Abstract
Background
Germline mutations in DNA repair genes and KLK3 have been associated with adverse prostate cancer (PCa) outcomes in separate studies but never jointly. The objective of this study is to simultaneously assess these two types of germline mutations.
Methods
Germline rare pathogenic mutations (RPMs) in 9 commonly tested DNA repair genes and KLK3 variants were tested for their associations with PCa progression in two PCa cohorts: (1) hospital-based PCa patients treated with radical surgery at the Johns Hopkins Hospital (JHH, N = 1943), and (2) population-based PCa patients in the UK Biobank (UKB, N = 10,224). Progression was defined as metastasis and/or PCa-specific death (JHH) and PCa-specific death (UKB). RPMs of DNA repair genes were annotated using the American College of Medical Genetics recommendations. Known KLK3 variants were genotyped. Associations were tested using a logistic regression model adjusting for genetic background (top ten principal components).
Results
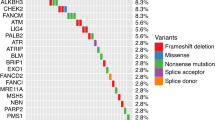
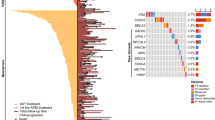
In the JHH, 3.2% (59/1,843) of patients had RPMs in 9 DNA repair genes; odds ratio (OR, 95% confidence interval) for progression was 2.99 (1.6–5.34), P < 0.001. In comparison, KLK3 I179T mutation was more common; 9.7% (189/1,943) carried the mutation, OR = 1.6 (1.05–2.37), P = 0.02. Similar results were found in the UKB. Both types of mutations remained statistically significant in multivariable analyses. In the combined cohort, compared to patients without any mutations (RPMs−/KLK3−), RPMs−/KLK3+ patients had modestly increased risk for progression [OR = 1.54 (1.15–2.02), P = 0.003], and RPMs+/KLK3+ patients had greatly increased risk for progression [OR = 5.41 (2.04–12.99), P < 0.001]. Importantly, associations of mutations with PCa progression were found in patients with clinically defined low- or intermediate risk for disease progression.
Conclusions
Two different cohorts consistently demonstrate that KLK3 I179T and RPMs of nine commonly tested DNA repair genes are complementary for predicting PCa progression. These results are highly relevant to PCa germline testing and provide critical information for KLK3 I179T to be considered in guidelines.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33.
Klotz L, Vesprini D, Sethukavalan P, Jethava V, Zhang L, Jain S, et al. Long-term follow-up of a large active surveillance cohort of patients with prostate cancer. J Clin Oncol. 2015;33:272–7.
Nyame YA, Stephenson AJ, Klein E. Re: long-term follow-up of a large active surveillance cohort of patients with prostate cancer. Eur Urol. 2015;68:906–7.
Cooperberg MR. Long-term active surveillance for prostate cancer: answers and questions. J Clin Oncol. 2015;33:238–40.
Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–53.
Na R, Zheng SL, Han M, Yu H, Jiang D, Shah S, et al. Germline mutations in ATM and BRCA1/2 distinguish risk for lethal and indolent prostate cancer and are associated with early age at death. Eur Urol. 2017;71:740–7.
Wu Y, Yu H, Li S, Wiley K, Zheng SL, LaDuca H, et al. Rare germline pathogenic mutations of DNA repair genes are most strongly associated with grade group 5 prostate cancer. Eur Urol Oncol. 2020;3:224–30.
Maier C, Herkommer K, Luedeke M, Rinckleb A, Schrader M, Vogel W. Subgroups of familial and aggressive prostate cancer with considerable frequencies of BRCA2 mutations. Prostate. 2014;74:1444–51.
Mijuskovic M, Saunders EJ, Leongamornlert DA, Wakerell S, Whitmore I, Dadaev T, et al. Rare germline variants in DNA repair genes and the angiogenesis pathway predispose prostate cancer patients to develop metastatic disease. Br J Cancer. 2018;119:96–104.
Darst BF, Dadaev T, Saunders E, Sheng X, Wan P, Pooler L, et al. Germline sequencing DNA repair genes in 5,545 men with aggressive and non-aggressive prostate cancer. J Natl Cancer Inst. 2020;113:616–25.
Momozawa Y, Iwasaki Y, Hirata M, Liu X, Kamatani Y, Takahashi A, et al. Germline pathogenic variants in 7636 Japanese patients with prostate cancer and 12 366 controls. J Natl Cancer Inst. 2020;112:369–76.
Mohler JL, Antonarakis ES, Armstrong AJ, D’Amico AV, Davis BJ, Dorff T, et al. Prostate cancer, version 2.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2019;17:479–505.
Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21.
Conti DV, Darst BF, Moss LC, Saunders EJ, Sheng X, Chou A, et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat Genet. 2021;53:65–75.
Ahn J, Berndt SI, Wacholder S, Kraft P, Kibel AS, Yeager M, et al. Variation in KLK genes, prostate-specific antigen and risk of prostate cancer. Nat Genet. 2008;40:1032–4.
Parikh H, Wang Z, Pettigrew KA, Jia J, Daugherty S, Yeager M, et al. Fine mapping the KLK3 locus on chromosome 19q13.33 associated with prostate cancer susceptibility and PSA levels. Hum Genet. 2011;129:675–85.
Kote-Jarai Z, Amin Al Olama A, Leongamornlert D, Tymrakiewicz M, Saunders E, Guy M, et al. Identification of a novel prostate cancer susceptibility variant in the KLK3 gene transcript. Hum Genet. 2011;129:687–94.
Xu J, Isaacs SD, Sun J, Li G, Wiley KE, Zhu Y, et al. Association of prostate cancer risk variants with clinicopathologic characteristics of the disease. Clin Cancer Res. 2008;14:5819–24.
Gudmundsson J, Besenbacher S, Sulem P, Gudbjartsson DF, Olafsson I, Arinbjarnarson S, et al. Genetic correction of PSA values using sequence variants associated with PSA levels. Sci Transl Med. 2010;2:62ra92.
Helfand BT, Roehl KA, Cooper PR, McGuire BB, Fitzgerald LM, Cancel-Tassin G, et al. Associations of prostate cancer risk variants with disease aggressiveness: results of the NCI-SPORE Genetics Working Group analysis of 18,343 cases. Hum Genet. 2015;134:439–50.
Gallagher DJ, Vijai J, Cronin AM, Bhatia J, Vickers AJ, Gaudet MM, et al. Susceptibility loci associated with prostate cancer progression and mortality. Clin Cancer Res. 2010;16:2819–32.
Pomerantz MM, Werner L, Xie W, Regan MM, Lee GS, Sun T, et al. Association of prostate cancer risk Loci with disease aggressiveness and prostate cancer-specific mortality. Cancer Prev Res. 2011;4:719–28.
Shui IM, Lindstrom S, Kibel AS, Berndt SI, Campa D, Gerke T, et al. Prostate cancer (PCa) risk variants and risk of fatal PCa in the National Cancer Institute Breast and Prostate Cancer Cohort Consortium. Eur Urol. 2014;65:1069–75.
Sullivan J, Kopp R, Stratton K, Manschreck C, Corines M, Rau-Murthy R, et al. An analysis of the association between prostate cancer risk loci, PSA levels, disease aggressiveness and disease-specific mortality. Br J Cancer. 2015;113:166–72.
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–9.
Shi Z, Platz EA, Wei J, Na R, Fantus RJ, Wang CH, et al. Performance of three inherited risk measures for predicting prostate cancer incidence and mortality: a population-based prospective analysis. Eur Urol. 2020;79:419–26.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Strande NT, Riggs ER, Buchanan AH, Ceyhan-Birsoy O, DiStefano M, Dwight SS, et al. Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the clinical genome resource. Am J Hum Genet. 2017;100:895–906.
Carter HB, Helfand B, Mamawala M, Wu Y, Landis P, Yu H, et al. Germline mutations in ATM and BRCA1/2 are associated with grade reclassification in men on active surveillance for prostate cancer. Eur Urol. 2019;75:743–9.
Cooperberg MR, Carroll PR, Dall’Era MA, Davies BJ, Davis JW, Eggener SE, et al. The state of the science on prostate cancer biomarkers: the San Francisco Consensus Statement. Eur Urol. 2019;76:268–72.
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–72.
Acknowledgements
This study was partially supported by grants from Department of Defense (W81XWH-16-1-0764, W81XWH-16-1-0765, and W81XWH-16-1-0766) and the National Cancer Institute (P30CA006973). We are grateful to the Ellrodt-Schweighauser, Chez and Melman families for establishing Endowed Chairs of Cancer Genomic Research and Personalized Prostate Cancer Care at NorthShore University HealthSystem in support of JX and BTH. Likewise, the support of W.T. Gerrard, Mario Duhon, Jennifer and John Chalsty is gratefully acknowledged by WBI. The authors gratefully acknowledge the generous support from donors to The Patrick C. Walsh Hereditary Prostate Cancer Research Program at The Brady Urological Institute.
Author information
Authors and Affiliations
Contributions
Conception and design: JX, WBI, KAC, and BTH. Acquisition of data: JX, WBI, and SLZ. Analysis and interpretation of data: ZS, JW, RN, C-HW, JX, WBI, KAC, and BTH. Drafting of manuscript: JX. Critical revision of the manuscript for important intellectual content: ZS, JW, RN, and C-HW, WKR, CS, MH, SLZ, BTH, KAC, WBI, and JX. Statistical analysis: ZS, JW, RN, C-HW. Obtaining funding: JX, WBI, and KAC. Administrative, technical, or material support: WKR and CS. Supervision: JX and WBI.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xu, J., Shi, Z., Wei, J. et al. KLK3 germline mutation I179T complements DNA repair genes for predicting prostate cancer progression. Prostate Cancer Prostatic Dis 25, 749–754 (2022). https://doi.org/10.1038/s41391-021-00466-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41391-021-00466-6