Abstract
Background
Non-alcoholic fatty liver disease (NAFLD) is one of the most common liver diseases in the U.S. and worldwide. The roles of early postnatal life stress (EPLS) and the fatty acid translocase (CD36) on the pathogenesis of adult-onset NAFLD remain unknown. We hypothesized that EPLS, in the form of neonatal maternal separation (NMS), would predispose mice towards developing adult NAFLD, increase hepatic CD36 expression, and differentially methylate Cd36 promoter concurrently.
Methods
NMS was performed on mice from postnatal day 1 to 21 and a high-fat/high-sucrose (HFS) diet was started at 4 weeks of age to generate four experimental groups: Naive-control diet (CD), Naive-HFS, NMS-CD, and NMS-HFS.
Results
NMS alone caused NAFLD in adult male mice at 25 weeks of age. The effects of NMS and HFS were generally additive in terms of NAFLD, hepatic Cd36 mRNA levels, and hepatic Cd36 promoter DNA hypomethylation. Cd36 promoter methylation negatively correlated with Cd36 mRNA levels. Two differentially methylated regions (DMRs) within Cd36 promoter regions appeared to be vulnerable to NMS in the mouse.
Conclusions
Our findings suggest that NMS increases the risk of an individual, particularly male, towards NAFLD when faced with a HFS diet later in life.
Impact
-
The key message of this article is that neonatal maternal separation and a postweaning high-fat/high-sucrose diet increased the risk of an individual, particularly male, towards NAFLD in adult life.
-
What this study adds to the existing literature includes the identification of two vulnerable differentially methylated regions in hepatic Cd36 promoters whose methylation levels very strongly negatively correlated with Cd36 mRNA.
-
The impact of this article is that it provides an early-life environment-responsive gene/promoter methylation model and an animal model for furthering the mechanistic study on how the insults in early-life environment are “transmitted” into adulthood and caused NAFLD.
Similar content being viewed by others
Introduction
Experiencing stress or adversity during early development increases the likelihood of developing chronic health disorders such as obesity1,2,3 and insulin resistance4,5 later in adulthood in both humans and animal models. As a result, a plethora of studies now exist exploring how early-life stress initiates the pathogenesis of obesity and insulin resistance. In contrast, the influence of early life stress on the pathogenesis of non-alcoholic fatty liver disease (NAFLD) remains largely unknown and receives relatively little attention.
NAFLD stands as the most common liver disease in the United States and worldwide. NAFLD is characterized primarily by excessive triglycerides (TG) in hepatocytes.6,7,8 The pathogenesis of NAFLD is multifactorial and remain elusive. Studies in recent years suggest that fatty acid translocase or cluster of differentiation 36 (CD36) plays a causal role in the pathogenesis of NAFLD.
CD36 is a multifunctional membrane receptor involved in long-chain fatty acid uptake, lipid metabolism, and inflammation.9,10,11,12 Hepatocytes normally express low levels of CD36 though CD36 expression increases with lipid-rich diets in humans and mouse models.13,14,15,16,17,18 Upregulation of CD36 membrane protein in liver elevates cellular uptake of fatty acids and positively correlates with hepatic steatosis.14,19,20,21 Liver-specific CD36 knockout attenuates steatosis in mouse models of NAFLD.22,23
Previously, our group demonstrated that an adverse early-life environment consisting of late pregnancy maternal stress and a maternal high-fat/high-sucrose (HFS) diet upregulated hepatic CD36 in a mouse model of adult NAFLD. The NAFLD occurred predominantly in male offspring, and a postweaning HFS diet further exacerbated the hepatic steatosis and upregulation of CD36.24 The increases in CD36 expression in this model occurred within the context of differentially methylated regions of the Cd36 promoter that correlated directly with hepatic NAFLD.24 A question exists as to whether other mouse models of early-life stress exposure will induce Cd36 promoter differential DNA CpG methylation, CD36 upregulation, and adult NAFLD, and, if so, this would suggest a potentially common conserved mechanism for the pathogenesis of NAFLD.
To answer this question, we used a previously established mouse model of neonatal maternal separation (NMS), that demonstrates increased susceptibility to weight gain, particularly on a HFS diet.5 We hypothesized that NMS mice would have greater incidence of NAFLD in adulthood. We also hypothesized that these phenotypical changes would be associated with Cd36 promoter differential DNA CpG methylation and CD36 upregulation.
Methods
Animals
All animal procedures were approved by the University of Kansas Medical Center Institutional Animal Care and Use Committee in compliance with the National Institute of Health Guide for the Care and Use of Laboratory Animals. Male and female C57Bl/6 mice (Charles River, Wilmington, MA) were housed in the Research Support Facility at the University of Kansas Medical Center. Mice were housed at 22 °C on a 12-h light cycle (600–1800) and received water and food ad libitum.
The NMS mouse model has been published preciously,5,25 with some modifications in this study. Briefly, pregnant C57Bl/6 dams were delivered to the animal facility between 14 and 16 days of gestation. Litters were divided equally into NMS and naive groups. From postnatal day (P) 1 until P21, all pups from a single cohort of pregnant dams were used for this study. NMS pups were removed en masse and placed in a clean glass beaker with bedding from their home cage for 180 min (11 a.m.–2 p.m.). The beaker was placed in an incubator maintained at 33 °C and 50% humidity. Naive mice remained undisturbed in their home cage except for normal animal husbandry. All mice were weaned on P22 and pair-housed with same sex litter mates and ad libitum access to water and a control diet (CD) composed of 20% kcal protein, 70% kcal carbohydrate (3.5% sucrose), and 10% kcal fat (Research Diets, Inc. New Brunswick, NJ, Cat. No. D12110704).
At 4 weeks of age, half of the naive and NMS groups were randomly placed on a high-fat/high-sucrose (HFS) diet, consisting of 20% kcal protein, 35% kcal carbohydrate (15% sucrose), and 45% kcal fat (4.73 kcal/g; Research Diets Cat. No. D12451, Supplementary Table 1) to mimic the higher fat and simple sugar content in a western-style diet.
At week 25 of life, mice were overdosed with inhaled isoflurane (>5%). Liver was dissected and weighed. Half of the liver was flash frozen in liquid nitrogen and stored at −80 °C. Half was fixed in 10% formalin.
Hepatic histology
Formalin-fixed livers were paraffin embedded. Slices (4 µm) were stained by hematoxylin and eosin and Masson’s trichrome. The NAFLD activity score (NAS) was evaluated by a pathologist blinded to the experimental groups by using the Kleiner scoring system.26 Briefly, the score is defined as the unweighted sum of the scores for steatosis (0–3), lobular inflammation (0–3), and ballooning (0–2). Fibrosis was recorded separately. A minimum of 5% steatosis (NAS score 1) was used for the operational minimal definition of histological NAFLD. NAS score 1–2 were largely considered mild NAFLD and considered “not nonalcoholic steatohepatitis (NASH)”. A score of ≥5 is interpreted as NASH.26
Hepatic TG quantification
Liver tissues were ground in liquid nitrogen. A portion of the grinds was weighed followed by TG isolation and quantification using Triglyceride Quantification Kit (MAK266, Sigma-Aldrich) following the manufacturer’s manual. The hepatic TG levels were expressed as mg/g of liver tissue.
Membrane protein extraction and immunoblotting
Liver membrane protein was extracted using a Mem-PER Plus Membrane Protein Extraction Kit (Thermo Scientific) and quantified using a Pierce BCA Protein Assay Kit (Thermo Scientific). Quantification of CD36 membrane protein was done using capillary immunoassay using Wes Simple Western system (Proteinsimple) as described previously.24 1:100 dilution of anti-CD36 antibody (ab133625, abcam) was used. 1:2000 pan Cadherin (ab51043, abcam) was used as an internal control.
RNA isolation and real-time reverse transcriptase (RT)–PCR
Total RNA isolation was performed by using RNeasy Mini Kit (74904, Qiagen) following the manufacturer’s instructions, including DNase I treatment. RNA was quantified spectrophotometrically. The integrity of RNA was assessed with an Agilent 2100 bioanalyzer in conjunction with the RNA 6000 Nano kit (Agilent). cDNA was synthesized using a High-capacity cDNA Reverse Transcription Kit (4368814, Thermo Fisher Scientific). Real-time RT-PCR was performed as described earlier.24 Target primers and probes for Cd36 total mRNA, Cd36 transcripts initiated from promoter 1 (P1 transcripts) and promoter 2 (P2 transcripts) were described previously.24 Transcripts initiated from promoter 3 (P3 transcripts) were calculated by subtracting P1 and P2 transcripts from total Cd36 since no specific assays can be designed for P3 transcripts.24 Primer efficiencies for these three sets of Cd36 primers/probes were tested and calculated from the slope using the formula Efficiency (%) = (10–1/slope − 1) × 100. Peptidylprolyl isomerase A (Ppia) was chosen as an internal control after assessing Ppia, glyceraldehyde-3-phosphate dehydrogenase (Gapdh), beta-actin (Actb), beta-2 microglobulin (B2m), beta-glucuronidase (Gusb), hydroxymethylbilane synthase (Hmbs), and hypoxanthine guanine phosphoribosyl transferase (Hprt) as candidate housekeeping genes. PCR conditions and calculations of mRNA expression were performed as demonstrated previously.24
Bisulfite pyrosequencing
Genomic DNA was isolated from liquid nitrogen-ground liver powder by using the DNeasy Blood & Tissue Kit (Qiagen) including RNase treatment. DNA quantity and purity were estimated spectrophotometrically. Bisulfite treatment of genomic DNA was performed using an EpiTect Plus Bisulfite Kit (Qiagen) as instructed in the manual.
For each PCR, bisulfite-treated DNA equivalent to 20 ng of the DNA prior to bisulfite treatment was used. Primers for PCR and sequencing were designed by using PyroMark Assay Design 2.0 software (Qiagen). Primer sequences, PCR conditions, and pyrosequencing for the three promoters were published previously.24 Promoter 2 has total 7 CpG sites within the proximal promoter. No specific assays can be designed for CpGs (-558, -556, and -241). Besides CpGs (-845, -740) studied previously, CpGs (-664, -645) were also added to this study by using the forward primer 5’TGAGTGAATAGAGAGATTGTTGTGGGATA and the reverse primer 5’Biosg/ACACACACCCCAAAAACAAA. PCR condition was 95 °C for 10 min, followed by 50 cycles of 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 30 s. Pyrosequencing was performed using Q48 Autoprep (Qiagen) with sequencing primer 5’AGATTGTTGTGGGATAT.
Statistics
Statistical analysis was performed using GraphPad Prism 8 software (GraphPad Software). Chi-square tests were performed to evaluate the diagnosis and severity of NAFLD between groups. To evaluate the severity of NAFLD, Chi-square tests were performed by subcategorizing NAS scores into NAS 0 (no steatosis), NAS 1–2 (steatosis, no NASH), NAS 3–4 (possible NASH or NASH), and NAS 5 and above (NASH) for evaluating the severity of NAFLD. Data are expressed as mean ± standard deviation (SD). Data were analyzed for normality or lognormality first and then analyzed by using 2-way ANOVA to assess main and interaction effects, with NMS and diet as independent variables. When significant main or interaction effect was detected, Bonferroni post hoc testing was used to identify the means that differ. An additive effect was reported when significant p values of both NMS-HFS compared with NMS-CD and NMS-HFS compared with Naive-HFS were found, but p interaction was not significant.24,27 Correlation analyses were computed using Pearson correlation coefficients. The level of significance was set at p < 0.05 for all statistical tests.
Results
NMS and HFS significantly increased the body fat percentage
Both NMS and HFS significantly increased body fat percentage in both male (Supplementary Fig. 1) and female mice28 at 14 and 25 weeks of life. In this model, NMS significantly decreased serum corticosterone (CORT) in males29 and resulted a trend toward increased serum CORT in female mice.28
NMS and HFS significantly increased the prevalence and severity of hepatic steatosis in adult mice
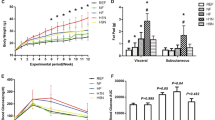
Hepatic steatosis was diagnosed based on NAS scores, with the criteria of 5% steatosis as the operational minimal definition of histological NAFLD.26 In males, Chi-square analysis revealed that all the three experimental groups had significantly more hepatic steatosis diagnosed compared to Naive-CD mice (Naive-HFS: p = 0.0004, NMS-CD: p = 0.0117, NMS-HFS: p = 0.0004) (Fig. 1a). In females, NMS-HFS mice had a significantly higher incidence of steatosis compared with Naive-CD (p = 0.0455) and Naive-HFS mice (p = 0.0455), but not with NMS-CD mice (p = 0.0721) (Fig. 1b). Only a few mild fibrosis cases were observed among the groups with no statistical significance detected (data not shown).
NAFLD activity scores in male (a) and female (b) mice. Labeled groups without a common letter differ (steatosis severity A < B < C < D), p < 0.05. Hepatic TG levels in male (c) and female (d) mice. Values are means ± SDs. n = 8. § and * denote significant NMS and diet effects, respectively. &, &&, &&& p < 0.05, 0.01, 0.001 HFS vs CD. CD control diet, HFS high-fat/high-sucrose diet, Naive no stress control group, NMS neonatal maternal separation, NAS NAFLD activity score, TG triglycerides.
Next, the impact of NMS and HFS diet on the severity of steatosis was determined. Chi-square analyses were performed by subcategorizing NAS scores into NAS 0 (no steatosis), NAS 1–2 (steatosis, no NASH), NAS 3–4 (possible NASH or NASH), and NAS 5 and above (NASH).26 In males, NMS-CD mice had more severe steatosis than Naive-CD mice (p = 0.0209) and NMS-HFS mice had significantly more cases of severe steatosis than Naive-HFS mice (p = 0.0389) (Fig. 1a). These data indicated that NMS significantly worsened the development of severe NAFLD in male mice, regardless of diet. However, in females, most of the observed steatosis was mild (only 2 with NAS 3–4 and no NAS 5) and only NMS-HFS mice had more severe steatosis compared to Naive-CD (p = 0.0455) or Naive-HFS mice (p = 0.0455) (Fig. 1b).
To further confirm these histological findings, hepatic TG content was quantified. Both NMS and HFS diet had significant main effects on increasing hepatic TG contents in male livers (p = 0.0154 and p < 0.0001, respectively) (Fig. 1c). Only a significant diet effect was found to increase hepatic TG levels in females (p = 0.0047) (Fig. 1d).
HFS diet significantly upregulated hepatic CD36 membrane protein in both sexes
HFS diet had a significant effect on increasing hepatic CD36 membrane protein levels in male and female mice (p = 0.0021 and 0.0024, respectively) (Fig. 2a, b). NMS did not significantly impact hepatic CD36 membrane protein levels in either sex (p > 0.05); however, NMS-HFS mice had significantly higher CD36 membrane protein levels compared to NMS-CD in both male and female mice (p = 0.025 and 0.0405, respectively) (Fig. 2a, b).
CD36 membrane protein expression and immunoblot image (a, b) and correlation analysis (c, d) in livers of 25-week-old male and female mice from the Naive and NMS groups and on the postweaning diet of CD and HFS. a, b Values are means ± SDs. n = 8. *Denotes significant diet effect. &p < 0.05 HFS vs CD. CD control diet, HFS high-fat/high-sucrose diet, Naive no stress control group, NMS neonatal maternal separation, R Pearson’s r, TG triglycerides.
A significant positive correlation was found between the levels of CD36 membrane protein and hepatic TG contents in male mice (p = 0.0227) (Fig. 2c), with a similar trend in female mice (p = 0.116) (Fig. 2d).
NMS and HFS diet synergistically upregulated Cd36 total mRNA and P2-initiated transcripts in male livers
Three different promoters ((P)1, P2, or P3) initiate transcription of the Cd36 gene, with P3 serving as the primary promoter under “normal” conditions.24 To determine whether NMS or HFS diet impacted transcriptional initiation of Cd36, specific primers/probes were designed for RT-PCR.24 The assay efficiencies for the total Cd36 mRNA, P2 transcripts, and P1 transcripts were 91.89%, 90.84%, and 91.75%, respectively, allowing for comparisons of the impact of NMS and HFS on the transcriptional activity between promoters.
As expected, in the male Naive-CD mice, P3-initiated transcripts were the major mRNA variant species in the liver (Fig. 3a and Supplementary Fig. 2a). However, P3- and P2-initiated transcripts were similarly expressed in female Naive-CD livers (Fig. 3b and Supplementary Fig. 3a). P1 transcripts were negligibly expressed in both sexes indicating that promoter 1 is minimally used for transcription in mouse livers (Fig. 3), which is consistent with our previous data.24
Hepatic mRNA expression of Cd36 total mRNA, P1, P2, and P3 transcripts in male (a) and female (b) livers of 25-week-old mice from the Naive and NMS groups and on the postweaning diet of CD and HFS. Values are mRNA expression levels relative to internal control Ppia and expressed as means ± SDs. n = 8. §, *, and ‡ denote significant effects of NMS, diet, and a NMS/diet interaction, respectively. &, &&, &&& p < 0.05, 0.01, 0.001 HFS vs CD. ### p < 0.001 NMS-HFS vs Naive-HFS. CD control diet, HFS high-fat/high-sucrose diet, Naive no stress control group, NMS neonatal maternal separation, P promoter.
In male mice, both NMS and HFS diet had significant main effects on the upregulation of Cd36 total mRNA (p < 0.0001 and p = 0.0003, respectively) and P2 transcripts (p < 0.0001 and p = 0.0003, respectively) (Fig. 3a). Importantly, NMS and HFS diet synergistically upregulated both Cd36 total mRNA and P2 transcripts (p = 0.0417 and 0.0333, respectively) with significantly higher levels in NMS-HFS mice compared to Naive-HFS mice (p = 0.0009 and 0.0007, respectively). Only a significant effect of HFS diet was found on P3 transcripts upregulation (p < 0.0001) (Fig. 3a). No significant difference was found for P1 transcripts between groups (Fig. 3a). Strikingly, in comparison to Naive-CD male livers, NMS-HFS increased the expression of P2 transcripts by 21.63-folds (0.173 ± 0.065 vs 0.008 ± 0.009), of P3 by 2.36-folds (0.066 ± 0.015 vs 0.028 ± 0.008), and of total Cd36 mRNA by 6.21-folds (0.242 ± 0.067 vs 0.039 ± 0.017). Altogether, NMS and HFS diet shifted the major promoter from P3 to P2 in male livers (Fig. 3a and Supplementary Fig. 2b).
In female mice, only a significant effect of HFS diet was found on the upregulation of total Cd36 mRNA (p < 0.0001), P2-initiated transcripts (p = 0.0005), and P3 transcripts (p < 0.0001) (Fig. 3b). Relative to Naive-CD mice, exposure to NMS and HFS diet increased the expression of P2 transcripts by 3.39-folds (0.073 ± 0.044 vs 0.022 ± 0.013), of P3 transcripts by 1.56-folds (0.047 ± 0.010 vs 0.030 ± 0.007), and of total Cd36 mRNA by 2.26-folds (0.123 ± 0.049 vs 0.054 ± 0.019). Both P2 and P3 were the main transcription initiating promoters without significant difference between the two (Fig. 3b and Supplementary Fig. 3a, b).
Cd36 total mRNA, P2- and P3-initiated transcripts were significantly positively correlated with the hepatic TG contents
Expression levels of Cd36 total mRNA and the P2 and P3 transcripts were correlated with hepatic TG contents. As with the protein expression data, the Cd36 total mRNA, P2 transcripts, and P3 transcripts were significantly positively correlated with the hepatic TG contents in both male and female livers (Fig. 4). P1 transcript levels were not correlated with hepatic TG (Fig. 4).
Correlations between hepatic TG contents and Cd36 total mRNA, P1, P2, and P3 transcripts in 25-week-old male (a) and female (b) mice from the Naive and NMS groups and on the postweaning diet of CD or HFS. CD control diet, HFS high-fat/high-sucrose diet, Naive no stress control group, NMS neonatal maternal separation, R Pearson’s r, TG triglycerides.
NMS and HFS significantly hypomethylated promoters 2 and 3 of Cd36 in male liver
DNA hypomethylation has previously been associated with transcriptional upregulation of hepatic Cd36 promoters.24 To determine if NMS and/or HFS diet-increased expression of P2 and P3 was related to DNA hypomethylation, bisulfite pyrosequencing of all three promoter regions was carried out. As expected, in the male livers, both NMS and HFS diet had significant main effects on the hypomethylation of two (CpGs (-845) and (-740)) out of four CpG sites studied around P2 and all six CpG sites around P3 (Fig. 5a). Importantly, additive effects of NMS and HFS diet were found on the hypomethylation of CpG (-740) in P2 and five (CpGs (-846, -835, -286, -269, -254)) out of the six CpG sites in P3, with NMS-HFS mice having significantly lower methylation compared to either Naive-HFS or NMS-CD mice (p < 0.05) (Fig. 5a). We defined the regions around CpG (-845, -740) of P2 and the regions around all the six CpG sites as differentially methylated regions (DMRs), which were sensitive to both NMS and postweaning HFS diet.
Percent of DNA CpG methylation of Cd36 promoters 1, 2, and 3 in the livers of 25-week-old male (a) and female (b) mice from the Naive and NMS groups and on the postweaning diet of CD or HFS. The negative number below each CpG site indicates the number of base pair upstream relative to the transcription start site of the corresponding promoter, respectively. Values are means ± SDs. n = 8. §, *, and ‡ denote significant effects of NMS, diet, and a NMS/diet interaction, respectively. &, &&, &&&, &&&& p < 0.05, 0.01, 0.001, 0.0001 HFS vs CD. #, ## p < 0.05, 0.01 NMS-HFS vs Naive-HFS. CD control diet, HFS high-fat/high-sucrose diet, Naive no stress control group, NMS neonatal maternal separation.
Paralleling the more moderate impacts of NMS and HFS diet on the expression of Cd36 mRNA, female liver exhibited less DNA hypomethylation, compared to the male mice (Fig. 5b). Only a NMS effect was observed for two (CpG (-845) and (-740)) of the four CpGs in P2 (p = 0.0132 and 0.0136, respectively) (Fig. 5b). NMS and HFS diet had significant main effects on the hypomethylation of five (-846, -835, −804, -269, -254) out of the six CpGs in P3. A synergistic effect between NMS and HFS was found on CpG (-269) and CpG (-254) (p = 0.0266 and 0.0197, respectively) (Fig. 5b). Altogether, the environment sensitive DMRs identified in male livers were similarly impacted by NMS and HFS diet in female livers, although to a lesser extent.
Cd36 total mRNA, P2- and P3-initiated transcripts were strongly and negatively correlated with promoter methylation in a CpG- and RNA variant-specific manner
Correlation analysis revealed strong negative correlations between the methylation of promoters and the levels of Cd36 total mRNA or variant transcripts in both sexes (Fig. 6). Specifically, CpG (-845) and (-740) of P2 and all six CpGs of P3 were very strongly and negatively correlated with total Cd36 mRNA and P2 transcripts, and moderately, with P3 transcripts (Fig. 6 and Supplementary Table 2). Importantly, these correlations were CpG site specific because the methylation status of the two studied CpGs in P1 and CpG (-664) and (-645) in P2 was not correlated with expression of any mRNA species of Cd36. Similarly, these correlations were Cd36 mRNA variant specific because the levels of P1-initiated transcripts were not correlated with methylation of any CpG site tested across the three promoters.
Pearson correlation score obtained from comparisons of promoter CpG methylation with Cd36 total mRNA and promoter transcripts in male (a) and female (b) livers at 25 weeks of age. The negative number below the graphs indicates the number of base pair upstream relative to the transcription start site of the corresponding exon, respectively. Values are Pearson’s r. P promoter.
These results, together with our previous findings in an independently conducted study in a mouse model of different early life environment (Table 1),24 indicated that the DMRs in P2 and P3 have conserved transcriptional regulatory roles in mouse liver that are sensitive to early life environments.
Discussion
The core findings of this study suggest that NMS in mice decreases DNA CpG methylation in the hepatic Cd36 promoter, increases hepatic Cd36 mRNA and protein levels, and causes NAFLD in adult male mice. Specific findings of this study include (1) the observation that postweaning HFS diet exacerbates the impact of NMS on NAFLD, and the putative associated mechanisms included in this study; (2) the positive correlation of Cd36 mRNA and protein levels with hepatic TG content; and (3) the identification of two vulnerable DMRs in the Cd36 promoter whose methylation levels very strongly negatively correlated with Cd36 mRNA in a mRNA variant- and CpG site-specific manner. The findings in males parallel similar findings from a previous model involving a maternal adverse early-life environment conducted at another institution (Table 1). Considering the methodological differences between the two model, we speculate that hypomethylation of the hepatic Cd36 DMRs represents a conserved mechanism through which adverse early life event and stressors initiate the pathogenesis of later life NAFLD.
A link between early life events and the development of NAFLD later in life does exist in the literature. For example, prenatal hypoxia followed by subsequent hypoxia at 6 months of life led to NALFD in male rats.30 This model of NAFLD is striking as it occurred in the absence of an obesogenic diet.30 Our previous study demonstrating a link between early life events and NAFLD described the impact of an adverse maternal environment (AME) consisting of both maternal chronic stress during late pregnancy, induced by non-invasive environmental perturbation such as reduced bedding, and maternal HFS diet from 5 weeks preconception through lactation.24 AME-exposed male offspring developed more frequent hepatic steatosis than the controls from this model despite consuming a low-fat control diet for 14 weeks after weaning. Interestingly, a postweaning HFS diet exacerbated the hepatic steatosis phenotype in the AME male offspring. In this previous study, the influence of prenatal stress on adult-onset NAFLD could not be determined because early-life exposure to obesogenic diet alone has been shown to cause the NAFLD phenotype.31,32 In contrast, the current study demonstrates that NMS alone increased the risk of developing NAFLD later in adulthood in a sex-specific manner.
A link between CD36 and adult NAFLD has been well-established in prior studies. Upregulation of CD36 membrane protein positively correlates with elevated hepatocyte uptake of fatty acids and hepatic steatosis in models of NAFLD originating in adulthood.14,19,20 Conversely, hepatocyte-specific deletion of CD36 attenuates steatosis in rodent models of adult NAFLD.22,23,33 Our previous study and the current data further support a role of CD36 in NAFLD development, as both showed a strong correlation between hepatic Cd36 mRNA and protein levels with TG content, as well as increased steatosis. The similarities of the findings between the two disparate models of early life stress suggest that upregulation of CD36 may provide a conserved mechanism underlying the pathogenesis of NAFLD. Further work is needed to determine the necessity of CD36 in early life stress-mediated NAFLD development, particularly in the absence of a HFS diet.
Epigenetic modifications are also associated with early life stress exposure and the development of negative health outcomes.34 As such, the two models of early life stress appear to share a conserved mechanism of increasing CD36 through the hypomethylation of the differentially methylated regions (DMRs) of the hepatic Cd36 promoters 2 and 3 (Table 1). Both P2 and P3 of Cd36 function as active promoters that nature conserves between mouse and human.22,35,36,37,38 Literature focusing upon transcriptional regulation of P2 is rare. Multiple transcription factor response elements have been reported within promoter 3.22,33,39,40 These transcription factors include liver X receptor (LXR), prognane X receptor (PXR), peroxisome proliferator activated receptor (PPAR) gamma, and the aryl hydrocarbon receptor (AhR).22,33,39,40 These TFs bind to their cognate cis-regulatory elements around −230 bp for PPAR, −400bp for PXR, −1080 bp for LXR, and −1250 bp for AhR upstream of the P3 transcription start site (TSS), respectively.22,33,39,40 Binding of these transcription factors to their respective response elements upregulates total Cd36 mRNA.
Functional studies of these two promoters suggest that they operate somewhat independently. For example, a diet containing a PPARα ligand upregulates hepatic P2-initiated transcripts, but not P3 transcripts.36,37 We specifically found that exposure to both NMS and HFS diet increased P2 transcripts approximately 22 (male) and 3.4 (female) fold, whereas increased P3 transcripts only 2.4 (male) and 1.6 (female) fold. Of importance, methylation levels of the identified DMRs in P2 and P3 significantly correlated with P2 transcript levels. These significant changes and correlations suggest that the methylation status of these DMRs are responsive to environmental exposures, such as stress and diet, and play a key role in the transcriptional regulation of hepatic Cd36 gene.41 Further studies are needed to elucidate other pathways by which the early postnatal life environment mediate changes in Cd36 gene expression and epigenetics, including how promoter 2 is activated.
Cd36 promoters 2 and 3 are CpG sparse with most of the CpG sites intermediately methylated. Genome-wide studies have shown that the genomic regions with intermediate DNA methylation (IM) is a conserved chromatin signature of genome regulation.42 These IM regions have average 57% methylation and are allele-independent. The IM signature enriches for regions with multiple indicators of regulatory functions, particularly those associated with enhancers, however, the precise function of intermediate DNA methylation states is not clear.42 Sae-Lee et al. have shown that the IM regions had binding specificity to certain DNMT isoforms over others using overexpression experiments in a human cell line.43 Importantly, DMRs in IM regions have been reported to be enriched among those associated with human diseases, such as allergic sensitization, in human association studies.44 Our methylation data of the mouse Cd36 promoters 2 and 3 had the feature of IM [except the CpGs (-664 and -665 of P2), which are inside the simple repeats and to be expected to be highly methylated] and responsive to early-life environments. Therefore, our animal/disease/gene model provides a unique opportunity to study epigenetic regulation of IM chromatin signature.
Consistent with human observations and previous rodent models, male mice in the study had more severe outcomes compared to the female mice.45,46 Both showed similar trends in terms of NAFLD prevalence/score, CD36 levels, and methylation changes, as well as hypomethylation at the same DMRs. Also, the severity of hepatic steatosis positively and significantly correlated with hepatic CD36 expression and negatively correlated with DNA CpG methylation of hepatic Cd36 P2 and P3, suggesting a conserved mechanism between the sexes. Future work will be necessary to understand how sex is influencing the impact of early life stress and HFS diet on hepatic outcomes, particularly considering that previous studies of the same NMS model have shown similar changes in urogenital hypersensitivity and function in male and female mice.47,48
Caution is always necessary when attempting to apply data from mouse models to human pathophysiology. Studies have shown that the development and severity of induced NAFLD is dependent on strain and species of the mice.49 Further studies are needed to see if the findings in the current and the comparator studies are applicable to other species, especially in humans.
In summary, exposure to NMS in mice resulted in adult-onset NAFLD, concurrent with upregulation of hepatic CD36 expression and hypomethylation of Cd36 promoter DMRs. The remarkable similarity of these findings to that of another model suggest that this may be a conserved response to early life stress. Regardless, these findings highlight the importance of the early postnatal life environment on the health in later life and provide a gene/animal model tool to further understanding the mechanisms at play.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Williamson, D. F., Thompson, T. J., Anda, R. F., Dietz, W. H. & Felitti, V. Body weight and obesity in adults and self-reported abuse in childhood. Int. J. Obes. Relat. Metab. Disord. 26, 1075–1082 (2002).
Lissau, I. & Sorensen, T. I. Parental neglect during childhood and increased risk of obesity in young adulthood. Lancet 343, 324–327 (1994).
Danese, A. & Tan, M. Childhood maltreatment and obesity: systematic review and meta-analysis. Mol. Psychiatry 19, 544–554 (2014).
Kaufman, D. et al. Early-life stress and the development of obesity and insulin resistance in juvenile bonnet macaques. Diabetes 56, 1382–1386 (2007).
Eller, O. C., Morris, E. M., Thyfault, J. P. & Christianson, J. A. Early life stress reduces voluntary exercise and its prevention of diet-induced obesity and metabolic dysfunction in mice. Physiol. Behav. 223, 113000 (2020).
Friedman, S. L., Neuschwander-Tetri, B. A., Rinella, M. & Sanyal, A. J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24, 908–922 (2018).
Ruhl, C. E., Clark, J. M. & Everhart, J. E. in Diabetes in America, 3rd edn (eds Cowie, C. C. et al.) Ch. 26 (NIDDK, 2018).
Bellentani, S. The epidemiology of non-alcoholic fatty liver disease. Liver Int. 37, 81–84 (2017).
Samovski, D. et al. Regulation of AMPK activation by CD36 links fatty acid uptake to beta-oxidation. Diabetes 64, 353–359 (2015).
Nassir, F., Adewole, O. L., Brunt, E. M. & Abumrad, N. A. CD36 deletion reduces VLDL secretion, modulates liver prostaglandins, and exacerbates hepatic steatosis in Ob/Ob mice. J. Lipid Res. 54, 2988–2997 (2013).
Li, Y. et al. Cd36 plays a negative role in the regulation of lipophagy in hepatocytes through an AMPK-dependent pathway. J. Lipid Res. 60, 844–855 (2019).
Su, X. & Abumrad, N. A. Cellular fatty acid uptake: a pathway under construction. Trends Endocrinol. Metab. 20, 72–77 (2009).
Bechmann, L. P. et al. Apoptosis is associated with CD36/fatty acid translocase upregulation in non-alcoholic steatohepatitis. Liver Int. 30, 850–859 (2010).
Miquilena-Colina, M. E. et al. Hepatic fatty acid translocase CD36 upregulation is associated with insulin resistance, hyperinsulinaemia and increased steatosis in non-alcoholic steatohepatitis and chronic hepatitis C. Gut 60, 1394–1402 (2011).
Greco, D. et al. Gene expression in human NAFLD. Am. J. Physiol. Gastrointest. Liver Physiol. 294, G1281–G1287 (2008).
Zhao, L. et al. CD36 palmitoylation disrupts free fatty acid metabolism and promotes tissue inflammation in non-alcoholic steatohepatitis. J. Hepatol. 69, 705–717 (2018).
Sheedfar, F. et al. Increased hepatic CD36 expression with age is associated with enhanced susceptibility to nonalcoholic fatty liver disease. Aging 6, 281–295 (2014).
Hajri, T., Zaiou, M., Fungwe, T. V., Ouguerram, K. & Besong, S. Epigenetic regulation of peroxisome proliferator-activated receptor gamma mediates high-fat diet-induced non-alcoholic fatty liver disease. Cells 10, 1355 (2021).
Koonen, D. P. et al. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes 56, 2863–2871 (2007).
Luiken, J. J. et al. Increased rates of fatty acid uptake and plasmalemmal fatty acid transporters in obese Zucker rats. J. Biol. Chem. 276, 40567–40573 (2001).
Buque, X. et al. A subset of dysregulated metabolic and survival genes is associated with severity of hepatic steatosis in obese Zucker rats. J. Lipid Res. 51, 500–513 (2010).
Zhou, J. et al. Hepatic fatty acid transporter CD36 is a common target of LXR, PXR, and Ppargamma in promoting steatosis. Gastroenterology 134, 556–567 (2008).
Wilson, C. G. et al. Hepatocyte-specific disruption of CD36 attenuates fatty liver and improves insulin sensitivity in HFD-fed mice. Endocrinology 157, 570–585 (2016).
Fu, Q. et al. Adverse maternal environment and postweaning western diet alter hepatic CD36 expression and methylation concurrently with nonalcoholic fatty liver disease in mouse offspring. J. Nutr. 151, 3102–3112 (2021).
Eller, O. C. et al. An omega-3-rich anti-inflammatory diet improved widespread allodynia and worsened metabolic outcomes in adult mice exposed to neonatal maternal separation. Neuroscience 468, 53–67 (2021).
Kleiner, D. E. et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321 (2005).
Roell, K. R., Reif, D. M. & Motsinger-Reif, A. A. An introduction to terminology and methodology of chemical synergy-perspectives from across disciplines. Front. Pharm. 8, 158 (2017).
Frick, J. M. et al. High-fat/high-sucrose diet worsens metabolic outcomes and widespread hypersensitivity following early-life stress exposure in female mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 324, R353–R367 (2023).
Fuentes, I. M. et al. Voluntary wheel running improves outcomes in an early life stress-induced model of urologic chronic pelvic pain syndrome in male mice. Pain 162, 1681–1691 (2021).
Cao, L. et al. Hepatic insulin signaling changes: possible mechanism in prenatal hypoxia-increased susceptibility of fatty liver in adulthood. Endocrinology 153, 4955–4965 (2012).
McCurdy, C. E. et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J. Clin. Investig. 119, 323–335 (2009).
Bruce, K. D. et al. Maternal high-fat feeding primes steatohepatitis in adult mice offspring, involving mitochondrial dysfunction and altered lipogenesis gene expression. Hepatology 50, 1796–1808 (2009).
Lee, J. H. et al. A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology 139, 653–663 (2010).
Demetriou, C. A. et al. Biological embedding of early-life exposures and disease risk in humans: a role for DNA methylation. Eur. J. Clin. Invest. 45, 303–332 (2015).
Barclay, J. L. et al. GH-dependent Stat5 signaling plays an important role in hepatic lipid metabolism. Endocrinology 152, 181–192 (2011).
Sato, O., Kuriki, C., Fukui, Y. & Motojima, K. Dual promoter structure of mouse and human fatty acid translocase/CD36 genes and unique transcriptional activation by peroxisome proliferator-activated receptor alpha and gamma ligands. J. Biol. Chem. 277, 15703–15711 (2002).
Sato, O., Takanashi, N. & Motojima, K. Third promoter and differential regulation of mouse and human fatty acid translocase/CD36 genes. Mol. Cell. Biochem. 299, 37–43 (2007).
Mikkelsen, T. S. et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell 143, 156–169 (2010).
Zhang, C. et al. Osteoprotegerin promotes liver steatosis by targeting the ERK-PPAR-Gamma-CD36 pathway. Diabetes 68, 1902–1914 (2019).
Zhou, J. et al. A novel Pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J. Biol. Chem. 281, 15013–15020 (2006).
Stricker, S. H., Koferle, A. & Beck, S. From profiles to function in epigenomics. Nat. Rev. Genet. 18, 51–66 (2017).
Elliott, G. et al. Intermediate DNA methylation is a conserved signature of genome regulation. Nat. Commun. 6, 6363 (2015).
Sae-Lee, C. et al. Genomic targets and selective inhibition of DNA methyltransferase isoforms. Clin. Epigenet. 14, 103 (2022).
Morin, A. et al. A functional genomics pipeline to identify high-value asthma and allergy CpGs in the human methylome. J. Allergy Clin. Immunol. 151, 1609–1621 (2023).
Kang, D. et al. Perceived stress and non-alcoholic fatty liver disease in apparently healthy men and women. Sci. Rep. 10, 38 (2020).
Schwimmer, J. B., McGreal, N., Deutsch, R., Finegold, M. J. & Lavine, J. E. Influence of gender, race, and ethnicity on suspected fatty liver in obese adolescents. Pediatrics 115, e561–e565 (2005).
Pierce, A. N. et al. Urinary bladder hypersensitivity and dysfunction in female mice following early life and adult stress. Brain Res. 1639, 58–73 (2016).
Fuentes, I. M., Pierce, A. N., Di Silvestro, E. R., Maloney, M. O. & Christianson, J. A. Differential influence of early life and adult stress on urogenital sensitivity and function in male mice. Front. Syst. Neurosci. 11, 97 (2017).
Jacobs, A., Warda, A. S., Verbeek, J., Cassiman, D. & Spincemaille, P. An overview of mouse models of nonalcoholic steatohepatitis: from past to present. Curr. Protoc. Mouse Biol. 6, 185–200 (2016).
Funding
The authors thank the Department of Research Administration at the Children’s Mercy Hospital and Department of Anatomy and Cell Biology, School of Medicine, University of Kansas Medical Center for their support and guidance. No extramural source of financial assistance was received in support of the study.
Author information
Authors and Affiliations
Contributions
R.H.L., J.A.C., and Q.F.: substantial contributions to conception and design; Q.F., J.M.F., M.F.O., O.C.E., E.M.M., and J.P.T.: acquisition of data, or analysis and interpretation of data; Q.F., R.H.L., and J.A.C.: drafting the article or revising it critically for important intellectual content; R.H.L.: final approval of the version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fu, Q., Frick, J.M., O’Neil, M.F. et al. Early-life stress perturbs the epigenetics of Cd36 concurrent with adult onset of NAFLD in mice. Pediatr Res 94, 1942–1950 (2023). https://doi.org/10.1038/s41390-023-02714-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-023-02714-y