Abstract
Big data has the capacity to transform both pediatric healthcare delivery and research, but its potential has yet to be fully realized. Curation of large multi-institutional datasets of high-quality data has allowed for significant advances in the timeliness of quality improvement efforts. Improved access to large datasets and computational power have also paved the way for the development of high-performing, data-driven decision support tools and precision medicine approaches. However, implementation of these approaches and tools into pediatric practice has been hindered by challenges in our ability to adequately capture the heterogeneity of the pediatric population as well as the nuanced complexities of pediatric diseases such as sepsis. Moreover, there are large gaps in knowledge and definitive evidence demonstrating the utility, usability, and effectiveness of these types of tools in pediatric practice, which presents significant challenges to provider willingness to leverage these solutions. The next wave of transformation for pediatric healthcare delivery and research through big data and sophisticated analytics will require focusing efforts on strategies to overcome cultural barriers to adoption and acceptance.
Impact
-
Big data from EHRs can be used to drive improvement in pediatric clinical care.
-
Clinical decision support, artificial intelligence, machine learning, and precision medicine can transform pediatric care using big data from the EHR.
-
This article provides a review of barriers and enablers for the effective use of data analytics in pediatric clinical care using pediatric sepsis as a use case.
-
The impact of this review is that it will inform influencers of pediatric care about the importance of current trends in data analytics and its use in improving outcomes of care through EHR-based strategies.
Similar content being viewed by others
Background
It has been estimated that approximately 30% of the world’s data volume is being generated by healthcare.1 This has led to the availability of big data, with characteristics shown in Table 1.2,3
Medicine and healthcare ultimately benefit from the insights provided by data analytics for healthcare delivery. Improving outcomes in medicine with big data requires the ability to effectively and innovatively process and analyze data such as individual and group data from medical and health informatics (including electronic health records (EHRs), biomedical (including imaging) data, and omics data (see Table 2)).
The ramifications of large and increasing volumes of health care data are astounding for both clinical care delivery as well as implications for generating new knowledge through research but requires the adoption of both technical and cultural uses of data analytics. In this paper, we will discuss the following potential use of big data in clinical care and in improvement: how big data can facilitate better improvement in local quality improvement (QI) initiatives, how big data is used to scale up improvement in collaborative learning, the uses of big data in clinical prediction and decision support, the value of artificial intelligence (AI) and machine learning (ML) in clinical practice, and personalized medicine through the use of big data. We will use relevant case examples and best practices in the diagnosis and management of pediatric sepsis to illustrate the concepts described.
-
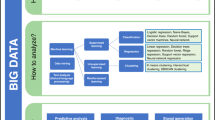
Two key concepts that will underpin the discussion (data analytics and clinical decision support) are explained in Table 3. These concepts aid in describing an organizational journey into advanced analytics capabilities (see Fig. 1).4,5
Improved patient outcomes can be realized for an organization as it progress through the various phases of analytics maturity, which are depicted here in grey boxes, with maturity level increasing from left to right. Below each phase of maturity, examples of relevant activities and data products for a sepsis case study are listed.
Using analytics for QI in pediatric sepsis
In order to illustrate the use of big data for QI (either in a local clinical care improvement project or in the use of collated big data that can drive change collectively), we illustrate a case example of pediatric sepsis. Sepsis is a potentially life-threatening complication of an infection in the blood and occurs when the body’s efforts at fighting infection trigger an inflammatory response throughout the body.6 It is one of the leading causes of death in children worldwide.6,7 Timely recognition and management of children with suspected sepsis has been demonstrated to improve outcomes and has led to national efforts at improving key processes and developing new interventions through QI efforts.8,9 Although QI approaches can accelerate the time to improved outcomes through a variety of methods, critical to approaches for rapid cycle process improvement is the use of data to understand the impact of a local improvement. These data inform the QI team to determine whether the intervention should be adopted as is, should be abandoned, or should be iterated. This approach to Plan Do Study Act cycles of health care interventions therefore requires timely data to inform decisions for the next iteration of work10 (Fig. 2).
The crux of QI approaches is a rapid iterative approach to change implementation involving four phases: (1) Plan—state what you will do; (2) Do—put your stated plan into action; (3) Study—evaluate the results/impact of your implemented plan; (4) Act—adapt, adopt, or abandon your plan based on what you learn.
For example, to improve time to first bolus of intravenous (IV) fluid in fluid resuscitation, a known best practice for the management of sepsis, a healthcare system may use a push–pull methodology to infuse fluids more rapidly than those delivered by gravity on a hanging IV bag. The system may undergo educational training and improve outcomes for some percentage of the time hitting the target of first bolus in 20 min. However, the use of an automated clinical decision tool in the EHR that recognizes a low blood pressure and comorbid conditions may trigger an EHR-based pediatric sepsis order set. This may further improve compliance with the effort to improve timeliness of first bolus. The local data systems are necessary to determine the impact of efforts (percentage of patients for whom target was achieved), its relationship to the intervention, and its impact on mortality. Therefore, one of the biggest barriers for healthcare systems to engage in QI work is having access to high-quality data (valid, reliable, and timely).
Using analytics from collective big data in pediatric sepsis
An investment in a local analytics infrastructure is important, but alone may not be sufficient to address relatively low prevalence outcomes (such as mortality from pediatric sepsis) in a reasonable period of time to drive improvement. Therefore, pooling of data across institutions can overcome the challenges of research and QI in improving outcomes for rare events through QI collaborations where large data sets from thousands of episodes of an event (e.g., pediatric sepsis) has been amassed, transformed, and informed interventions that serve as best practices. Successful QI collaborations have been described to have high performing data collection, sharing, and transformation capabilities with timely visualizations of processes and outcomes to drive rapid cycle process improvement by demonstrating the interventions that are and those that are not successful at improving processes and outcomes.9,11,12 These have included the Improving Pediatric Sepsis Outcomes collaborative with now over 300,000 episodes of pediatric sepsis-related cases captured.9,12 This sepsis collaborative gathers data from all participating hospitals to assess performance pertaining to identified standards of care for key processes in improving both timely diagnosis and effective management of pediatric sepsis to identify and learn from best practices. The collaborative transparently shares performance data to enhance learning from practice variation between centers. QI methodology accelerates the uptake of best practices in order to improve processes of care, and ultimately outcomes for patients. Requisite to the work is the sophisticated collection, curation, and transformation of big data to drive improvement.
Clinical prediction for decision support in pediatric sepsis
Clinical prediction rules, also referred to as prediction models, decision models, or risk scores, form the basis of many clinical decision support (CDS) systems. Clinical prediction rules can be broadly defined as “proceduralized efforts that assess the current or historical characteristics of a patient in order to derive an estimate of the future risk of target outcomes (prognostic), the likelihood of a current specified disease state (diagnostic), or likely response to treatment (therapeutic).”13 Following either knowledge-based (rule-based) or non-knowledge based (data-driven) types of CDS, the prediction of risk may be based on decision rules or data-driven models. In pediatric sepsis, several high-performing predictive tools have been developed for a wide range of sepsis definitions and patient populations.14,15,16 A continuous, automated EHR-based sepsis screening algorithm to identify severe sepsis among children in the inpatient and emergency department settings is an example of rule-based prediction designed by experts to provide decision support for early detection of sepsis.15 With increasing accessibility to large healthcare datasets and computational power, more data-driven approaches for prediction of sepsis have emerged by learning from an institution’s sepsis data to refine a prediction tool with weighted predictors that may not have been included or existing predictors that had different weighting in the initial rule-based model. Most of these rely on a combination of expert-informed selection of predictors and application of a statistical model or learning algorithm,14,16 such as the PELOD-2 score that has been applied to the task of pediatric sepsis detection.17,18 More recent models have relied more on knowledge gleaned from data instead of experts by leveraging advanced AI and ML algorithms.19,20,21,22 That has led to the description of high-performing models for detecting early onset sepsis in neonatal patients that leveraged statistical and algorithmic approaches to learn a model from sociodemographic, laboratory, blood pressure, and body measurement data without the potential bias of experts deciding on the model.19
Despite the development of high-performing clinical prediction rules in the last two decades, a majority are not routinely used in clinical practice and studies of their uptake in clinical practice remain low.13,23,24 Successful uptake has been associated with three key considerations: (1) utility—the usefulness to providers, (2) credibility—degree to which providers trust and believe in the predictions, and (3) usability—perceived difficulty of using the system.13 Influencing factors and successful strategies for addressing each of these concepts are described in Table 4.
In both adult and pediatric sepsis, predictive performance has been the primary focus of most evaluation studies.14,16,25,26 High performance in sepsis prediction is critical in order to prevent missed cases while minimizing over-diagnosis that may lead to unintended consequences (e.g., alarm fatigue or inappropriate or overuse of antibiotics);25,27 however, little attention has been paid to other factors that may influence adoption and acceptance into clinical practice. There is evidence that implementation of pediatric sepsis prediction tools may be associated with decreases in acute kidney injuries, hospital and ICU lengths of stay, mortality rates, and durations of organ dysfunction.25 Unfortunately, impact assessments on outcomes are challenging due to difficulties in isolating the effects of sepsis prediction tools from larger QI programs and wide variability in definitions for sepsis affecting the inclusion of cases considered in those studies.14,25 One pediatric sepsis prediction system providing continuous predictive analytics monitoring of ECG data to detect neonatal sepsis assessed both impact on outcomes and provider perceptions and adoption after implementation into practice.28,29 A significant reduction in sepsis-related mortality for very low birth weight neonates was demonstrated in a multi-site randomized control trial.28,29 Actionability of information, integration into workflow, provider clinical background, and clinical leader use of system were critical factors influencing its use and adoption in one hospital.29
In the era of big data, clinical prediction for decision support will likely see a drastic increase in the availability of high-performing, data-driven predictive models. These models may be complex and present knowledge unfamiliar to providers, which will present unique challenges in assuring the actionability and face validity of the information provided by these models. More focus will need to be on how to successfully implement these models into clinical practice as decision support tools. This could be enabled by adopting user-centered approaches to CDS system development, validation, implementation, and refinement. In the case of a sepsis CDS tool, this would include involving clinical providers (e.g., nurse, physician, trainee) who will be the end users of the tool in every stage of the tool’s lifecycle. There will be an increased need for more rigorous utility and usability assessments of these types of systems to better understand factors that may influence their adoption and acceptance.
The role of AI and ML
As one considers the future of advanced analytics, it is impossible to envision it without AI and ML. AI is not one thing but encompasses a wide variety of activities that use computerized technology to perform tasks commonly associated with human tasks, such as playing strategic games, translating language, or computer-based image recognition. It has been used to develop a variety of innovative CDS systems. It has shown great potential in predicting the clinical condition of patients and assisting in clinical decision-making. AI-derived algorithms can be applied to multiple stages of disease diagnosis and management such as those related to sepsis: as early prediction (a sepsis alert tool), prognosis assessment (a prediction flag in the EHR that computes a likely sepsis outcome that could include mortality), and optimal management (that may be based on comorbid conditions such as cancer and immunosuppression, thus recommending a specific class of antibiotics).
ML is a subset of AI involving the use of algorithms to analyze historical data to predict new output values. The results of that learning are used to make predictions and decisions about events in real health care settings. ML can be classified according to different learning methods such as supervised learning (learning from labeled data or data where there is prior knowledge of an expected output value), unsupervised learning (learning from unlabeled data), and semi-supervised learning (learning from a combination of labeled and unlabeled data).30,31 As an example, a ML approach for creating a prediction model to predict which patients may have sepsis 6 h in advance may use computer technology and software to input several variables from the EHR, including laboratory values, vital signs, and comorbid conditions (beyond those variables that subject matter experts would have included). Similarly, ML could be used to develop a prediction model to predict sepsis in a pediatric ICU using multiple EHR variables and include other inputs such as bedside monitor data for mean arterial pressure, heart rate and oxygen saturation levels. This model could be converted into CDS to alert clinicians as to the risk, and link to sepsis management guidelines.
The definition of what constitutes a ML model can be quite broad, and it is better conceptualized as a spectrum between fully human-guided versus fully machine-guided data analysis.32 Several ML models for early detection of neonatal sepsis that were developed by applying learning algorithms to a set of predictors selecting through expert and literature input fall quite low on the “ML spectrum.”33 On the other hand, a model to predict early sepsis in neonates that leveraged statistical approaches to identify predictors and a neural network algorithm to learn a predictive model would fall much higher on the “ML spectrum.”19 With the advent of big data and increased computational power, models that sit higher on the “ML spectrum” will likely form the basis of many future CDS systems. However, as noted in the prior section, adoption and acceptance of these models in clinical practice may be challenging due to their complexity. This is especially true for models traditionally considered “black boxes,” or models that lack interpretability (also described as the ability of a human to understand factors contributing to a model’s behavior).34 With increasing societal concerns and regulations on intelligent algorithms,34 recognition of the importance of incorporating providers’ and domain knowledge in modeling processes,32,34,35 and provider demand for model explanations,24 interpretability will be vital to the future successful application of ML models in healthcare.
The application of ML algorithms has been described in a variety of pediatric diseases such as bronchiolitis,31 but ML in more complex and less defined diseases such as sepsis (with less concurrence on prospective gold-standard sepsis definitions) has had limited positive results, but has become more promising with larger data sets and more expansive capabilities for processing big data.14,36 These approaches to developing models to predict some relationship or event have been applied to research (e.g., omics) and health care delivery (e.g., sepsis) and have exhibited high degrees of performance on complex problems in medicine.
Integrating concepts into personalized medicine in pediatrics
Pediatric advancements have been largely hindered by great heterogeneity within the age spectrum seen in the specialty. Although the quality and complexity of care has improved in invasive and noninvasive monitoring devices, laboratory tests, imaging modalities, and therapeutics, the field’s ability to garner precision or personalized medicine approaches amid a backdrop of well-established “normals” has been lacking.37 Furthermore, this heterogeneity within the pediatric populations have made meaningfully powered randomized control trials exceedingly difficult to attain. Thus, the field has become reliant on extrapolation of adult studies, which furthermore compound a lack of age-specific refined diagnostics and therapeutics.
Several factors can explain why adult trials struggle to translate into the pediatric population. Adult patients are affected by several comorbidities associated with aging and lifelong behaviors such as poor diet and smoking causing atherosclerosis, coronary artery disease, atrial fibrillation, chronic obstructive pulmonary disease, obesity, diabetes mellitus, chronic kidney disease, and liver cirrhosis. These common pathologic entities create a significantly more homogenous critically ill adult population when compared with acutely ill children. Children, on the other hand, have age-specific physiological changes, lack the same degree of comorbidities, and suffer from different degrees of multiorgan dysfunction.38,39 Additionally, variability in the host immune response to infection impacts individual mortality risk, and the recent advances in immunophenotyping children with sepsis show great promise in improving outcomes through precision therapy.40 Likewise, adult critical care trials also most often target mortality as the primary outcome. Since children often have a much lower rate of mortality than adults for any given organ system failure, these modest changes in a large adult trial are often diluted to insignificant when evaluating a pediatric cohort of patients.41,42 Trials of targeted therapies will undeniably continue to yield equivocal or negative results until we can improve classification of disease.
Similar challenges have been seen in the development of targeted therapeutic approaches in sepsis, where 50 years of trials have been unsuccessful leading to a sole focus on supportive care.43,44,45 These hindrances in sepsis may be secondary to trials that apply therapies in a “one size fits all” strategy without a clear understanding of different clinical and pathophysiologic phenotypes. For decades, researchers and clinicians have focused on trying to unravel the complex response of the immune system to sepsis. Despite >100 trials targeted at improving outcomes from sepsis, sepsis remains a leading cause of death in both the United States and the developing world.46,47,48 Thus, specific definitions of sepsis based on a careful understanding of different immune, hemostatic, or physiologic alteration in phenotype are difficult. This is very similar for the field of pediatric acute care where a lack of clear definitions of pediatric-specific disease entities that account for heterogeneity has hampered the ability to appropriately study specific therapies.
As an example, although precision therapy for cancer patients can map each patient’s unique tumor mutation profile and therapies for autoimmune diseases can identify and target individual cell type and/or cytokine dysregulation, there remains a void in patient phenotyping for many diseases such as sepsis that would allow for similar individualized therapies. To underscore the critical need for a method to phenotype these patients with sepsis, we have to look no further than the coronavirus disease 2019 (COVID-19) pandemic. While many COVID-19 patients were being treated with drug therapies that block cytokine signaling or suppress immune effector cell function, other COVID-19 patients were being treated with drugs that enhance or restore the immune response.49 Thus, diametrically opposing therapies were being used in identical COVID-19 cohorts without any approach that could reveal their immunologic endotype. For the development of precision application of new immunomodulatory therapies in sepsis to succeed, there is a critical need for a diagnostic modality that can both determine the functional state of the patient’s immune system in a quantifiable manner as well as evaluate the effectiveness of potential immune restorative therapies.
To improve the field’s understanding and specificity in refining definitions of pediatric disease, new paradigm shifting approaches in ML, predictive modeling, functional immunophenotyping, and AI may enable opportunities to overcome these challenges. With the rapid growth both in computing power and data storage that has enabled a wide range of applications for ML and AI within medicine, AI and ML have impacted drug discovery, personalized diagnostics, therapeutics, and medical imaging.50,51 However, leveraging these advances is challenged by ways to improve the AI output from existing phenotype or endotype development. For instance, immuno-adjuvant therapy to boost host immunity has been proposed as a potential adjunctive powerful weapon in the arsenal for sepsis, but the inability to carefully appropriate these therapies has been hindered by a paucity in functional understanding of an individual’s innate and adaptive immune function against underlying epigenetic alterations, environmental cues, and other pathophysiology temporal to the acute disease and its evolution. It is challenging to identify clinical “cues” that may inform clinicians which patients will require closer surveillance for clinical decline. The fundamental development of predictive modeling initiated after robust scientific characterization at the bench and integrated with clinical signs could establish an accurate reflection of host protective immune status in pediatric septic patients in theranostic application.
The ultimate goal of personalized medicine is to be able to accurately decipher which patient has a disease, what is the impact of that disease on relevant clinical outcomes, and identify best treatments for that patient based on unique physiologic, genetic, immunologic, and hematologic responses. The utilization of AI and ML has been challenging in sepsis as sepsis is a disease that is dynamic with changes in immunologic and hematologic perturbations; any technology must take into account that a patient changes by the minute. Although AI and ML have assisted with early prediction of septic shock, improving the accuracy of diagnosis, shorter time to antibiotics, and in development of sepsis bundles, this has not translated to the individual patient in management decisions separate from supportive adjunctives.30,42,52,53 Furthermore, predictive models to predict sepsis have not performed better than clinicians and may have issued false alarms on patients without evidence of sepsis.54 Sadly in sepsis, the field has not developed the ability to assess in real time individual patient pathophysiological alterations over time.
The use of data analytics will likely become only helpful in management of patients after the different functional pathophysiologic responses and understanding of pattern recognition of various clinical parameters have established specific patient endotypes that can be applied to real-time clinical variables. Presently, the field is utilizing a multiomic evaluation of children with sepsis in a number of studies which will greatly enhance the development of these predictive models and application in AI and ML. Secondly, hospitals will need to better equip each hospital bed with continuous monitoring for key vital signs so that timely acquisition of these variables can be integrated into modeling strategies. The ability to mine sensor-generated data streams for physio-markers against functional “omic” changes is beholden to successful acquisition of data point collection en masse and in a timely fashion. Lastly, the lack of success of electronic medical record AI technologies that grossly underperformed their publicized hype has created a barrier among clinicians to readily adopt and implement these strategies into clinical practice. Although many barriers and enablers to improving pediatric health care delivery with robust data analytics for the EHR exist, among the greatest limiting factor will be adoption by caregivers thus institutions should embrace the enablers described herein (Table 5).
Conclusion
We have described the use of big data to drive QI in local clinical settings as well as its use in collated form to drive collective learnings and improvements across systems through collaborative learning. We have also described the use of big data as the foundation for clinical prediction and decision support, inclusive of its use in AI and ML. All of these factors can be leveraged to drive personalized medicine. However, the complexity of pediatric diseases such as sepsis provides ongoing challenges to the optimization of advanced analytics in delivering personalized solutions in healthcare. Perhaps the greatest challenge to adoption of sophisticated analytics in research and healthcare delivery is cultural and requires substantive adaptive change by caregivers to understand and adopt the power of bench-to-bedside to computer-to-bedside approaches to improving diagnostic accuracy and therapeutic effectiveness.
References
Coughlin, S., Roberts, D., O’Neill, K. & Brooks, P. Looking to tomorrow’s healthcare today: a participatory health perspective. Intern. Med. J. 48, 92–96 (2018).
Raghupathi, W. & Raghupathi, V. Big data analytics in healthcare: promise and potential. Health Inf. Sci. Syst. 2, 3 (2014).
Ristevski, B. & Chen, M. Big data analytics in medicine and healthcare. J. Integr. Bioinform. 15 20170030 (2018).
Richardson, J. E. et al. Multiple perspectives on the meaning of clinical decision support. AMIA Annu Symp. Proc. 2010, 1427–1431 (2010).
Macias, C. G., Bartley, K. A., Rodkey, T. L. & Russell, H. V. Creating a clinical systems integration strategy to drive improvement. Curr. Treat. Options Pediatr. 1, 334–346 (2015).
Graetz, T. J. & Hotchkiss, R. S. Sepsis: preventing organ failure in sepsis - the search continues. Nat. Rev. Nephrol. 13, 5–6 (2017).
Black, R. E. et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet 375, 1969–1987 (2010).
Evans, I. V. R. et al. Association between the New York sepsis care mandate and in-hospital mortality for pediatric sepsis. JAMA 320, 358–367 (2018).
Larsen, G. Y. et al. Development of a quality improvement learning collaborative to improve pediatric sepsis outcomes. Pediatrics 147, e20201434 (2021).
Provost, L. P. & Murray, S. K. The Health Care Data Guide: Learning from Data for Improvement 2nd edn (Wiley, 2022).
Britto, M. T. et al. Using a network organisational architecture to support the development of learning healthcare systems. BMJ Qual. Saf. 27, 937–946 (2018).
Paul, R. et al. A quality improvement collaborative for pediatric sepsis: lessons learned. Pediatr. Qual. Saf. 3, e051 (2018).
Kennedy, G. & Gallego, B. Clinical prediction rules: a systematic review of healthcare provider opinions and preferences. Int. J. Med. Inf. 123, 1–10 (2019).
Persad, E. et al. Neonatal sepsis prediction through clinical decision support algorithms: a systematic review. Acta Paediatr. 110, 3201–3226 (2021).
Eisenberg, M., Madden, K., Christianson, J. R., Melendez, E. & Harper, M. B. Performance of an automated screening algorithm for early detection of pediatric severe sepsis. Pediatr. Crit. Care Med. 20, e516–e523 (2019).
Sahu, P. et al. Prediction modelling in the early detection of neonatal sepsis. World J. Pediatr. 18, 160–175 (2022).
Leteurtre, S. et al. Pelod-2: an update of the pediatric logistic organ dysfunction score. Crit. Care Med. 41, 1761–1773 (2013).
Schlapbach, L. J., Straney, L., Bellomo, R., MacLaren, G. & Pilcher, D. Prognostic accuracy of age-adapted SOFA, SIRS, PELOD-2, and QSOFA for in-hospital mortality among children with suspected infection admitted to the intensive care unit. Intensive Care Med. 44, 179–188 (2018).
López-Martínez, F., Núñez-Valdez, E. R., Lorduy Gomez, J. & García-Díaz, V. A neural network approach to predict early neonatal sepsis. Computers Electr. Eng. 76, 379–388 (2019).
Helguera-Repetto, A. C. et al. Neonatal sepsis diagnosis decision-making based on artificial neural networks. Front. Pediatr. 8, 525 (2020).
Leon, C., Carrault, G., Pladys, P. & Beuchee, A. Early detection of late onset sepsis in premature infants using visibility graph analysis of heart rate variability. IEEE J. Biomed. Health Inf. 25, 1006–1017 (2021).
Xiao, Y., Griffin, M. P., Lake, D. E. & Moorman, J. R. Nearest-neighbor and logistic regression analyses of clinical and heart rate characteristics in the early diagnosis of neonatal sepsis. Med. Decis. Mak. 30, 258–266 (2010).
Dekker, F. W., Ramspek, C. L. & van Diepen, M. Con: Most clinical risk scores are useless. Nephrol. Dial. Transplant. 32, 752–755 (2017).
Shah, N. D., Steyerberg, E. W. & Kent, D. M. Big data and predictive analytics: recalibrating expectations. JAMA 320, 27–28 (2018).
Eisenberg, M. A. & Balamuth, F. Pediatric sepsis screening in US hospitals. Pediatr. Res. 91, 351–358 (2022).
Islam, M. M. et al. Prediction of sepsis patients using machine learning approach: a meta-analysis. Comput. Methods Prog. Biomed. 170, 1–9 (2019).
Fleuren, L. M. et al. Machine learning in intensive care medicine: ready for take-off? Intensive Care Med. 46, 1486–1488 (2020).
Kitzmiller, R. R. et al. Diffusing an innovation: clinician perceptions of continuous predictive analytics monitoring in intensive care. Appl. Clin. Inf. 10, 295–306 (2019).
Moorman, J. R. et al. Predictive monitoring for early detection of subacute potentially catastrophic illnesses in critical care. Annu. Int. Conf. IEEE Eng. Med Biol. Soc. 2011, 5515–5518 (2011).
Wu, M., Du, X., Gu, R. & Wei, J. Artificial intelligence for clinical decision support in sepsis. Front. Med. 8, 665464 (2021).
Raita, Y. et al. Machine learning-based prediction of acute severity in infants hospitalized for bronchiolitis: a multicenter prospective study. Sci. Rep. 10, 10979 (2020).
Beam, A. L. & Kohane, I. S. Big data and machine learning in health care. JAMA 319, 1317–1318 (2018).
Masino, A. J. et al. Machine learning models for early sepsis recognition in the neonatal intensive care unit using readily available electronic health record data. PLoS ONE 14, e0212665 (2019).
Vellido, A. Societal issues concerning the application of artificial intelligence in medicine. Kidney Dis. 5, 11–17 (2019).
Johnson, T. L. et al. Augmenting predictive modeling tools with clinical insights for care coordination program design and implementation. EGEMS 3, 1181 (2015).
Iregbu, K. et al. Global health systems’ data science approach for precision diagnosis of sepsis in early life. Lancet Infect. Dis. 22, e143–e152 (2022).
Epstein, D. & Brill, J. E. A history of pediatric critical care medicine. Pediatr. Res. 58, 987–996 (2005).
Watson, R. S., Crow, S. S., Hartman, M. E., Lacroix, J. & Odetola, F. O. Epidemiology and outcomes of pediatric multiple organ dysfunction syndrome. Pediatr. Crit. Care Med. 18, S4–S16 (2017).
Villeneuve, A. et al. Multiple organ dysfunction syndrome in critically ill children: clinical value of two lists of diagnostic criteria. Ann. Intensive Care 6, 40 (2016).
Atreya, M. R. & Wong, H. R. Precision medicine in pediatric sepsis. Curr. Opin. Pediatr. 31, 322–327 (2019).
Heneghan, J. A. & Pollack, M. M. Morbidity: changing the outcome paradigm for pediatric critical care. Pediatr. Clin. North Am. 64, 1147–1165 (2017).
Shah, N. & Said, A. S. Extracorporeal support prognostication-time to move the goal posts? Membranes 11, 537 (2021).
Weiss, S. L. et al. Surviving Sepsis Campaign International Guidelines for the Management of Septic Shock and Sepsis-Associated Organ Dysfunction in Children. Pediatr. Crit. Care Med. 21, e52–e106 (2020).
Sankar, J., Ismail, J., Sankar, M. J., C P, S. & Meena, R. S. Fluid bolus over 15-20 versus 5-10 min each in the first hour of resuscitation in children with septic shock: a randomized controlled trial. Pediatr. Crit. Care Med. 18, e435–e445 (2017).
Maitland, K. et al. Mortality after fluid bolus in african children with severe infection. N. Engl. J. Med. 364, 2483–2495 (2011).
Prout, A. J. et al. Epidemiology of readmissions after sepsis hospitalization in children. Hosp. Pediatr. 9, 249–255 (2019).
Heldens, M. et al. Sepsis incidence and mortality are underestimated in Australian intensive care unit administrative data. Med J. Aust. 209, 255–260 (2018).
Weiss, S. L. et al. Global epidemiology of pediatric severe sepsis: the sepsis prevalence, outcomes, and therapies study. Am. J. Respir. Crit. Care Med. 191, 1147–1157 (2015).
Remy, K. E. et al. Immunotherapies for Covid-19: lessons learned from sepsis. Lancet Respir. Med. 8, 946–949 (2020).
Gutierrez, G. Artificial intelligence in the intensive care unit. Crit. Care 24, 101 (2020).
Lovejoy, C. A., Buch, V. & Maruthappu, M. Artificial intelligence in the intensive care unit. Crit. Care 23, 7 (2019).
Beaulieu-Jones, B. K. et al. Machine learning for patient risk stratification: standing on, or looking over, the shoulders of clinicians. NPJ Digital Med. 4, 62 (2021).
Tarabichi, Y. et al. Improving timeliness of antibiotic administration using a provider and pharmacist facing sepsis early warning system in the emergency department setting: a randomized controlled quality improvement initiative. Crit. Care Med. 50, 418–427 (2022).
Wong, A. et al. External validation of a widely implemented proprietary sepsis prediction model in hospitalized patients. JAMA Intern. Med. 181, 1065–1070 (2021).
Cecchin, E. & Stocco, G. Pharmacogenomics and personalized medicine. Genes 11, 679 (2020).
Aslam, B., Basit, M., Nisar, M. A., Khurshid, M. & Rasool, M. H. Proteomics: technologies and their applications. J. Chromatogr. Sci. 55, 182–196 (2017).
Berner, E. S. Health Informatics Ch. XI (Springer International Publishing, 2016).
Sutton, R. T. et al. An overview of clinical decision support systems: benefits, risks, and strategies for success. npj Digital Med. 3, 17 (2020).
Kappen, T. H. et al. Barriers and facilitators perceived by physicians when using prediction models in practice. J. Clin. Epidemiol. 70, 136–145 (2016).
Elish, M. C. The stakes of uncertainty: developing and integrating machine learning in clinical care. Ethnogr. Praxis Ind. Conf. Proc. 2018, 364–380 (2018).
Garg, A. X. et al. Effects of computerized clinical decision support systems on practitioner performance and patient outcomes: a systematic review. JAMA 293, 1223–1238 (2005).
Kilsdonk, E., Peute, L. W. & Jaspers, M. W. Factors influencing implementation success of guideline-based clinical decision support systems: a systematic review and gaps analysis. Int. J. Med. Inf. 98, 56–64 (2017).
Van Belle, V. & Van Calster, B. Visualizing risk prediction models. PLoS ONE 10, e0132614 (2015).
Edwards, A., Elwyn, G. & Mulley, A. Explaining risks: turning numerical data into meaningful pictures. BMJ 324, 827–830 (2002).
Langley, G. J. et al. The Improvement Guide: A Practical Approach to Enhancing Organizational Performance 2nd edn (Jossey-Bass, 2009).
Author information
Authors and Affiliations
Contributions
All three authors fully participated in researching evidence, conceptualization, drafting, development, editing and refining this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
As this is a review manuscript, patient consent was not required for this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Macias, C.G., Remy, K.E. & Barda, A.J. Utilizing big data from electronic health records in pediatric clinical care. Pediatr Res 93, 382–389 (2023). https://doi.org/10.1038/s41390-022-02343-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-022-02343-x
This article is cited by
-
Emerging role of artificial intelligence, big data analysis and precision medicine in pediatrics
Pediatric Research (2023)
-
The Opportunities and Challenges for Artificial Intelligence to Improve Sepsis Outcomes in the Paediatric Intensive Care Unit
Current Infectious Disease Reports (2023)