Abstract
Background
Pathogenic GATA6 variants have been associated with congenital heart disease (CHD) and a spectrum of extracardiac abnormalities, including pancreatic agenesis, congenital diaphragmatic hernia, and developmental delay. However, the comprehensive genotype-phenotype correlation of pathogenic GATA6 variation in humans remains to be fully understood.
Methods
Exome sequencing was performed in a family where four members had CHD. In vitro functional analysis of the GATA6 variant was performed using immunofluorescence, western blot, and dual-luciferase reporter assay.
Results
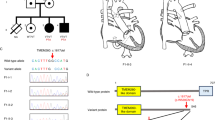
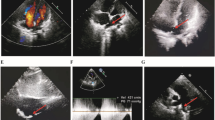
A novel, heterozygous missense variant in GATA6 (c.1403 G > A; p.Cys468Tyr) segregated with affected members in a family with CHD, including three with persistent truncus arteriosus. In addition, one member had childhood onset diabetes mellitus (DM), and another had necrotizing enterocolitis (NEC) with intestinal perforation. The p.Cys468Tyr variant was located in the c-terminal zinc finger domain encoded by exon 4. The mutant protein demonstrated an abnormal nuclear localization pattern with protein aggregation and decreased transcriptional activity.
Conclusions
We report a novel, familial GATA6 likely pathogenic variant associated with CHD, DM, and NEC with intestinal perforation. These findings expand the phenotypic spectrum of pathologic GATA6 variation to include intestinal abnormalities.
Impact
-
Exome sequencing identified a novel heterozygous GATA6 variant (p.Cys468Tyr) that segregated in a family with CHD including persistent truncus arteriosus, atrial septal defects and bicuspid aortic valve. Additionally, affected members displayed extracardiac findings including childhood-onset diabetes mellitus, and uniquely, necrotizing enterocolitis with intestinal perforation in the first four days of life.
-
In vitro functional assays demonstrated that GATA6 p.Cys468Tyr variant leads to cellular localization defects and decreased transactivation activity.
-
This work supports the importance of GATA6 as a causative gene for CHD and expands the phenotypic spectrum of pathogenic GATA6 variation, highlighting neonatal intestinal perforation as a novel extracardiac phenotype.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 14 print issues and online access
$259.00 per year
only $18.50 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data presented in current publication have been deposited in and are available in dbGaP database under dbGaP accession phs002010.v1.p1. Some restrictions apply for dbGaP, as data is available to researchers who meet access criteria. Data access is through the dbGaP website and researchers apply for data access. Supplied accession number should be used to search the dataset on the website: https://www.ncbi.nlm.nih.gov/gap/.
References
Tremblay, M., Sanchez-Ferras, O. & Bouchard, M. Gata transcription factors in development and disease. Development 145, dev164384 (2018).
Peterkin, T., Gibson, A., Loose, M. & Patient, R. The roles of Gata-4, -5 and -6 in vertebrate heart development. Semin Cell Dev. Biol. 16, 83–94 (2005).
Afouda, B. A. Towards understanding the gene-specific roles of gata factors in heart development: Does Gata4 lead the way? Int J. Mol. Sci. 23, 5255 (2022).
Pierpont, M. E. et al. Genetic basis for congenital heart disease: Revisited: A Scientific Statement from the American Heart Association. Circulation 138, e653–e711 (2018).
Yasuhara, J. & Garg, V. Genetics of congenital heart disease: A narrative review of recent advances and clinical implications. Transl. Pediatr. 10, 2366–2386 (2021).
Garg, V. et al. Gata4 mutations cause human congenital heart defects and reveal an interaction with Tbx5. Nature 424, 443–447 (2003).
Tomita-Mitchell, A., Maslen, C. L., Morris, C. D., Garg, V. & Goldmuntz, E. Gata4 sequence variants in patients with congenital heart disease. J. Med Genet 44, 779–783 (2007).
LaHaye, S. et al. Utilization of whole exome sequencing to identify causative mutations in familial congenital heart disease. Circ. Cardiovasc Genet 9, 320–329 (2016).
Yang, B. et al. Protein-altering and regulatory genetic variants near gata4 implicated in bicuspid aortic valve. Nat. Commun. 8, 15481 (2017).
Dixit, R. et al. Functionally significant, novel gata4 variants are frequently associated with tetralogy of fallot. Hum. Mutat. 39, 1957–1972 (2018).
Musfee, F. I. et al. Rare deleterious variants of Notch1, Gata4, Smad6, and Robo4 are enriched in bav with early onset complications but not in bav with heritable thoracic aortic disease. Mol. Genet Genom. Med. 8, e1406 (2020).
Padang, R., Bagnall, R. D., Richmond, D. R., Bannon, P. G. & Semsarian, C. Rare non-synonymous variations in the transcriptional activation domains of Gata5 in bicuspid aortic valve disease. J. Mol. Cell Cardiol. 53, 277–281 (2012).
Wei, D. et al. Gata5 loss-of-function mutations underlie tetralogy of fallot. Int J. Med. Sci. 10, 34–42 (2013).
Jiang, J. Q. et al. Prevalence and spectrum of Gata5 mutations associated with congenital heart disease. Int J. Cardiol. 165, 570–573 (2013).
Bonachea, E. M. et al. Rare Gata5 sequence variants identified in individuals with bicuspid aortic valve. Pediatr. Res. 76, 211–216 (2014).
Shi, L. M. et al. Gata5 loss-of-function mutations associated with congenital bicuspid aortic valve. Int J. Mol. Med. 33, 1219–1226 (2014).
Xin, M. et al. A threshold of Gata4 and Gata6 expression is required for cardiovascular development. Proc. Natl. Acad. Sci. USA 103, 11189–11194 (2006).
Nemer, G. & Nemer, M. Transcriptional activation of Bmp-4 and regulation of Mammalian organogenesis by Gata-4 and -6. Dev. Biol. 254, 131–148 (2003).
Molkentin, J. D. The zinc finger-containing transcription factors Gata-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 275, 38949–38952 (2000).
Lepore, J. J. et al. Gata-6 regulates semaphorin 3c and is required in cardiac neural crest for cardiovascular morphogenesis. J. Clin. Invest 116, 929–939 (2006).
Kodo, K. et al. Gata6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc. Natl. Acad. Sci. USA 106, 13933–13938 (2009).
Lin, X. et al. A novel Gata6 mutation in patients with tetralogy of fallot or atrial septal defect. J. Hum. Genet 55, 662–667 (2010).
Maitra, M., Koenig, S. N., Srivastava, D. & Garg, V. Identification of Gata6 sequence variants in patients with congenital heart defects. Pediatr. Res. 68, 281–285 (2010).
Kodo, K. et al. Genetic analysis of essential cardiac transcription factors in 256 patients with non-syndromic congenital heart defects. Circ. J. 76, 1703–1711 (2012).
Wang, J. et al. Novel Gata6 mutations associated with congenital ventricular septal defect or tetralogy of fallot. DNA Cell Biol. 31, 1610–1617 (2012).
Huang, R. T., Xue, S., Xu, Y. J. & Yang, Y. Q. Somatic mutations in the Gata6 gene underlie sporadic tetralogy of fallot. Int J. Mol. Med. 31, 51–58 (2013).
Zhang, E. et al. Targeted sequencing identifies novel Gata6 variants in a large cohort of patients with conotruncal heart defects. Gene 641, 341–348 (2018).
Škorić-Milosavljević, D. et al. Gata6 mutations: Characterization of two novel patients and a comprehensive overview of the gata6 genotypic and phenotypic spectrum. Am. J. Med Genet A 179, 1836–1845 (2019).
Gharibeh, L. et al. Gata6 regulates aortic valve remodeling, and its haploinsufficiency leads to right-left type bicuspid aortic valve. Circulation 138, 1025–1038 (2018).
Williams, S. G., Byrne, D. J. F. & Keavney, B. D. Rare Gata6 variants associated with risk of congenital heart disease phenotypes in 200,000 Uk biobank exomes. J. Hum. Genet 67, 123–125 (2022).
Allen, H. L. et al. Gata6 haploinsufficiency causes pancreatic agenesis in humans. Nat. Genet 44, 20–22 (2011).
Chao, C. S. et al. Novel Gata6 mutations in patients with pancreatic agenesis and congenital heart malformations. PLoS One 10, e0118449 (2015).
Du, Y. T., Moore, L., Poplawski, N. K. & De Sousa, S. M. C. Familial Gata6 mutation causing variably expressed diabetes mellitus and cardiac and renal abnormalities. Endocrinol. Diabetes Metab. Case Rep. 2019, 19–0022 (2019).
Sanchez-Lechuga, B. et al. Case report: Adult onset diabetes with partial pancreatic agenesis and congenital heart disease due to a de novo Gata6 mutation. BMC Med Genet 21, 70 (2020).
Raghuram, N., Marwaha, A., Greer, M. C., Gauda, E. & Chitayat, D. Congenital hypothyroidism, cardiac defects, and pancreatic agenesis in an infant with Gata6 mutation. Am. J. Med Genet A 182, 1496–1499 (2020).
De Franco, E. et al. Gata6 mutations cause a broad phenotypic spectrum of diabetes from pancreatic agenesis to adult-onset diabetes without exocrine insufficiency. Diabetes 62, 993–997 (2013).
Yu, L. et al. Whole exome sequencing identifies de novo mutations in Gata6 associated with congenital diaphragmatic hernia. J. Med Genet 51, 197–202 (2014).
Gordon, D. M. et al. Exome sequencing in multiplex families with left-sided cardiac defects has high yield for disease gene discovery. PLoS Genet 18, e1010236 (2022).
Manivannan, S. N. et al. Novel frameshift variant in myl2 reveals molecular differences between dominant and recessive forms of hypertrophic cardiomyopathy. PLoS Genet 16, e1008639 (2020).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17, 405–424 (2015).
Ittisoponpisan, S. et al. Can predicted protein 3d structures provide reliable insights into whether missense variants are disease associated? J. Mol. Biol. 431, 2197–2212 (2019).
Rodrigues, C. H. M., Pires, D. E. V. & Ascher, D. B. Dynamut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci. 30, 60–69 (2021).
Edelheit, O., Hanukoglu, A. & Hanukoglu, I. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol. 9, 61 (2009).
Klickstein, J. A., Mukkavalli, S. & Raman, M. Aggrecount: An unbiased image analysis tool for identifying and quantifying cellular aggregates in a spatially defined manner. J. Biol. Chem. 295, 17672–17683 (2020).
Collett, R. W. & Edwards, J. E. Persistent truncus arteriosus; a classification according to anatomic types. Surg. Clin. North Am. 29, 1245–1270 (1949).
Kelly, B. J. et al. Churchill: An ultra-fast, deterministic, highly scalable and balanced parallelization strategy for the discovery of human genetic variation in clinical and population-scale genomics. Genome Biol. 16, 6 (2015).
Ramensky, V., Bork, P. & Sunyaev, S. Human non-synonymous Snps: Server and survey. Nucleic Acids Res. 30, 3894–3900 (2002).
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. & Kircher, M. Cadd: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–d894 (2019).
Garber, M. et al. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 25, i54–i62 (2009).
Shihab, H. A. et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 34, 57–65 (2013).
Siepel, A. & Haussler, D. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol. Biol. Evol. 21, 468–488 (2004).
Davydov, E. V. et al. Identifying a high fraction of the human genome to be under selective constraint using Gerp++. PLoS Comput Biol. 6, e1001025 (2010).
Ioannidis, N. M. et al. Revel: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet 99, 877–885 (2016).
Quang, D., Chen, Y. & Xie, X. Dann: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 31, 761–763 (2015).
Yang, A. et al. Chdgene: A curated database for congenital heart disease genes. Circ. Genom. Precis Med 15, e003539 (2022).
Zheng, G. F. et al. A novel Gata6 mutation associated with congenital ventricular septal defect. Int J. Mol. Med. 29, 1065–1071 (2012).
Homsy, J. et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350, 1262–1266 (2015).
Jin, S. C. et al. Contribution of rare inherited and de novo variants in 2871 congenital heart disease probands. Nat. Genet 49, 1593–1601 (2017).
Sharma, A. et al. Gata6 mutations in hipscs inform mechanisms for maldevelopment of the heart, pancreas, and diaphragm. Elife 9, e53278 (2020).
Trainor, C. D., Ghirlando, R. & Simpson, M. A. Gata Zinc finger interactions modulate DNA binding and transactivation. J. Biol. Chem. 275, 28157–28166 (2000).
Bates, D. L., Chen, Y., Kim, G., Guo, L. & Chen, L. Crystal structures of multiple gata zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by gata. J. Mol. Biol. 381, 1292–1306 (2008).
Yau, D. et al. Case report: Maternal mosaicism resulting in inheritance of a novel gata6 mutation causing pancreatic agenesis and neonatal diabetes mellitus. Diagn. Pathol. 12, 1 (2017).
Slesnick, T. C., Sachdeva, R., Kreeger, J. R., Pernetz, M. A. & Border, W. L. in Echocardiography in Pediatric and Congenital Heart Disease 492–507 (2021).
Patel, R. M. & Denning, P. W. Intestinal microbiota and its relationship with necrotizing enterocolitis. Pediatr. Res 78, 232–238 (2015).
Guthrie, S. O. et al. Necrotizing enterocolitis among neonates in the United States. J. Perinatol. 23, 278–285 (2003).
McElhinney, D. B. et al. Necrotizing enterocolitis in neonates with congenital heart disease: Risk factors and outcomes. Pediatrics 106, 1080–1087 (2000).
Spinner, J. A. et al. Necrotizing enterocolitis and associated mortality in neonates with congenital heart disease: A Multi-Institutional Study. Pediatr. Crit. Care Med 21, 228–234 (2020).
Petrosyan, M., Guner, Y. S., Williams, M., Grishin, A. & Ford, H. R. Current concepts regarding the pathogenesis of necrotizing enterocolitis. Pediatr. Surg. Int 25, 309–318 (2009).
Kodo, K. et al. Regulation of Sema3c and the interaction between cardiac neural crest and second heart field during outflow tract development. Sci. Rep. 7, 6771 (2017).
Jiang, X. et al. Variants in a cis-regulatory element of Tbx1 in conotruncal heart defect patients impair Gata6-mediated transactivation. Orphanet J. Rare Dis. 16, 334 (2021).
Fenix, A. M. et al. Gain-of-function cardiomyopathic mutations in Rbm20 rewire splicing regulation and re-distribute ribonucleoprotein granules within processing bodies. Nat. Commun. 12, 6324 (2021).
Liu, J. et al. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. FASEB J. 20, 362–364 (2006).
Rajasekaran, N. S. et al. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell 130, 427–439 (2007).
Su, H. et al. Cop9 signalosome controls the degradation of cytosolic misfolded proteins and protects against cardiac proteotoxicity. Circ. Res 117, 956–966 (2015).
Rauch, R. et al. Comprehensive genotype-phenotype analysis in 230 patients with tetralogy of fallot. J. Med Genet 47, 321–331 (2010).
Walker, E. M., Thompson, C. A. & Battle, M. A. Gata4 and Gata6 regulate intestinal epithelial cytodifferentiation during development. Dev. Biol. 392, 283–294 (2014).
Walker, E. M., Thompson, C. A., Kohlnhofer, B. M., Faber, M. L. & Battle, M. A. Characterization of the developing small intestine in the absence of either Gata4 or Gata6. BMC Res Notes 7, 902 (2014).
Aronson, B. E., Stapleton, K. A. & Krasinski, S. D. Role of gata factors in development, differentiation, and homeostasis of the small intestinal epithelium. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G474–G490 (2014).
McMillan, T., Girgis, R. & Sellers, E. A. Neonatal diabetes and protein losing enteropathy: A case report. BMC Med. Genet 17, 32 (2016).
Kim, J. H., Sampath, V. & Canvasser, J. Challenges in diagnosing necrotizing enterocolitis. Pediatr. Res. 88, 16–20 (2020).
Sampath, V. et al. Sigirr genetic variants in premature infants with necrotizing enterocolitis. Pediatrics 135, e1530–e1534 (2015).
Härtel, C. et al. Nod2 loss-of-function mutations and risks of necrotizing enterocolitis or focal intestinal perforation in very low-birth-weight infants. Inflamm. Bowel Dis. 22, 249–256 (2016).
Cuna, A., George, L. & Sampath, V. Genetic predisposition to necrotizing enterocolitis in premature infants: current knowledge, challenges, and future directions. Semin Fetal Neonatal Med. 23, 387–393 (2018).
Bein, A., Eventov-Friedman, S., Arbell, D. & Schwartz, B. Intestinal tight junctions are severely altered in nec preterm neonates. Pediatr. Neonatol. 59, 464–473 (2018).
Högberg, N., Stenbäck, A., Carlsson, P. O., Wanders, A. & Lilja, H. E. Genes regulating tight junctions and cell adhesion are altered in early experimental necrotizing enterocolitis. J. Pediatr. Surg. 48, 2308–2312 (2013).
Laudisi, F. et al. Gata6 deficiency leads to epithelial barrier dysfunction and enhances susceptibility to gut inflammation. J. Crohns Colitis 16, 301–311 (2022).
Acknowledgements
We thank the family members for study participation, Gloria Zender in Cell Line Core and Samantha Fichtner and Jade Hayden in Heart Center Clinical Research Core.
Funding
This work was supported in part by funding from National Institutes of Health (R01 HL109758) to V.G., K.L.M, and P.W.
Author information
Authors and Affiliations
Contributions
J.Y. and V.G. conceived and designed the study. K.M, C.S., K.L.M, and V.G. enrolled patients. J.Y., M.A., K.M., and C.S. collected clinical data. J.Y., S.N.M., and U.M. performed and acquired experimental data. J.Y., S.N.M., D.M.G., P.J.L, and P.W. performed bioinformatics analyses. J.Y., S.N.M., U.M., D.M.G., P.J.L, K.L.M., P.W., and V.G. analyzed and interpreted the data. J.Y. and V.G. drafted the manuscript. C.S., A.M.B., M.G., H.Y., K.L.M., P.W., and V.G. critically reviewed the manuscript. All authors read and approved the final version of the manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
Statement of written consent was obtained as described in the Ethics statement.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yasuhara, J., Manivannan, S.N., Majumdar, U. et al. Novel pathogenic GATA6 variant associated with congenital heart disease, diabetes mellitus and necrotizing enterocolitis. Pediatr Res 95, 146–155 (2024). https://doi.org/10.1038/s41390-023-02811-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-023-02811-y