Abstract
Background
Abiraterone (Abi) is an androgen receptor signaling inhibitor that significantly improves patients’ life expectancy in metastatic prostate cancer (PCa). Despite its beneficial effects, many patients have baseline or acquired resistance against Abi. The aim of this study was to identify predictive serum biomarkers for Abi treatment.
Methods
We performed a comparative proteome analysis on three Abi sensitive (LNCaPabl, LAPC4, DuCaP) and resistant (LNCaPabl-Abi, LAPC4-Abi, DuCaP-Abi) PCa cell lines using liquid chromatography tandem mass spectrometry (LC-MS/MS) technique. Two bioinformatic selection workflows were applied to select the most promising candidate serum markers. Serum levels of selected proteins were assessed in samples of 100 Abi-treated patients with metastatic castration-resistant disease (mCRPC) using ELISA. Moreover, FSCN1 serum concentrations were measured in samples of 69 Docetaxel (Doc) treated mCRPC patients.
Results
Our proteome analysis identified 68 significantly, at least two-fold upregulated proteins in Abi resistant cells. Using two filtering workflows four proteins (AMACR, KLK2, FSCN1 and CTAG1A) were selected for ELISA analyses. We found high baseline FSCN1 serum levels to be significantly associated with poor survival in Abi-treated mCRPC patients. Moreover, the multivariable analysis revealed that higher ECOG status (>1) and high baseline FSCN1 serum levels (>10.22 ng/ml by ROC cut-off) were independently associated with worse survival in Abi-treated patients (p < 0.001 and p = 0.021, respectively). In contrast, no association was found between serum FSCN1 concentrations and overall survival in Doc-treated patients.
Conclusions
Our analysis identified baseline FSCN1 serum levels to be independently associated with poor survival of Abi-treated, but not Doc-treated mCRPC patients, suggesting a therapy specific prognostic value for FSCN1.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) is one of the most frequently diagnosed solid cancers among men worldwide [1]. Abiraterone is a selective and irreversible inhibitor of the enzyme CYP17A1, which is a crucial factor during androgen biosynthesis [2]. Abi has demonstrated improved overall and progression-free survival in both hormone sensitive and castration-resistant metastatic PCa both in chemotherapy-naïve and post-chemotherapy settings [3,4,5,6]. However, many patients show primary resistance or develop secondary resistance against this therapy. Currently further therapies with various mechanisms of action are increasingly becoming available providing reasonable options for metastatic castration-resistant prostate cancer (mCRPC) patients. Therefore, predictive biomarkers such as pathologic BRCA1/2 mutations for PARP inhibitors and PSMA-uptake for PSMA radioligand therapy are needed for improved therapeutic decision-making.
In the last years, several Abi resistance mechanisms have been described, which can be classified into two groups; androgen receptor (AR) signaling related and non-AR-related mechanisms [7]. The most common AR-related mechanism is AR copy number gain, which leads to enhanced AR expression resulting in reduced sensitivity to Abi [8]. A further AR-related resistance mechanism is related to specific activating point mutations or splice variants (such as AR-V7) of the AR [9, 10]. In addition, different non-AR-related mechanisms may contribute to Abi insensitivity, e.g. the alterations of DNA repair genes and neuroendocrine transdifferentiation [11, 12]. Additionally, our group has recently found elevated serum ALCAM levels to be associated with shorter survival of Abi- but not Doc-treated patients [13].
The aim of the present study was to identify therapy predictive biomarkers of Abi resistance of PCa. First, we performed comparative proteome analysis on Abi-sensitive and resistant PCa cell lines. This analysis identified 68 significantly, at least two-fold upregulated proteins in Abi resistant cells. Then, we used two different bioinformatics workflows in order to select the most promising candidates. Serum concentrations of selected proteins were determined in Abi-treated mCRPC patients’ samples by using the ELISA method. As Fascin-1 (FSCN1) serum levels are associated with poor survival in Abi-treated patients, its serum concentrations were assessed also in samples of Doc-treated mCRPC men.
Materials and methods
Cell culture and reagents
For in vitro experiments, we used the LNCaPabl, LAPC4, and DuCaP human PCa cell lines and their Abi-resistant sublines (LNCaPabl-Abi, LAPC4-Abi, and DuCaP-Abi). Abi-resistant cells were generated by treatment with increasing concentrations of Abi, as described by Puhr et al. [14]. LAPC4 and DuCaP were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (Biowest, Nuaillé, France), 1% Glutamax (Thermo Fisher Scientific, Darmstadt, Germany) and 1% penicillin/streptomycin (Lonza, Basel, Switzerland). For the LAPC4 cell culture, 1 nM dihydrotestosterone was applied. LNCaPabl was cultured in RPMI 1640 medium supplemented with 10% charcoal stripped FBS (HyClone, GE Healthcare), 1% Glutamax (Thermo Fisher Scientific), and 1% penicillin/streptomycin (Lonza, Basel, Switzerland). Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2. The identity of all cell lines was confirmed by short tandem repeat analysis. All experiments were performed with mycoplasma-free cells. Abi (MedChemExpress) was dissolved in EtOH as a 100 mM stock solution and stored at −80 °C.
Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis
In order to identify differentially expressed proteins between Abi-sensitive and resistant cell lines, proteome analyses were done using the LC-MS/MS technique. Six technical replicates for each cell line were used for the analysis. Details on LC-MS/MS and protein identification are described in Supplementary Materials and Methods.
Biomarker selection
In Abi-resistant cells, proteins quantified with minimum two unique peptides and those passing the applied significance thresholds (FDR-corrected p-value ≤ 0.05, fold change ≥2) were considered. Two different bioinformatic workflows were used in order to identify the most promising proteins.
First, we applied a workflow, which used the existing prediction programs (SignalP 4.1, SecretomeP 2.0, TargetP 1.1, TMHMM 2.0) and databases (Uniprot, Human Protein Atlas, NCBI, ExoCarta) for the prediction of potentially secreted proteins.
Second, we applied a selection method by scoring the proteins based on their known oncological role and their molecular interactions (number of edges) according to the STRING database. STRING database was used as follows: we conducted a multiple protein search with those proteins that were significantly, at least two-fold upregulated in Abi resistant compared to parental sensitive PCa cells. We investigated which of these proteins have the highest number of interactions with each other. For this, we considered the “known interactions” based on the STRING, which included the 1) interactions between proteins from curated databases and 2) the experimentally determined interactions. In addition, text-mining edges were also considered, if the co-mention in the reference articles raised the functional or physical relationship of proteins. Based on these, we scored our protein list, with 1 as the lowest, and 3 being the highest link numbers.
In addition, we considered the availability of ELISA assays for later serum analyses.
Patient cohort and sample
Serum samples were collected within one day before Abi treatment between 11/2008 and 05/2015 from 100 mCRPC patients. In addition, serum samples at 3 months after therapy start were also available for 40 Abi-treated patients. Serum samples were collected at the Department of Urology at the Medical University of Vienna and at the Semmelweis University, Budapest. As one of the selected proteins was associated with survival in Abi-treated patients, its serum levels were determined also in serum samples of 69 mCRPC patients who received Doc chemotherapy between 1/2013 and 04/2019. The study was performed in accordance with the ethical standards of the Helsinki Declaration and was approved by the ethical boards of the hospitals (TUKEB 55/2014, ECS 1986/2017). PSA response was defined, according to the Prostate Cancer Clinical Trials Working Group Criteria (PCWG) II, as at least 50% PSA decline from baseline during therapy [15].
Serum ELISA analyses
Serum concentrations of FSCN1, KLK2 (Kallikrein-2), AMACR (alpha-methylacyl-CoA racemase), and CTAG1A (cancer testis antigen 1A) were measured in 100 Abi-treated patients by using ELISA kits (Aviva System Biology Corp, San Diego, USA) according to the manufacturer’s instructions. Absorbance was quantified at 450 nm by a Multiscan FC Microplate Photometer (Thermo Fisher Scientific).
Statistical analysis
Statistical tests were performed with the SPSS 26.0 (IBM, Chicago, IL) software. For paired comparisons between groups, the nonparametric, 2-sided Wilcoxon rank-sum test was applied. We applied the nonparametric receiver operating characteristics (ROC) curves to determine the optimal cut-off value with the highest sensitivity and specificity for the prediction of death within 24 months. Survival analyses were done using Kaplan–Meier curves, log-rank test, and univariable Cox proportional hazards regression analysis. For multivariable analysis, Cox regression models were used including parameters with a p-value of 0.05 in the univariable analysis. Investigators were blinded to clinical group assignments during the analyses.
Results
Identification of proteins with differential expression between Abi sensitive and resistant PCa cells
We identified 413 (LNCaP vs LNCaP-Abi), 588 (LAPC4 vs LAPC4-Abi) and 172 (DUCAP vs DUCAP-Abi) significantly differentially regulated proteins by at least 2 unique peptides using LC-MS/MS analysis (Supplementary Tables 1–3). Of these above identified proteins, we filtered those which were significantly upregulated in Abi resistant cells and showed at least two-fold higher expression in resistant compared to the parental sensitive PCa cell lines. This step resulted in 25, 38, 5 proteins in LNCaP-Abi, LAPC4-Abi, and DuCaP-Abi cell line pairs. In order to further select the most promising candidate proteins, we used two different bioinformatics workflows. The first, “secreted protein” workflow identified KLK2, while the second “protein scoring” workflow selected FSCN1, CTAG1A and AMACR for further ELISA analyses. FSCN1 reached high score by the “protein scoring” workflow because of its known role in oncological processes. CTAG1A had a higher score as it showed the strongest (25.58-fold) upregulation in LAPC4-Abi-resistant cells. AMARC reached a high score because its molecular interactions (number of edges) according to the STRING database (Fig. 1, Supplementary Fig. 1).
Selected protein levels in patients’ samples
Patients’ characteristics
The patients’ characteristics are given in Table 1.
In the Abi cohort, the median age was 70 (range: 52–90) years, the median pre-treatment PSA value was 66.5 ng/ml. Eighty-seven patients had bone, 17 had lymph node and 10 had visceral metastases. Sixty-nine patients died within a median follow-up period of 19 months.
In the Doc cohort, the median age was 70 years (range: 43–86), the median pre-treatment PSA level was 73 ng/ml. Sixty-four patients had bone, 7 had lymph node and 24 had visceral metastases. Fifty-five of 69 patients died within 24 months.
Associations of clinicopathological data with serum FSCN1, KLK2, CTAG1A and AMACR baseline levels
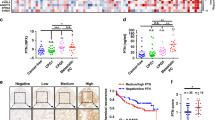
In the Abi cohort, we found no associations between baseline FSCN1 levels and patients’ clinicopathological parameters (Supplementary Table 4). CTAG1A serum levels were significantly lower in patients who showed a 30% or 50% PSA response to Abi (p = 0.016, p = 0.047, respectively). KLK2 levels were significantly higher in patients who had pain (p = 0.012) (Supplementary Table 5). In the Doc cohort, we found significantly lower FSCN1 serum levels in those patients who underwent primary local therapy (radiation or RPE) (p = 0.038). Moreover, FSCN1 serum levels were significantly lower in men who had visceral metastases (p = 0.027) (Supplementary Table 4). AMACR was undetectable in patients’ serum samples.
Survival analyses
In the Abi cohort, high ECOG status (>1) was associated with shorter overall survival (OS) (p < 0.001). High baseline PSA and FSCN1 levels (>9.39 and 10.22 ng/ml by ROC cut-off) were significantly associated with poor OS (p < 0.001, p = 0.022, p = 0.002; respectively) (Table 2). Multivariable analysis revealed that high ECOG status (>1), and high baseline FSCN1 serum levels are independently associated with poor OS in Abi-treated patients (Table 3). Kaplan–Meier OS curve revealed that higher baseline FSCN1 serum levels are significantly associated with poor OS (p = 0.001) (Fig. 2). Cancer-specific survival was available only in the Abi cohort. By using this endpoint, high pretreatment FSCN1 serum levels proved to be associated with shorter cancer-specific survival both in the univariable and multivariable analyses (Supplementary Tables 6 and 7) (Supplementary Fig. 2).
In the Doc cohort, we found no associations between the analyzed parameters and patients’ OS, while cancer-specific survival was not available (Table 2, Fig. 2).
Prognostic value of FSCN1 level changes during Abi therapy
FSCN1 serum concentrations were measured at 3 months after therapy start and were dichotomized as any increase, at least 20% increase, any decrease and at least 20% decrease. We found no correlations between FSCN1 level changes and OS (Supplementary Table 8).
Discussion
In the present study, we performed a comparative proteome analysis on three Abi resistant and corresponding parental Abi sensitive PCa cell line pairs. From 68 identified proteins in used Abi resistant cell lines, 4 were selected (FSCN1, KLK2, AMACR, CTAG1A) and further assessed by serum analyses in samples of Abi-treated mCRPC patients. This revealed a significant and independent association between high FSCN1 serum concentrations and poor OS. In contrast, FSCN1 concentrations were not associated with OS in Doc-treated patients. These results suggest that FSCN1 is a potentially predictive serum marker for Abi-treatment in mCRPC.
Our comparative proteome profiling method identified a large number of proteins potentially involved in Abi resistance. Interestingly, when comparing the significant at least two-fold upregulated proteins identified in the three PCa cell line pairs, we found no overlap, which suggests a multifactorial background of Abi resistance. As a consequence, probably rather a larger panel then a single marker will be able to cover all possible resistance mechanisms and so adequately predict Abi resistance.
Using the “secreted protein” selection workflow we selected KLK2 protein. Human KLK2 is a member of the kallikrein protein family, which is involved in several biological processes [16]. Many studies reported that high serum KLK2 levels are associated with high Gleason score and early biochemical recurrence in PCa [17]. In contrast, loss of KLK2 expression in PCa tissue was shown to be associated with the presence of aggressive PCa [16]. Tjon-Kon-Fat et al. analyzed the association of platelet-bound biomarkers for their predictive value in Abi- and in Doc-treated CRPC patients and found high KLK2 levels to be associated with OS and progression-free survival in Abi but not Doc-treated patients [18]. Accordingly, our proteome analysis identified KLK2 as a 3.41-fold upregulated protein in Abi resistant (LAPC4-Abi) cells. In addition, our serum KLK2 analysis in pretreatment samples of Abi-treated patients identified a trend between higher KLK2 serum levels and shorter OS of Abi-treated patients, however this correlation missed to reach the significance level (p = 0.071).
The “protein scoring” workflow identified three potential biomarkers. AMACR is an enzyme that is involved in bile acid synthesis and beta-oxidation of branched fatty acids [19]. Previous studies found AMACR protein and mRNA tissue expression to be specific to PCa and therefore suggested AMACR as a highly sensitive diagnostic marker for prostate adenocarcinoma [20, 21]. AMARC may also be associated with Doc resistance of PCa. Yoshizawa et al. showed that combined Doc treatment and AMACR inhibition caused decreased cell proliferation in AR-V7 positive PCa cell line [22]. However, the role of AMACR in Abi resistance is yet to be described. Based on our proteome analysis, AMACR showed a 2.84-fold upregulation in Abi-resistant (LAPC4-Abi) cells, but in our ELISA analyses it was not detectable in patients’ serum samples.
The other protein identified by the “protein scoring” workflow was CTAG1A, which is a cancer testis antigen. The exact biological function of this protein is not well established, although some studies suggest its participation in cell cycle regulation and apoptosis [23]. Grupp et al. analyzed tissue samples of more than 11,000 patients and found high CTAG1A protein expression as an independent predictor of prognosis in ERG-positive PCa [24]. Moreover, it was shown that in ERG-negative cancer, CTAG1A expression is significantly associated with PTEN deletion [24]. Based on this, CTAG1A was suggested as a hallmark marker for a separate molecular subgroup of PCa. However, the role of CTAG1A in Abi resistance is unknown. In the present study, we found that CTAG1A is 28.58-fold overexpressed in Abi-resistant (LNCaPabl-Abi) cells, but we did not find a significant correlation between serum CTAG1A concentrations and OS in Abi-treated patients.
The third protein identified by the “protein scoring” workflow was Fascin-1 (FSCN1), which is an actin-binding protein. In vitro study showed that FSCN1 enhanced the migratory capacity of PCa cells [25]. Moreover, an in vivo PCa mouse xenograft study found that inhibition of FSCN1 effectively blocked tumor progression [25]. Based on a systematic review and meta-analysis, high FSCN1 expression is associated with an increased risk of progression in colorectal, breast and esophageal cancer and with the presence of metastatic lesions in gastric and colorectal cancer [26]. In PCa, Darnel et al. analyzed the tissue sample of 196 men who underwent radical prostatectomy and found epithelial FSCN1 expression to be higher in localized and castration resistant PCa compared to benign prostate tissue while no correlation was found between FSCN1 epithelial expression and surgical margins, stage and Gleason score [25]. Furthermore, another study assessed FSCN1 immunostaining in 211 prostate tumors and found that only 8% of the tumors had >10% FSCN1 positive cells. Moreover, they found no significant correlation between FSCN1 expression of tumor cells and pathological stage, Gleason score and PSA levels. However, high stromal FSCN1 expression was significantly associated with high Gleason score [27]. In addition, Tataru et al. compared serum levels of FSCN1 between PCa patients and healthy controls and found no diagnostic value for FSCN1 [28].
FSCN1 may be involved in the development of systemic therapy resistance. FSCN1 plays a crucial role in doxorubicin resistance by facilitating the epithelial-mesenchymal transition (EMT) in hepatocellular carcinoma cells [29]. Similarly, Pan et al. found that FSCN1 participates in EMT and enhances the development of Doc resistance in lung adenocarcinoma cell lines [30]. However, the role of FSCN1 in Abi resistance has not yet been investigated so far. Our comparative proteome analysis revealed a 6,18-fold upregulation of FSCN1 expression in Abi-resistant (LAPC4-Abi) cells. Our ELISA analysis identified baseline FSCN1 serum levels to be independently associated with poor OS of Abi-treated patients. In contrast to Abi-treated patients, we found no correlations between FSCN1 levels and shorter OS in Doc-treated patients, suggesting a therapy specific prognostic value for FSCN1. Based on these results patients with high serum FSCN1 levels may less benefit from Abi than from Doc treatment. Our results however, need further confirmation in larger prospective patient cohorts. Furthermore, the analysis of the potential Abi predictive role of FSCN1 serum levels in metastatic hormone sensitive cases is necessary to assess its value also in this therapeutic setting.
Our study has some limitations inherent from its retrospective nature (e.g. missing data points based on clinical documentation). In addition, only one cohort for Abi and one cohort for Doc treatment was available for analysis, which did not allow us to perform independent validation of our results. Therefore, independent validation preferably in a prospectively collected patient cohort is necessary, before implementing our results in the clinical routine. Moreover, cause of death data was available only for the Abi cohort and none of the Doc cohort and thus cancer-specific survival could only be used as an endpoint in Abi-treated patients. However, if applying cancer-specific survival as an endpoint high serum FSCN1 level was found to be and independent risk-factor. Finally, as we did not perform analyses of already identified resistance markers (e.g. AR-V7) we cannot concurrently evaluate the predictive value of FSCN1. In addition, our in vitro model is rather representative for acquired than for de novo resistance mechanisms, while the assessed serum samples were collected before treatment start (and are therefore rather representative for de novo resistance markers), which is a methodological limitation for this study. However, acquired resistance mechanisms and markers may overlap with those of de novo resistance, therefore our approach is most probably able to identify predictive markers in the pretreatment serum samples.
In conclusion, our results revealed, for the first time, an independent prognostic value of FSCN1 for Abi- but not Doc-treated patients. Based on these, FSCN1 serum level is a promising predictive biomarker for the identification of mCRPC patients with baseline resistance to Abi.
Data availability
The raw mass spectrometry data generated in this study have been deposited in the PRIDE database with accession number PXD038697 (http://www.ebi.ac.uk/pride). Other data that support the findings of this study are available from the corresponding author upon request.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Rehman Y, Rosenberg JE. Abiraterone acetate: oral androgen biosynthesis inhibitor for treatment of castration-resistant prostate cancer. Drug Des Devel Ther. 2012;6:13–8.
Fizazi K, Scher HI, Molina A, Logothetis CJ, Chi KN, Jones RJ, et al. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2012;13:983–92.
Ryan CJ, Smith MR, Fizazi K, Saad F, Mulders PF, Sternberg CN, et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castrationresistant prostate cancer (COU-AA-302): final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2015;16:152–60.
Fizazi K, Tran N, Fein L, Matsubara N, Rodriguez-Antolin A, Alekseev BY, et al. Abiraterone plus prednisone in metastatic, castration-sensitive prostate cancer. N Engl J Med. 2017;377:352–60.
James ND, de Bono JS, Spears MR, Clarke NW, Mason MD, Dearnaley DP, et al. Abiraterone for prostate cancer not previously treated with hormone therapy. N Engl J Med. 2017;377:338–51.
Csizmarik A, Hadaschik B, Kramer G, Nyirady P, Szarvas T. Mechanisms and markers of resistance to androgen signaling inhibitors in patients with metastatic castration-resistant prostate cancer. Urol Oncol. 2021;39:728.e13–728.e24.
Tolmeijer SH, Boerrigter E, Schalken JA, Geerlings MJ, van Oort IM, van Erp NP, et al. A systematic review and meta-analysis on the predictive value of cell-free DNA-based androgen receptor copy number gain in patients with castration-resistant prostate cancer. JCO Precis Oncol. 2020;4:714–29.
Conteduca V, Wetterskog D, Sharabiani MTA, Grande E, Fernandez-Perez MP, Jayaram A, et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: a multi-institution correlative biomarker study. Ann Oncol. 2017;28:1508–16.
Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Zhu Y, et al. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first-and second-line abiraterone and enzalutamide. J Clin Oncol. 2017;35:2149–56.
Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8:444–57.
Szarvas T, Csizmarik A, Fazekas T, Hüttl A, Nyirády P, Hadaschik B, et al. Comprehensive analysis of serum chromogranin A and neuron- specific enolase levels in localized and castration resistant prostate cancer. BJU Int. 2021;127:44–55.
Csizmarik A, Keresztes D, Nagy N, Bracht T, Sitek B, Witzke K, et al. Proteome profiling of enzalutamide-resistant cell lines and serum analysis identified ALCAM as marker of resistance in castration-resistant prostate cancer. Int J Cancer. 2022;151:1405–19.
Puhr M, Hoefer J, Eigentler A, Ploner C, Handle F, Schaefer G, et al. The glucocorticoid receptor is a key player for prostate cancer cell survival and a target for improved antiandrogen therapy. Clin Cancer Res. 2018;24:927–38.
Scher HI, Halabi S, Tannock I, Morris M, Sternberg CN, Carducci MA, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol. 2008;26:1148–59.
Bonk S, Kluth M, Jansen K, Hube-Magg C, Makrypidi-Fraune G, Höflmayer D, et al. Reduced KLK2 expression is a strong and independent predictor of poor prognosis in ERG-negative prostate cancer. Prostate. 2020;80:1097–107.
Lawrence MG, Lai J, Clements JA. Kallikreins on steroids: structure, function, and hormonal regulation of prostate-specific antigen and the extended kallikrein locus. Endocr Rev. 2010;31:407–46.
Tjon-Kon-Fat LA, Lundholm M, Schröder M, Wurdinger T, Thellenberg-Karlsson C, Widmark A, et al. Platelets harbor prostate cancer biomarkers and the ability to predict therapeutic response to abiraterone in castration resistant patients. Prostate. 2018;78:48–53.
Fu P, Bu C, Cui B, Li N, Wu J. Screening of differentially expressed genes and identification of AMACR as a prognostic marker in prostate cancer. Andrologia. 2021;53:e14067.
Walsh PC. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. J Urol. 2001;168:1635.
Stephen N, Badhe BA. Diagnostic utility of immunohistochemical markers alpha methyl acyl coA racemase (AMACR) and Ets related gene (ERG) in prostate cancer. Int J Clin Exp Pathol. 2022;15:364–72.
Yoshizawa A, Takahara K, Saruta M, Zennami K, Nukaya T, Fukaya K, et al. Combined α-methylacyl-CoA racemase inhibition and docetaxel treatment reduce cell proliferation and decrease expression of heat shock protein 27 in androgen receptor-variant-7-positive prostate cancer cells. Prostate Int. 2021;9:18–24.
Thomas R, Al-Khadairi G, Roelands J, Hendrickx W, Dermime S, Bedognetti D, et al. NY-ESO-1 Based Immunotherapy of Cancer: Current Perspectives. Front Immunol. 2018;9:947.
Grupp K, Ospina-Klinck D, Tsourlakis MC, Koop C, Wilczak W, Adam M, et al. NY-ESO-1 expression is tightly linked to TMPRSS2-ERG fusion in prostate cancer. Prostate. 2014;74:1012–22.
Darnel AD, Behmoaram E, Vollmer RT, Corcos J, Bijian K, Sircar K, et al. Fascin regulates prostate cancer cell invasion and is associated with metastasis and biochemical failure in prostate cancer. Clin Cancer Res. 2009;15:1376–83.
Tan VY, Lewis SJ, Adams JC, Martin RM. Association of fascin-1 with mortality, disease progression and metastasis in carcinomas: a systematic review and meta-analysis. BMC Med. 2013;11:52.
Jefferies MT, Pope CS, Kynaston HG, Clarke AR, Martin RM, Adams JC. Analysis of Fascin-1 in relation to gleason risk classification and nuclear ETS-related gene status of human prostate carcinomas: an immunohistochemical study of clinically annotated tumours from the Wales Cancer Bank. Biomark Cancer. 2017;9:1179299X17710944.
Tătaru OS, Martha O, Crocetto F, Barone B, Voidazan S, Borda A, et al. Fascin-1 and its role as a serological marker in prostate cancer: a prospective case-control study. Future Sci OA. 2021;7:FSO745.
Zhang Y, Lu Y, Zhang C, Huang D, Wu W, Zhang Y, et al. FSCN‑1 increases doxorubicin resistance in hepatocellular carcinoma through promotion of epithelial-mesenchymal transition. Int J Oncol. 2018;52:1455–64.
Pan Y, Chen J, Tao L, Zhang K, Wang R, Chu X, et al. Long noncoding RNA ROR regulates chemoresistance in docetaxel-resistant lung adenocarcinoma cells via epithelial mesenchymal transition pathway. Oncotarget. 2018;8:33144–58.
Acknowledgements
This work was supported by the National Research, Development and Innovation Office – NKFIH / FK 124431. Tibor Szarvas was supported by a János Bolyai Research Scholarship of the Hungarian Academy of Sciences. Supported by the ÚNKP-21-5-SE-3, ÚNKP-21-3-II-SE-13 and ÚNKP-22-4-I-SE-25 New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund. A part of this study was funded by P.U.R.E. (Protein Research Unit Ruhr within Europe), Ministry of Innovation, Science and Research of North-Rhine Westphalia, Germany.
Funding
Open access funding provided by Semmelweis University.
Author information
Authors and Affiliations
Contributions
AC and TS were the project administrator and wrote the original manuscript draft. AC, NN, PN, BH, and TS concuptalized the study. AC, NN, DK, MV, TS, JL, IT, LT, GK, SS, and AM conducted the data curation. AC, NN, DK, TB, BS, KW, JL, IT and LT contributed to the methodology. AC, NN, DK, TB and TS created the visualiziations. DK, NN, MV, TB, BS, KW, JL, IT, LT, MP, GK, SS, AM, BH, PN and TS reviewed and edited the manuscript. AC, NN, DK and TS performed the formal analysis. TS and PN supervised the project. TS acquired funding.
Corresponding author
Ethics declarations
Competing interests
BH has had advisory roles for ABX, AAA/Novartis, Astellas, AstraZeneca, Bayer, Bristol Myers Squibb, Janssen R&D, Lightpoint Medical, Inc., and Pfizer; has received research funding from Astellas, Bristol Myers Squibb, AAA/Novartis, German Research Foundation, Janssen R&D, and Pfizer; and has received compensation for travel from Astellas, AstraZeneca, Bayer and Janssen R&D, all outside the submitted work. The other authors declare no potential conflicts of interest.
Ethics statement
The study was performed in accordance with the ethical standards of the Helsinki Declaration and was approved by the ethical boards of the hospitals (TUKEB 55/2014, ECS 1986/2017). All patients signed consent to an institutional review board-approved protocol before sample collection.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Csizmarik, A., Nagy, N., Keresztes, D. et al. Comparative proteome and serum analysis identified FSCN1 as a marker of abiraterone resistance in castration-resistant prostate cancer. Prostate Cancer Prostatic Dis (2023). https://doi.org/10.1038/s41391-023-00713-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41391-023-00713-y