Abstract
Background
The goal of precision medicine in prostate cancer (PCa) is to individualize the treatment according to the patient’s germline mutation status. PCa has a very high rate of genetic predisposition compared with other cancers in men, with an estimated rate of cancers ascribable to hereditary factors of 5–15%.
Methods
A systematic search (PubMed, Web of Science, and ClinicalTrials.gov) of English literature from 2000 to 2022, using the keywords “prostate cancer”, “germline mutations”, “family history”, and “inheritance” was conducted, according to the Preferred Reporting Items for Systematic Reviews and Meta-analyses guidelines.
Results
The search identified 980 publications. Of these, 200 papers were removed before screening (duplicates, non-English literature, and publication year before 2000) and 245 records were excluded after title/abstract screening. Finally, 50 articles were included in the final analysis. We analyze the latest evidence on the genetic basis of PCa predisposition and clinical implications for more personalized screening protocols and therapeutic management of this high-prevalent cancer.
Discussion
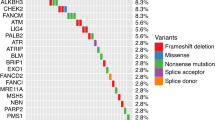
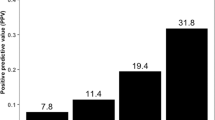
Emerging data show that germline mutations in homologous recombination genes (BRCA1/2, ATM, CHECK2), in mismatch repair genes (MLH1, MLH2, MSH6), and other additional genes are associated with the development and aggressiveness of PCa. Germline testing and genetic counseling have increasingly important implications in cancer screening and therapeutic decisions making for patients affected by PCa. Patients with localized PCa and some gene mutations are more likely to develop aggressive cancer, so active treatment may be preferable to active surveillance for these patients. Moreover, in patients with metastatic PCa, these gene alterations may be useful biomarkers for predicting response to specific therapy such as PARP inhibitors, recently approved for the treatment of metastatic castration-resistant PCa. The evidence supports recent guidelines and recommendations considering germline genetic testing for patients with a positive family history of PCa or men with high risk or metastatic disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. Ca Cancer J Clin. 2021;71:7–33. https://doi.org/10.3322/caac.21654
Bostwick DG, Burke HB, Djakiew D, Euling S, Ho S, Landolph J, et al. Human prostate cancer risk factors. Cancer. 2004;101:2371–2490. https://doi.org/10.1002/cncr.20408
Rawla P. Epidemiology of Prostate Cancer. World J Oncol. 2019;10(Apr):63–89. https://doi.org/10.14740/wjon1191
Hsing AW, Sakoda LC, Chua S. Obesity, metabolic syndrome, and prostate cancer. Am J Clin Nutr. 2007;86:s843–857. https://doi.org/10.1093/ajcn/86.3.843s
Bruner DW, Moore D, Parlanti A, Dorgan J, Engstorm P. Relative risk of prostate cancer for men with affected relatives: systematic review and meta- analysis. Int J Cancer. 2003;107:797–803. https://doi.org/10.1002/ijc.11466
Kiciński M, Vangronsveld J, Nawrot TS. An epidemiological reappraisal of the familial aggregation of prostate cancer: a meta-analysis. PLoS One. 2011;6:e27130 https://doi.org/10.1371/journal.pone.0027130
Sacco E, Prayer-Galetti T, Pinto F, Ciaccia M, Fracalanza S, Betto G, et al. Familial and hereditary prostate cancer by definition in an Italian surgical series: clinical features and outcome. Eur Urol. 2005;47:761–768. https://doi.org/10.1016/j.eururo.2005.01.016
Mikropoulos C, Goh C, Leongamornlert D, Kote-Jarai Z, Eeles R. Translating genetic risk factors for prostate cancer to clinic: 2013 and beyond. Future Oncol. 2014;10:1679–1694. https://doi.org/10.2217/fon.14.72
Dias A, Kote-Jarai Z, Mikropoulos C, Eeles R. Prostate Cancer Germline Variations and Implications for Screening and Treatment. Cold Spring Harb Perspect Med. 2018;8:a030379 https://doi.org/10.1101/cshperspect.a030379
Schumacher FR, Olama AAA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50:928–936. https://doi.org/10.1038/s41588-018-0142-8
Carter BS, Bova GS, Beaty TH, Steinberg GD, Childs B, Isaacs WB, et al. Hereditary prostate cancer: epidemiologic and clinical features. J Urol. 1993;150:797–802. https://doi.org/10.1016/s002-5347(17)35617-3
Rebbeck TR. Prostate Cancer Genetics: Variation by Race, Ethnicity, and Geography. Semin Radiat Oncol. 2017;27:3–10. https://doi.org/10.1016/j.semradonc.2016.08.002
Madersbacher S, Alcaraz A, Emberton M, Hammerer P, Ponholzer A, Schroder FH, et al. The influence of family history on prostate cancer risk: implications for clinical management. BJU Int. 2011;107:716–721. https://doi.org/10.1111/j.1464-410X.2010.10024.x
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71 https://doi.org/10.1136/bmj.n71
Barber L, Gerke T, Markt SC, Peisch SF, Wilson KM, Ahearn T, et al. Family History of Breast or Prostate Cancer and Prostate Cancer Risk. Clin Cancer Res. 2018;24:5910–5917. https://doi.org/10.1158/1078-0432.CCR-18-0370
Haraldsdottir S, Hampel H, Wei L, Wu C, Frankel W, Bekaii-Saab T, et al. Prostate cancer incidence in males with Lynch syndrome. Genet Med. 2014;16:553–557. https://doi.org/10.1038/gim.2013.193
Kote-Jarai Z, Easton DF, Stanford JL, Ostrander EA, Schleutker J, Ingles SA, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomark Prev. 2008;17:2052–2061. https://doi.org/10.1158/1055-9965.EPI-08-0317
Eeles RA, Olama AAA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45:385–391. https://doi.org/10.1038/ng.2560. 391e1-2
Conti DV, Darst BF, Moss LC, Saunders EJ, Sheng X, Chou A, et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat Genet. 2021;53:65–75. https://doi.org/10.1038/s41588-020-00748-0
Vietri MT, D’Elia G, Caliendo G, Resse M, Casamassimi A, Passariello L, et al. Hereditary Prostate Cancer: Genes Related, Target Therapy and Prevention. Int J Mol Sci. 2021;22:3753 https://doi.org/10.3390/ijms22073753
Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N. Engl J Med. 2016;375:443–453. https://doi.org/10.1056/nejmoa1603144
Leongamornlert D, Saunders E, Dadaev T, Tymrakiewicz M, Goh C, Jugurnauth-Little S, et al. Frequent germline deleterious mutations in DNA repair genes in familial prostate cancer cases are associated with advanced disease. Br J Cancer. 2014;110:1663–1672. https://doi.org/10.1038/bjc.2014.30
Lee YC, Lee YL, Li CY. BRCA Genes and Related Cancers: A Meta-Analysis from Epidemiological Cohort Studies. Med (Kaunas). 2021;57:905 https://doi.org/10.3390/medicina57090905
Silvestri V, Lesli G, Barnes DR, Agnarsson BA, Aittomaki K, Alducci E, et al. Characterization of the Cancer Spectrum in Men With Germline BRCA1 and BRCA2 Pathogenic Variants: Results From the Consortium of Investigators of Modifiers of BRCA1/2(CIMBA). JAMA Once. 2020;6:1–13. https://doi.org/10.1001/jamaoncol.2020.2134
Leongamornlert D, Mahmud N, Tymrakiewicz M, Saunders E, Dadaev T, Castro E, et al. Germline BRCA1 mutations increase prostate cancer risk. Br J Cancer. 2012;106:1697–1701. https://doi.org/10.1038/bjc.2012.146
Kote-Jarai Z, Leongamornlert D, Saunders E, Tymrakiewicz M, Castro E, Mahmud N et al. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: Implications for genetic testing in prostate cancer patients. Br J Cancer. 105:1230–1234. https://doi.org/10.1038/bjc.2011.383.
Gallagher DJ, Gaudet MM, Pal P, Kirchhoff T, Balistreri L, Vora K, et al. Germline BRCA mutations denote a clinicopathologic subset of prostate cancer. Clin Cancer Res. 2010;16:2115–2121. https://doi.org/10.1158/1078-0432.CCR-09-2871
Maier C, Herkommer K, Luedeke M, Rinckleb A, Schrader M, Vogel W. Subgroups of familial and aggressive prostate cancer with considerable frequencies of BRCA2 mutations. Prostate. 2014;74:1444–1451. https://doi.org/10.1002/pros.22860
Castro E, Romero-Laorden N, Del Pozo A, Lozano R, Medina A, Puente J, et al. PROREPAIR-B: A Prospective Cohort Study of the Impact of Germline DNA Repair Mutations on the Outcomes of Patients With Metastatic Castration-Resistant Prostate Cancer. J Clin Oncol. 2019;37:490–503. https://doi.org/10.1200/JCO.18.00358
Lang SH, Swift SL, White H, Misso K, Kleijnen J, Quek RGW. A systematic review of the prevalence of DNA damage response gene mutations in prostate cancer. Int J Oncol. 2019;55:597–616. https://doi.org/10.3892/ijo.2019.4842
Na R, Zheng SL, Han M, Yu H, Jiang H, Jiang D, et al. Germline Mutations in ATM and BRCA1/2 Distinguish Risk for Lethal and Indolent Prostate Cancer and are Associated with Early Age at Death. Eur Urol. 2017;71:740–747. https://doi.org/10.1016/j.eururo.2016.11.033
Carter HB, Helfand B, Mamawala M, Wu Y, Landis P, Yu H, et al. Germline Mutations in ATM and BRCA1/2 Are Associated with Grade Reclassification in Men on Active Surveillance for Prostate Cancer. Eur Urol. 2019;75:743–749. https://doi.org/10.1016/j.eururo.2018.09.021
Angele S, Falconer A, Edwards SM, Dork T, Bremer M, Moullan N, et al. ATM polymorphisms as risk factors for prostate cancer development. Br J Cancer. 2004;91:783–787. https://doi.org/10.1038/sj.bjc.6602007
Wang Y, Dai B, Ye D. CHEK2 mutation and risk of prostate cancer: a systematic review and meta-analysis. Int J Clin Exp Med. 2015;8:15708–15715.
Naslund-Koch C, Nordestgaard BG, Bojesen SE. Increased Risk for Other Cancers in Addition to Breast Cancer for CHEK2∗1100delC Heterozygotes Estimated From the Copenhagen General Population Study. J Clin Oncol. 2016;34:1208–1216. https://doi.org/10.1200/JCO.2015.63.3594
Cybulski C, Wokolorczyk D, Kluzniak W, Jakubowska A, Gorski B, Gronwald J, et al. An inherited NBN mutations is associated with poor prognosis prostate cancer. Br J Cancer. 2013;108:461–468. https://doi.org/10.1038/bjc.2012.486
Wu Y, Yu H, Zheng SL, Na R, Mamawala M, Landis T, et al. A comprehensive evaluation of CHEK2 germline mutations in men with prostate cancer. Prostate 2018;78:607–615. https://doi.org/10.1002/pros.23505
Southey MC, Teo ZL, Winship I. PALB2 and breast cancer: ready for clinical translation! Apple Clin Genet. 2013;6:43–52. https://doi.org/10.2147/TACG.S34116
Nicolosi P, Ledet E, Yang S, Michakski S, Freschi B, O’Leary E, et al. Prevalence of Germline Variants in Prostate Cancer and Implications for Current Genetic Testing Guidelines. JAMA Oncol. 2019;5:523–528. https://doi.org/10.1001/jamaoncol.2018.6760
Thompson ER, Boyle SE, Johnson J, Ryland GL, Sawyer S, Choong DYH, et al. Analysis of RAD51C germline mutations in high-risk breast and ovarian cancer families and ovarian cancer patients. Hum Mutat. 2012;33:95–99. https://doi.org/10.1002/humu.21625
Norris JD, Chang CY, Wittmann BM, Kunder RS, Cui H, Fan D, et al. The Homeodomain Protein HOXB13 Regulates the Cellular Response to Androgens. Mol Cell. 2019;36:405–416. https://doi.org/10.1016/j.molcel.2009.10.020
Cai Q, Wang X, Li X, Gong R, Guo X, Tang Y, et al. Germline HOXB13 p. Gly84Glu mutation and cancer susceptibility: A pooled analysis of 25 epidemiological studies with 145,257 participates. Oncotarget. 2015;6:42312–42321. https://doi.org/10.18632/oncotarget.5994
Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, et al. Germline Mutations in HOXB13 and Prostate-Cancer Risk. N. Engl J Med. 2012;366:141–149. https://doi.org/10.1056/NEJMoa1110000
Baretti M, Le DT. DNA mismatch repair in cancer. Pharm Ther. 2018;189:45–62. https://doi.org/10.1016/j.pharmthera.2018.04.004
Ponti G, Castellsagué E, Ruini C, Percesepe A, Tomasi A. Mismatch repair genes founder mutations and cancer susceptibility in Lynch syndrome. Clin Genet. 2015;87:507–516. https://doi.org/10.1111/cge.12529.
Ryan S, Jenkins MA, Win AK. Risk of prostate cancer in lynch syndrome: A systematic review and meta-Analysis. Cancer Epidemiol Biomark Prev. 2014;23:437–449. https://doi.org/10.1158/1055-9965.EPI-13-1165
Wu Y, Yu H, Li S, Wiley K, Zheng SL, LaDuca H, et al. Rare Germline Pathogenic Mutations of DNA Repair Genes Are Most Strongly Associated with Grade Group 5 Prostate Cancer. Eur Urol Oncol. 2020;3:224–230. https://doi.org/10.1016/j.euo.2019.12.003
Guedes LB, Antonarakis ES, Schweizer MT, Mirkheshti N, Almutairi F, Park JC, et al. MSH2 Loss in Primary Prostate Cancer. Clin Cancer Res. 2017;23:6863–6874. https://doi.org/10.1158/1078-0432.CCR-17-0955
Schaid DJ, McDonnel SK, FitzGerald LM, DeRycke L, Fogarty Z, Giles GG, et al. Two-stage Study of Familial Prostate Cancer by Whole-exome Sequencing and Custom Capture Identifies 10 Novel Genes Associated with the Risk of Prostate Cancer. Eur Urol. 2021;79:353–361. https://doi.org/10.1016/j.eururo.2020.07.038
Nguyen-Dumont T, Dowty JG, Maclnnis RJ, Steen JA, Riaz M, Dugué PA, et al. Rare Germline Pathogenic Variants Identified by Multigene Panel Testing and the Risk of Aggressive Prostate Cancer. Cancers. 2021;13:1945 https://doi.org/10.3390/cancers13071495
Giri VN, Knudsen KE, Kelly WK, Abida W, Andriole GL, Bangma CH, et al. Role of Genetic Testing for Inherited Prostate Cancer Risk: Philadelphia Prostate Cancer Consensus Conference 2017. J Clin Oncol. 2018;36:414–424. https://doi.org/10.1200/JCO.2017.74.1173
Doan DK, Schmidt KT, Chau CH, Figg WD, et al. Germline Genetics of Prostate Cancer: Prevalence of Risk Variants and Clinical Implications for Disease Management. Cancers. 2021;13:2154 https://doi.org/10.3390/cancers13092154
Allemailem KS, Almatroudi A, Alrumaihi F, Almansour NM, Aldakheel FM, Rather RA, et al. Single nucleotide polymorphisms (SNPs) in prostate cancer: its implications in diagnostics and therapeutics. Am J Transl Res. 2021;13:3868–3889.
Macinnis RJ, Antoniou AC, Eeles RA, Severi G, Olama AAA, McGuffog L, et al. A risk prediction algorithm based on family history and common genetic variants: application to prostate cancer with potential clinical impact. Genet Epidemiol. 2011;35:549–556. https://doi.org/10.1002/gepi.20605
Gronberg H, Adolfsson J, Aly M, Nordstrom T, Wiklund P, Brandberg Y, et al. Prostate cancer screening in men aged 50-69 years (STHLM3): a prospective population-based diagnostic study. Lancet Oncol. 2015;16:1667–1676. https://doi.org/10.1016/S1470-2045(15)00361-7
Castro E, Mikropoulos C, Bancroft EK, Dadaev T, Goh C, Taylor N, et al. The PROFILE Feasibility Study: Targeted Screening of Men With a Family History of Prostate Cancer. Oncologist 2016;21:716–722. https://doi.org/10.1634/theoncologist.2015-0336
Eeles RA. The PROFILE Study: Germline Genetic Profiling: Correlation With Targeted Prostate Cancer Screening and Treatment. https://www.clinicaltrials.gov/ct2/show/NCT02543905?term=nct02543905&draw=2&rank=1
R. A. Eeles, Institute of Cancer Research and Royal Marsden Hospital. “The IMPACT Study - Identification of Men With a Genetic Predisposition to Prostate Cancer. NCT00261456
Bancroft EK, Page EC, Brook MN, Thomas S, Taylor N, Pope J, et al. A prospective prostate cancer screening programme for men with pathogenic variants in mismatch repair genes (IMPACT): initiali results from an international prospective study. Lancet Oncol. 2021;22:1618–1631. https://doi.org/10.1016/S1470-2045(21)00522-2
Bancroft EK, Page EC, Castro E, Lilja H, Vickers A, Sjoberg D, et al. Targeted prostate cancer screening in BRCA1 and BRCA2 mutation carriers: results from the initial screening round of the IMPACT study. Eur Urol. 2014;66:489–499. 10.106/j.eururo.2014.01.003
National Comprehensive Cancer Network Guidelines Version 2.2021. Available online: https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf (accessed on February 2022).
European Association of Urology. Prostate Cancer guidelines. Available online: https://uroweb.org/guidelines/prostate-cancer (accessed on February 2022).
Parker C, Castro E, Fizazi K, Heindenreich A, Ost P, Procopio G et al. Prostate cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Available: https://www.annalsofoncology.org/action/showPdf?pii=S0923-7534%2820%2939898-7
Chen RC, Rumble BR, Loblaw DA, Finelli A, Ehdaie B, Cooperberg MR, et al. Active Surveillance for the Management of Localized Prostate Cancer (Cancer Care Ontario Guideline): American Society of Clinical Oncology Clinical Practice Guideline Endorsement. J Clin Oncol. 2016;34:2182–2190. https://doi.org/10.1200/JCO.2015.65.7759
Telang JM, Lane BR, Cher ML, Miller DC, Dupree JM. Prostate cancer family history and eligibility for active surveillance: a systematic review of the literature. BJU Int. 2017;120:464–467. https://doi.org/10.1111/bju.13862
Halstuch D, Ber Y, Kedar D, Golan S, Baniel J, Margel D. Short-term outcomes of active surveillance for low risk prostate cancer among men with germline DNA repair gene mutations. J Urol. 2020;204:707–713. https://doi.org/10.1097/ju.0000000000001027
De Bono J. TOPARP: A Phase II Trial of Olaparib in Patients With Advanced Castration Resistant Prostate Cancer. https://clinicaltrials.gov/ct2/show/NCT01682772
Mateo J, Porta N, Bianchini D, McGovern U, Elliott T, Jones R, et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP - B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21:162–174. https://doi.org/10.1016/S1470-2045(19)30684-9
De Bono J, Hussain M. Study of Olaparib (Lynparza™) Versus Enzalutamide or Abiraterone Acetate in Men With Metastatic Castration-Resistant Prostate Cancer (PROfound Study) https://clinicaltrials.gov/ct2/show/NCT02987543
A Study of Rucaparib in Patients With Metastatic Castration-resistant Prostate Cancer and Homologous Recombination Gene Deficiency (TRITON2). https://clinicaltrials.gov/ct2/show/NCT02952534
An Efficacy and Safety Study of Niraparib in Men With Metastatic Castration-Resistant Prostate Cancer and DNA-Repair Anomalies (Galahad). https://clinicaltrials.gov/ct2/show/NCT02854436
De Bono J. A Study of Talazoparib in Men With DNA Repair Defects and Metastatic Castration-Resistant Prostate Cancer. https://clinicaltrials.gov/ct2/show/NCT03148795?term=NCT03148795&draw=2&rank=1
Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N. Engl J Med. 2015;373:1697–1708. https://doi.org/10.1056/NEJMoa1506859
De Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med. 2020;382:2091–2102. https://doi.org/10.1056/NEJMoa1911440
Adiba W, Patnaik A, Campbell D, Shapiro J, Bryce AH, McDermott R, et al. Rucaparib in Men With Metastatic Castration Resistant Prostate Cancer Harboring a BRCA1 or BRCA2 Gene Alteration. J Clin Oncol. 2020;38:3763–3772. https://doi.org/10.1200/JCO.20.01035
Abida W, Campbell D, Patnaik A, Shapiro JD, Sautois B, Vogelzang NJ, et al. Non-BRCA DNA Damage Repair Gene Alterations and Response to the PARP Inhibitor Rucaparib in Metastatic Castration-Resistant Prostate Cancer: Analysis from the Phase II TRITON2 Study. Clin Cancer Res. 2020;26:2487–2496. https://doi.org/10.1158/1078-0432.CCR-20-0394
Smith MR, Scher HI, Sandhu S, Efstathiou E, Lara PN Jr. Yu EY, et al. Niraparib in patients with metastatic castration-resistant prostate cancer and DNA repair gene defects (GALAHAD); a multicentre, open-label, phase 2 trial. Lancet Oncol. 2022;23:362–373. https://doi.org/10.1016/S1470-2045(21)00757-9
De Bono JS, Mehra N, Scagliotti GV, Castro E, Dorff T, Stirling A, et al. Talazoparib monotherapy in metastatic castration-resistant prostate cancer with DNA repair alterations (TALAPRO-1): an open-label, phase 2 trial. Lancet Once. 2021;22:1250–1264. https://doi.org/10.1016/S1470-2045(21)00376-4
Le DT, Durham JN, Smith KN, Wang H, Barlett BR, Aulakh LK, et al. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. https://doi.org/10.1126/science.aan6733
Antonarakis ES, Shaukat F, Velho PI, Kaur H, Shenderov E, Pardoll DM, et al. Clinical Features and Therapeutic Outcomes in Men with Advanced Prostate Cancer and DNA Mismatch Repair Gene Mutations. Eur Urol. 2019;75:378–382. https://doi.org/10.1016/j.eururo.2018.10.009
Abida W, Cheng ML, Armenia J, Middha S, Autio KA, Vargas HA, et al. Analysis of the Prevalence of Microsatellite Instability in Prostate Cancer and Response to Immune Checkpoint Blockade. JAMA Once. 2019;5:471–478. https://doi.org/10.1001/jamaoncol.2018.5801
Barata P, Agarwal N, Nussenzveig R, Gerendash B, Jaeger E, Hatton W, et al. Clinical activity of pembrolizumab in metastatic prostate cancer with microsatellite instability high (MSI-H) detected by circulating tumor DNA. J Immunother Cancer. 2020;8:e001065. https://doi.org/10.1136/jitc-2020-001065
Mota JM, Barnett E, Nauseef JT, Nguyen B, Stopsack KH, Wibmer A, et al. Platinum-Based Chemotherapy in Metastatic Prostate Cancer With DNA Repair Gene Alterations. JCO Precis Oncol. 2020;4:355–366. https://doi.org/10.1200/po.19.00346
Author information
Authors and Affiliations
Contributions
Conception and design FM, ES; Acquisition of data FM, AT, CG, RB; Writing-original draft preparation FM, ES; Writing-review and editing AT, CG, RB, SM, FG; Critical revision of the paper for important intellectual content: RI, FP, PB, ES. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Marino, F., Totaro, A., Gandi, C. et al. Germline mutations in prostate cancer: a systematic review of the evidence for personalized medicine. Prostate Cancer Prostatic Dis 26, 655–664 (2023). https://doi.org/10.1038/s41391-022-00609-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41391-022-00609-3
This article is cited by
-
Gender-affirming hormone therapy in transgender women and risk of prostate cancer: pathophysiological mechanisms and clinical implications
Prostate Cancer and Prostatic Diseases (2024)
-
Germline Mutations and Ancestry in Prostate Cancer
Current Oncology Reports (2024)
-
Guidelines for genetic testing in prostate cancer: a scoping review
Prostate Cancer and Prostatic Diseases (2023)
-
Performance of clinical risk scores and prediction models to identify pathogenic germline variants in patients with advanced prostate cancer
World Journal of Urology (2023)