice aged 6–8 weeks were bought from the Animal Experimental Center of Guangzhou University of Chinese Medicine (Guangdong, China). Zbtb18 conditional knockout mice were generated by crossing Alb-Cre mice with Zbtb18fl/fl mice. Zbtbfl/fl mice were generated via embryo stem cells (ESCs) targeting technology (Cyagen Inc. Guangzhou, China). In the targeting vector, Exon 2 was selected as the conditional knockout region, the SdNeocassette was flanked by SDA (self-deletion anchor) sites, and the CKO region was flanked by loxp sites. Diphtheria toxoid (DTA) was used for the negative selection. The linearized vector was infected into C57BL/6 ESCs. After G418 selection, resistant clones were picked and identified by PCR and Southern blot to choose the correctly targeted ones. Targeted ES cell clones 2D10 and 1E12 were injected into C57BL/6 albino embryos, which were then re-implanted into CD-1 pseudo-pregnant females. Founder animals were identified by their coat color, and their germline transmission was confirmed by breeding with C57BL/6 females and subsequent genotyping of the offspring. The sdNeo cassette served as a self-excision cassette to generate selected-gene-free heterozygous flox mice from chimeras. Four female heterozygous targeted mice were generated from clone 2D10, one male and one female heterozygous targeted mouse were generated from clone 1E12, as final deliverables for this project. The genotyping primers were as follows: Forward primer, 5’-TACCTGCAGATCTTACCGC-3’; reverse primer, 5’-CAGGCAAAGTCCCACACAAA-3’. The positive founder mice and wild-type male mice were bred to get F1 Zbtb18 heterozygote mice.
Zbtb18 conditional knockin mice were generated by crossing Alb-Cre mice and Rosa26-Zbtb18LSL mice. Rosa26-Zbtb18LSL mice were generated by Suzhou Cyagen Co., Ltd (Suzhou, China) by applying the CRISPR/Cas9 system. Briefly, for Zbtb18 knock-in mice, a CAG-loxp-stop-loxp sequence followed by mouse Zbtb18 was inserted at Rosa26 locus (Rosa26-LSL-Zbtb18) using the CRISPR/Cas9 system. The sequence of the small guide RNA (sgRNA) was 5′-CTCCAGTCTTTCTAGAAGAT-GGG-3′. Zbtb18 knock-in mice were mated to Alb-Cre mice to generate hepatocyte-specific Zbtb18 knock-in mice (ROSA26-Zbtb18; Alb-Cre). FXR knockout mice were maintained in our lab as previously reported.22 All mice were given free access to food and water and were maintained a 12/12 h light-dark cycle at 25 °C ambiences. HFD 60% (D12492, Research Diets New Brunswick, NJ, USA) was used to establish a DIO mice model and MCD (Trophic Animal Feed High-Tech Co., Ltd, China) for four weeks to establish fibrosis models as previously reported.46 Animal care or experimental designs were following the guidelines of and approved by the Animal Ethics Committee of Guangzhou University of Chinese Medicine (approval NO. 20221207001).
Adeno-associated virus expressing Zbtb18 (AAV-Zbtb18), adeno-associated virus expressing green fluorescent protein (AAV-Gfp) and all plasmids were constructed and purchased from Hanbio Biotechnology Co. Ltd. (Shanghai, China). A total amount of 0.5–1 × 1011 v.g. diluted in 200 µl of PBS was injected into the tail vein of mice 4 weeks before the indicated experiments. Adenovirus expressing green fluorescent protein (Ad-Gfp), adenovirus expressing Zbtb18 (Ad-Zbtb18), adenovirus expressing FXR (Ad-FXR), and all plasmids were constructed and purchased from Shanghai Obio Technology Company (Shanghai, China). 1.0–1.5 × 109 active viral particles in 200 μL saline were injected into the tail vein of mice and 7–9 days after infection, mice were subjected to the designed experiments or fasted for 6 h and their livers and plasma samples were collected for further analysis.
Histological analysis, Immunofluorescence analysis, and Lipid Droplet staining
H&E and Oil red O staining were conducted as previously reported. [1] Briefly, liver tissues fixed with 10% neutral-buffered formalin were cut into 7 μm-thick sections, followed by routine H&E, Oil Red O, and Sirius staining. Histological analysis was performed by an experienced hepatopathologist from the First Affiliated Hospital of Chinese Medicine using a light microscope (Olympus). Immunofluorescence was conducted at Servicebio (Wuhan, China) according to the previous protocols. Liver sections or MPHs were incubated at 4 °C overnight with primary antibodies against F4/80 (Servicebio, China), CD11b (Servicebio, China), α-SMA (Abmart, China), FXR (Abcam, USA), Bsep (Abmart, China) and Zbtb18 (Proteintech, China), followed by incubation with indicated secondary antibodies. To detect lipid droplets, liver sections were incubated with BODIPY 493/503 (Thermofisher Scientific) for 30 min. All fluorescence images were obtained using a confocal laser scanning microscope (Leica) or a light microscope (Olympus).
Fatty acid oxidation detection
The assays were performed as previously described at Soochow Pukang Biotechnology Co., LTD.35,47 Briefly, primary hepatocytes infected with the indicated adenovirus were incubated with 0.4 μCi/ml [9,10-3H]oleic acid (Perkin Elmer Life Sciences) and 100 μm unlabeled oleic acid (conjugated with BSA) in Krebs-Ringer buffer (119 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2.6 mM MgSO4, 24.6 mM NaHCO3, 2.6 mM KH2PO4, 10 mM HEPES, pH 7.4) for 3 h at 37 °C. Chloroform was used to extract the unoxidized oleic acid. Supernatants were collected and incubated with 1.3 M perchloric acid. All the solutions were then centrifuged at 16,000 × g for 10 min. Supernatants were neutralized with 2 M KOH, 0.6 M MOPS. Next, 3 mL of scintillation liquid was then added and [3H] radioactivity was measured.
Tolerance tests
Tolerance tests were conducted as previously reported. Briefly, for the glucose tolerance test (GTT), mice were fasted overnight and ip. injected with D-glucose (1–2 g/kg), and blood glucose was assessed at 0, 15, 30, 45, 60, 90, and 120 min in the tail vein using a glucose monitor (OnCall EZIV, China). For the insulin tolerance test (ITT), mice were fasted for 6 h and ip. injected with insulin (0.5–0.75 U/kg) followed by the measurement of blood glucose at the same time points.
Metabolic cages and MRI
To monitor energy expenditure, mice were individually housed and adapted for 24 h in metabolic cages. Their heat production, respiratory exchange ratio (RER), carbon dioxide production (VCO2), oxygen consumption (VO2) and X-axis ambulation (XAMB) were recorded during the next 36 h by using a Promethion monitoring system (SABLE). Data of 24 h were shown as previously reported. An Echomri.Combo-700 was used to detect the body composition including fat mass and lean mass.
ELISA and determination of TGs and TCs
Serum ketone levels were assessed using ELISA kits (Ruixinbio, Quanzhou, China). Serum proinflammatory cytokines IL-1β, IL-6, or TNF-α were measured using ELISA kits (Ruixinbio, Quanzhou, China). Serum and hepatic TGs and TCs levels were quantified using reagent kits from Jiancheng Bioengineering Institute (Nanjing, China).
Luciferase reporter gene assay
HepG2 cells (American Type Culture Collection, Manassas, VA, USA) were cultured in 24-well plates using Dulbecco’s Modified Eagle Medium containing 10% (vol./vol.) FBS (Invitrogen). Luciferase reporter genes were then co-transfected into the cells together with the indicated expression plasmids. The Ramlila luciferase expression vector pCMV-RL-TK (Promega) was used as an internal control. After 48 h, cells were harvested and assessed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). Relative luciferase activity was corrected for Renilla-luciferase activity of pCMV-RL-TK and normalized to the activity of the controls. The 5’ end of the mouse FXR gene extending from position −1838 bp (relative to the transcription start site) to +47 was cloned into the pGL3-Basic (Promega) luciferase reporter plasmid with the MluI/XhoI sites. A series of 5’ deletions and corresponding mutation constructs of FXR (−1074Luc, −677Luc, mut) were prepared by PCR using −1838-Luc as a template. The primers used for plasmid construction are shown in the Supplemental Table.
Chromatin immunoprecipitation (ChIP) assay
Pulverized liver tissues from C57BL/6 J, HFD, db/m or db/db mice were lysed and sonicated. The protein–DNA complexes were immunoprecipitated with mouse IgG antibody (control) or anti-Zbtb18 antibody. The promoter region of FXR was amplified by PCR or qPCR using the following primer pair: 5’-TGAGACAGGATTTCTCTATG-3’ as the forward primer and 5’-CCTTTAATCCTAGCACTTA-3’ as the reverse primer.
RNA-sequencing
RNA sequencing was performed and analyzed at Berry Genomics Corporation (Beijing, China) as previously reported. Briefly, 1 µg RNA isolated from indicated samples were used as input material for the RNA sample preparations. Sequencing was conducted using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) according to the manufacturer’s recommendations, followed by adding an Index code in each sample to attribute sequences. A cBot Cluster Generation System was used to perform the clustering of the index-coded samples using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. The library preparations were sequenced on an Illumina NovaSeq platform and 150 bp paired-end reads were generated. HTSeq v0.6.1 was used for the subsequent analysis.
ChIP-sequencing and Peak finding
Input DNA and ChIP DNA were prepared for ChIP-seq via Illumina kit per the manufacturer’s protocol. Briefly, each sample was subjected to end repair, followed by the addition of an A base to the 3′ end. Adaptor ligated DNA fragments were size-selected (175–225 bp), PCR-amplified, and further size-selected (175–225 bp). ChIP-seq data was obtained via Illumina Genome Analyzer II. 36 base pairs were analyzed by Illumina’s pipeline software for quality filtering, and aligned to the Mm9 reference mouse genome. Only uniquely aligned reads were kept for subsequent bioinformatics analysis. To identify Zbtb18 peaks, 2e analyzed the Chip-seq data by the latest version of MACS2 software.
Western blot analysis
Protein samples isolated from indicated cells or liver samples were lysed, and homogenized, and their concentration was assessed by using RIPA lysis buffer and a bicinchoninic acid protein assay kit (Beyotime Biotechnology, Shanghai, China). Subsequently, 80–100 μg of protein was loaded onto a 10% SDS–PAGE gel, and the separated proteins were transferred to polyvinylidene difluoride membranes. Specific antibodies were used to conduct western blot assays, with rabbit anti-Zbtb18 from Proteintech (Wuhan, China), anti-FXR, anti-SHP, anti-BSEP, anti-AKT, anti-p-AKT, anti-GSK-3β, anti-p-GSK-3β, anti-NLRP3, anti-ASC, anti-Capase-1, anti-NF-кB and anti-p-NF-кB, which were purchased from Abmart (Shanghai China), and anti-β-actin bought from Abclonal (Wuhan, China) according to their manufacturers’ instructions.
Real-time quantitative PCR
Total RNA was isolated from indicated cells or liver samples using a Trizol reagent kit (Invitrogen), followed by a reverse-transcription to cDNA using a high-capacity cDNA reverse-transcription kit. Subsequent q-PCR was conducted with the resulting cDNA, using PowerUpTM SYBRTM Green Master Mix, and β-actin was used to normalize the expression of genes. The specific primer sequences are shown in Supplementary Table 1.
Cell culture and pretreatment
Mouse primary hepatocytes (MPHs) were isolated as previously reported and cultured in a RPMI-1640 medium (10% fetal bovine serum, 100 units/ml penicillin, and 0.1 mg/ml streptomycin). Cells were infected with Ad-Gfp, Ad-Zbtb18 or Ad-FXR for 12 h, and next exposed to OA (660 μmol/L) and PA (330 μmol/L) for another 24 h to be thereafter collected for subsequent analysis. BMDM cells were cultured in the cell-conditioned medium collected as previously described.
Data availability
The RNA-sequencing data have been deposited in NCBI under SRA accession numbers (Zbtb18 overexpressing MPHs: PRJNA994862; Clinical NAFLD patients: PRJNA994268). ChIP-seq data have been deposited in NCBI under SRA accession numbers (SRA: PRJNA998803). All data in this article are available upon reasonable request from the corresponding authors.
References
Tsuneki, H. et al. Hypothalamic orexin prevents non-alcoholic steatohepatitis and hepatocellular carcinoma in obesity. Cell Rep. 41, 111–497 (2022).
Parlati, L., Régnier, M., Guillou, H. & Postic, C. New targets for NAFLD. JHEP Rep. 3, 100346 (2021).
Ratziu, V., Francque, S. & Sanyal, A. Breakthroughs in therapies for NASH and remaining challenges. J. Hepatol. 76, 1263–1278 (2022).
Scorletti, E. & Carr, R. M. A new perspective on NAFLD: Focusing on lipid droplets. J. Hepatol. 76, 934–945 (2022).
Mameli, C. et al. An update on the assessment and management of metabolic syndrome, a growing medical emergency in paediatric populations. Pharm. Res. 119, 99–117 (2017).
Lozano, P. et al. Lipid screening in childhood and adolescence for detection of multifactorial dyslipidemia: evidence report and systematic review for the US preventive services task force. Jama 316, 634–644 (2016).
Zhao, X. Y. et al. Long noncoding RNA licensing of obesity-linked hepatic lipogenesis and NAFLD pathogenesis. Nat. Commun. 9, 29–86 (2018).
Zhang, L. et al. S100A11 Promotes Liver Steatosis via FOXO1-Mediated Autophagy and Lipogenesis. Cell Mol. Gastroenterol. Hepatol. 11, 697–724 (2021).
Park, H. S. et al. TXNIP/VDUP1 attenuates steatohepatitis via autophagy and fatty acid oxidation. Autophagy 17, 2549–2564 (2021).
Moore, M. P. et al. Compromised hepatic mitochondrial fatty acid oxidationand reduced markers of mitochondrial turnover in human NAFLD. Hepatology 76, 1452–1465 (2022).
Powell, E. E., Wong, V. W. & Rinella, M. Non-alcoholic fatty liver disease. Lancet 397, 2212–2224 (2021).
Brenner, C., Galluzzi, L., Kepp, O. & Kroemer, G. Decoding cell death signals in liver inflammation. J. Hepatol. 59, 583–594 (2013).
Mridha, A. R. et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J. Hepatol. 66, 1037–1046 (2017).
Gaul, S. et al. Hepatocyte pyroptosis and release of inflammasome particles induce stellate cell activation and liver fibrosis. J. Hepatol. 74, 156–167 (2021).
Pafili, K. & Roden, M. Nonalcoholic fatty liver disease (NAFLD) from pathogenesis to treatment concepts in humans. Mol. Metab. 50, 101–122 (2021).
Cave, M. C. et al. Nuclear receptors and nonalcoholic fatty liver disease. Biochim. Biophys. Acta 1859, 1083–1099 (2016).
Mazzini, G. S. et al. Concomitant PPARα and FXR Activation as a Putative Mechanism of NASH Improvement after Gastric Bypass Surgery: a GEO Datasets Analysis. J. Gastrointest. Surg. 23, 51–57 (2019).
Clifford, B. L. et al. FXR activation protects against NAFLD via bile-acid-dependent reductions in lipid absorption. Cell Metab. 33, 1671–1684.e1674 (2021).
Han, C. Y. et al. FXR Inhibits Endoplasmic Reticulum Stress-Induced NLRP3 Inflammasome in Hepatocytes and Ameliorates Liver Injury. Cell Rep. 24, 2985–2999 (2018).
Huang, S. et al. A new mechanism of obeticholic acid on NASH treatment by inhibiting NLRP3 inflammasome activation in macrophage. Metabolism 120, 154–197 (2021).
Kremoser, C. FXR agonists for NASH: How are they different and what difference do they make? J. Hepatol. 75, 12–15 (2021).
Liu, C. et al. Hepatic SIRT6 Modulates Transcriptional Activities of FXR to Alleviate Acetaminophen-induced Hepatotoxicity. Cell Mol. Gastroenterol. Hepatol. 14, 271–293 (2022).
Chen, L., Wang, L., Qin, J. & Wei, D. S. CtBP2 interacts with Zbtb18 to promote malignancy of glioblastoma. Life Sci. 262, 118477 (2020).
Fedele, V. et al. Epigenetic Regulation of Zbtb18 Promotes Glioblastoma Progression. Mol. Cancer Res. 15, 998–1011 (2017).
Wen, H. et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 12, 408–415 (2011).
Wan, Z. et al. Mononuclear phagocyte system blockade improves therapeutic exosome delivery to the myocardium. Theranostics 10, 218–230 (2020).
Li, B. et al. SIRT6-regulated macrophage efferocytosis epigenetically controls inflammation resolution of diabetic periodontitis. Theranostics 13, 231–249 (2023).
Caballero-Díaz, D. et al. Clathrin switches transforming growth factor-β role to pro-tumorigenic in liver cancer. J. Hepatol. 72, 125–134 (2020).
Younossi, Z. et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 15, 11–20 (2018).
Friedman, S. L., Neuschwander-Tetri, B. A., Rinella, M. & Sanyal, A. J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24, 908–922 (2018).
Meroni, M., Longo, M., Rustichelli, A. & Dongiovanni, P. Nutrition and genetics in NAFLD: The perfect binomium. Int. J. Mol. Sci. 21, 2986 (2020).
Zeigerer, A. NAFLD - A rising metabolic disease. Mol. Metab. 50, 101274 (2021).
Sun, N. et al. Hepatic Krüppel-like factor 16 (KLF16) targets PPARα to improve steatohepatitis and IR. Gut 70, 2183–2195 (2021).
Zhang, H. et al. Mouse KLF11 regulates hepatic lipid metabolism. J. Hepatol. 58, 763–770 (2013).
Cui, A. et al. Dexamethasone-induced Krüppel-like factor 9 expression promotes hepatic gluconeogenesis and hyperglycemia. J. Clin. Invest. 129, 2266–2278 (2019).
Wilson, B. C. et al. Intellectual disability-associated factor Zbtb11 cooperates with NRF-2/GABP to control mitochondrial function. Nat. Commun. 11, 5469 (2020).
Liu, G. et al. Regulation of hepatic lipogenesis by the zinc finger protein Zbtb20. Nat. Commun. 8, 14824 (2017).
Chen, S. et al. Control of hepatic gluconeogenesis by the promyelocytic leukemia zinc finger protein. Mol. Endocrinol. 28, 1987–1998 (2014).
Ding, L., Yang, L., Wang, Z. & Huang, W. Bile acid nuclear receptor FXR and digestive system diseases. Acta Pharm. Sin. B 5, 135–144 (2015).
Xu, W. et al. Hepatocellular cystathionine γ lyase/hydrogen sulfide attenuates nonalcoholic fatty liver disease by activating farnesoid X receptor. Hepatology 76, 1794–1810 (2022).
Hou, J. et al. TREM2 sustains macrophage-hepatocyte metabolic coordination in nonalcoholic fatty liver disease and sepsis. J. Clin. Invest. 131, 135197 (2021).
Kazankov, K. et al. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat. Rev. Gastroenterol. Hepatol. 16, 145–159 (2019).
Schuppan, D., Surabattula, R. & Wang, X. Y. Determinants of fibrosis progression and regression in NASH. J. Hepatol. 68, 238–250 (2018).
Raza, S., Rajak, S., Upadhyay, A., Tewari, A. & Anthony Sinha, R. Current treatment paradigms and emerging therapies for NAFLD/NASH. Front. Biosci. (Landmark Ed.) 26, 206–237 (2021).
Lee, S. U. & Maeda, T. POK/Zbtb proteins: an emerging family of proteins that regulate lymphoid development and function. Immunol. Rev. 247, 107–119 (2012).
Li, X. et al. Targeting ferroptosis alleviates methionine-choline deficient (MCD)-diet induced NASH by suppressing liver lipotoxicity. Liver Int. 40, 1378–1394 (2020).
Shen, L. et al. Hepatic differentiated embryo-chondrocyte-expressed gene 1 (Dec1) inhibits sterol regulatory element-binding protein-1c (Srebp-1c) expression and alleviates fatty liver phenotype. J. Biol. Chem. 289, 32–42 (2014).
Acknowledgements
This work was supported by National Natural Science Foundation of China (82370872, 82070891, 82160891, 82174319, 82370235); National Natural Science Foundation of Guangdong (2023A1515012618); Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine(ZYYCXTD-C-202208); Key projects of Guangdong Provincial Department of Education (2021ZDZX2010); Basic and Applied Basic Research Project of Guangzhou (202201011501, 202201010134, 202201011240); the Natural Science Foundation of Jiangsu Province (BK20211055); the Jiangsu Qing Lan Project; China Postdoctoral Science Foundation (2022M710120). Guangdong Provincial Key Laboratory of TCM Pathogenesis and Prescriptions of Heart and Spleen Diseases (2022B1212010012).
Author information
Authors and Affiliations
Contributions
Y.G. and Y.S.C. designed the study. L.Z., J.B.C., X.Y.Y., C.P.S, J.W.H., D.Z., N.H.L., Y.D.Z., Y.J.C., C.N.L, K.J.T., J.Y.G., T.Q.C., S.W.D., J.Y.L., J.B.C., and S.Y.H. performed the experiments and acquired the data. L.Z., D.Z., J.W.H., N.H.L., and X.Y.Y. analyzed the data. L.Z. and Y.G. generated the figures and wrote the manuscript. L.Z., Y.G., H.B.Z., H.F.P., X.Q.T., and Y.S.C. revised the manuscript and supervised this study. All authors have read and approved the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, L., Chen, J., Yang, X. et al. Hepatic Zbtb18 (Zinc Finger and BTB Domain Containing 18) alleviates hepatic steatohepatitis via FXR (Farnesoid X Receptor). Sig Transduct Target Ther 9, 20 (2024). https://doi.org/10.1038/s41392-023-01727-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41392-023-01727-7