Abstract
Background
Accelerated catch-up growth following intrauterine restriction increases the risk of developing visceral adiposity and metabolic abnormalities. However, the underlying molecular mechanisms of such metabolic programming are still poorly understood.
Methods
A Wistar rat model of catch-up growth following intrauterine restriction was used. A gene expression array was performed in the retroperitoneal adipose tissue sampled at postnatal day (PD) 42.
Results
Five hundred and forty-six differentially expressed genes (DEGs) were identified (adjusted p value < 0.05). Gene ontology enrichment analysis identified pathways related to immune and lipid metabolic processes, brown fat cell differentiation, and regulation of PI3K. Ccl21, Npr3, Serpina3n, Pnpla3, Slc2a4, and Serpina12 were validated to be upregulated in catch-up pups (all p < 0.01) and related to several fat expansion and metabolic parameters, including body weight at PD42, postnatal body weight gain, white and brown adipose tissue mass, plasma triglycerides, and insulin resistance index (all p < 0.05).
Conclusions
Genes related to immune and metabolic processes were upregulated in retroperitoneal adipose tissue following catch-up growth in juvenile rats and were found to be associated with fat expansion and metabolic parameters. Our results provide evidence for several dysregulated genes in white adipose tissue that could help develop novel strategies to prevent the metabolic abnormalities associated with catch-up growth.
Impact
-
Catch-up growth presents several dysregulated genes in white adipose tissue related to metabolic abnormalities.
-
Ccl21, Npr3, Serpina3n, Pnpla3, Slc2a4, and Serpina12 were validated to be upregulated in catch-up pups and related to visceral fat expansion and metabolic parameters.
-
Profiling and validation of these dysregulated genes in visceral adipose tissue could help develop novel strategies to prevent the metabolic abnormalities associated with catch-up growth.
Similar content being viewed by others
Introduction
Obesity is a relevant medical condition that has the dimension of a global epidemic and carries the risk of developing other noncommunicable diseases, such as type 2 diabetes (T2D) and coronary artery disease.1 Accumulating evidence reveals that nutrition has a critical role during crucial periods of development, including the intrauterine life.2,3 The concept of the “thrifty phenotype hypothesis” states that nutritional insults affecting intrauterine growth make the fetus adopt survival strategies, through metabolic adaptations, to optimize the use of nutrients and prioritize the growth of certain organs, such as the brain, at the expense of others, mostly peripheral organs.4 Although these adjustments can benefit the individual in the short term, they will be potentially detrimental under subsequent conditions of abundant nutrient supply since the organism remains programmed to scarcity conditions.5,6,7,8,9 Negative consequences of such metabolic programming include the development of obesity and the associated metabolic diseases later in life.5,8
Studies in low birth weight children have pointed out that early postnatal catch-up growth, rather than having reduced prenatal growth, increases the risk of developing cardiometabolic diseases later in life.10,11 Animal models showed that even before any catch-up growth is evident, prenatally restricted rats exhibit physiologic, morphologic, and transcriptomic defects that can be considered as the initial signs of metabolic programming, which can be worsened or readapted with catch-up growth.12 Several studies have reported that low birth weight children with catch-up growth manifest abnormalities already at school age, such as precocious puberty, overweight, augmented adiposity, insulin resistance, and low-grade inflammation.13,14,15,16,17 Despite the robust associations of these studies, the molecular mechanisms mediating the interaction between catch-up growth and metabolic-related conditions are still largely unknown.
Adipose tissue has been identified as one of the principal targets of such metabolic programming, as emerging evidence has linked catch-up growth to the excess of adipose tissue and impaired metabolism in animal models. Pigs that experienced early catch-up growth developed a remarkable increase in adiposity and an abnormal lipid profile after both chow diet feeding18 and high fat diet feeding.19 Studies in rats have come to similar results: restricted rats following high caloric or chow diets increased their fat storage and showed a deleterious metabolic phenotype.20
Previous results in our laboratory showed that catch-up growth following intrauterine restriction in rats increased the adipocyte size in the retroperitoneal fat pad, and presented a higher amount of circulating triglycerides and higher homeostatic model assessment of insulin resistance (HOMA-IR), indicating the role of catch-up in fat accumulation and metabolism alterations.21 Several studies of adult animals with intrauterine restriction and subsequent catch-up have been developed to describe the genes in the adipose tissue playing important roles in the development of catch-up-associated metabolic disorders and their comorbidities.22,23 However, little is known about the gene profiling at earlier stages, such as at school age. We hypothesize that the gene profile of retroperitoneal adipose tissue already at a juvenile time-point will be dysregulated in catch-up rats.
Herein, we aimed to study the gene expression profile in the retroperitoneal adipose tissue of juvenile rats with intrauterine restriction and postnatal catch-up growth compared to those without catch-up growth in order to better understand the molecular mechanisms involved in catch-up growth.
Materials and methods
Animal model
All studies were approved by the Animal Research Ethics Committee (C.E.E.A., #DAAM 6911) of the University of Barcelona according to the principles and procedures outlined in the Catalan Government guideline 214/97. Wistar rats (Janvier, Le Genest Saint Isle, France) were bred and maintained under control conditions (22 °C, 12:12 light–dark cycle, humidity 50%, free access to water) and fed a standard laboratory chow (Teklad Global Rodent Maintenance Diet, Harlan Laboratories Inc.; 14% protein, 4% fat, 2900 kcal/kg). A model of intrauterine restriction induced by caloric restriction throughout gestation was developed as described previously21 (Supplementary Fig. S1). Briefly, virgin females were housed with males overnight and mating was confirmed by observation of spermatozoa in vaginal smears. From day 1 of gestation, the dams were then housed individually. Dams were fed on a 50% calorie-restricted diet during gestation to obtain pups with low birth weight (R). During suckling, dams were fed with standard rat chow to obtain catch-up growth pups (R/C) or were kept on a calorie-restricted diet to obtain restricted pups (R/R). An additional control group (C/C) was also present to confirm both catch-up growth and restriction events.21 At postnatal day 22 (PD22), weaning pups were weighed, separated from their mothers, and provided with a chow diet ad libitum till PD42. Body weights from PD1 until PD42 were recorded. At the end of the experiment, pups that were fasted overnight were anesthetized with 3% isoflurane and euthanized by cardiac exsanguination. After dissection, tissues (retroperitoneal and interscapular brown adipose tissues) were weighed for assessment of body composition and stored at −80 °C until analysis.21 Plasma samples were obtained from blood extracted during the cardiac exsanguination, collected in heparinized syringes, and centrifuged at 3500 r.p.m. for 15 min at 4 °C. Glucose and triglycerides were determined by dry chemistry technique (Spotchem II, Spotchem EZ SP-4410, Menarini Diagnostics-Akray, Japan) and insulin and total adiponectin levels were measured by enzyme-linked immunosorbent assay (ELISA) using a commercial kit (Ultrasensitive Rat Insulin ELISA kit and Rat Adiponectin ELISA kit, Crystal Chem Inc.) as previously described.21 HOMA-IR was also calculated as published before.24 As sex differences in anthropometric or biochemical parameters were not found, both females and males were used in the study.
Gene expression array
Gene expression profiling was performed using retroperitoneal adipose tissue of eight R/C samples and eight R/R samples (four females and four males, randomly selected from different litters per each group) using Affymetrix Rat Expression Array (Clariom S Array, Affymetrix, California). Total RNA was extracted from homogenized retroperitoneal adipose tissue with Qiazol lysis reagent (Qiagen, Venlo, The Netherlands), subsequently purified by RNAeasy mini kit (Qiagen) and coupled with on-column DNase digestion by RNase-free DNase Set (Qiagen) according to the manufacturer’s instructions. RNA integrity was tested with the Agilent 2100 Bionanalyzer (Agilent Technologies, California).
Arrays were performed at the High Technology Unit in Vall d’Hebron Research Center (Barcelona, Spain) following the Affymetrix standard protocols. Raw data were extracted and analyzed using Affymetrix Transcriptome Analysis Console 4.0 software (TAC, Affymetrix, California). Briefly, the probe signal values were log transformed, background corrected, and quantile normalized. Comparison between catch-up and non-catch-up groups was set and differentially expressed genes (DEGs) were identified using the following criteria log2 fold change (FC) >3 and <−3, and false discovery rate (FDR)-adjusted p value < 0.05 to avoid false-positive issues. Gene Ontology (GO) enrichment analysis of DEGs was performed with Gene Ontology enRIchment anaLysis and visuaLizAtion tool (GOrilla, Multi Knowledge Project)25 considering an FDR-adjusted p value < 0.05. Afterwards, Reduce and Visualize Gene Ontology (REViGO) software26 was used to remove redundancy and cut down the number of significant pathways. Afterwards, the pathways containing any of the 20 top-ranked upregulated or downregulated DEGs were selected. Finally, all DEGs present in the selected pathways were chosen to be validated.
Real-time quantitative PCR (RT-qPCR) analysis
Genes selected for validation were analyzed in 16 rat samples per group (R/C, R/R, and C/C samples; 1/2 females and 1/2 males). Total RNA was isolated from adipose tissue as described above and was retro-transcribed to complementary DNA (cDNA) by High-capacity cDNA Reverse Transcription kit (Applied Biosystems, Thermo Fisher Scientific). Gene expression was assessed by RT-qPCR using the LightCycler 480 Real-Time PCR System (Roche Diagnostics, Indianapolis), according to the manufacturer’s protocol. Taqman Gene Expression Assays were purchased from Applied Biosystems for Adrb3 (Rn00565393_m1), Agt (Rn00593114_m1), Apod (Rn00562832_m1), Ccl19 (Rn01439563_m1), Ccl21 (Rn01764651_g1), Cd28 (Rn00565469_m1), Cd79b (Rn00591795_m1), Cr2 (Rn04244351_m1), Igf1 (Rn00710306_m1), Itk (Rn01496863_m1), Npr3 (Rn00563495_m1), Pck1 (Rn01529014_m1), Plac8 (Rn01450126_m1), Pnpla3 (Rn01502361_m1), Pparg (Rn00440945_m1), Slc2a4 (Rn00562597_m1), Sema4d (Rn01435039_m1), Serpina3n (Rn00755832_mH), Serpina12 (Rn01518409_m1), and Ucp1 (Rn00562126_m1) genes. Data obtained were normalized to the expression of housekeeping gene Gapdh (Rn01775763_g1). Relative gene expression was retrieved using the comparative threshold (Ct) method and is presented as relative expression (2−ΔCt).
Statistical analysis
SPSS 22.0 (SPSS software, IBM) was used for statistical analyses. For array analysis, p values were calculated by analysis of variance (ANOVA) using the TAC 4.0 software. For validation analysis, unpaired t test was used to establish significant differences between R/C and R/R groups. Differences with p value < 0.05 were considered significant. Spearman’s correlation was performed to analyze associations between gene expression and body composition and metabolic parameters. Bonferroni correction for multiple comparisons was made by applying a p value ≤ 0.01 (0.05 divided by 5 validated genes). Multiple linear regression analyses adjusting by sex were done to confirm significant correlations seen between gene expression and body composition or metabolic parameters. When necessary, logarithmic transformation was used to achieve the normal distribution of continuous variables.
Results
Animal model
In this catch-up animal model, previously used and characterized by our laboratory,21 R/C and R/R had similar birth weight at PD1 but body weight at PD22 and PD24, as well as body weight gain was higher in R/C compared to R/R (Supplementary Fig. S1), indicating that R/C was caught up in weight. Moreover, in a previous publication, we also showed that R/C pups presented accelerated catch-up growth in comparison to C/C control group, and R/R pups presented restriction in comparison to C/C control group.21 R/C pups also showed higher accumulation of retroperitoneal adipose tissue, were more insulin resistant (higher glucose, insulin, and HOMA-IR index), and had higher levels of plasma triglycerides and lower total adiponectin compared to R/R (Table 1).
Gene expression profile of retroperitoneal adipose tissue: array analysis
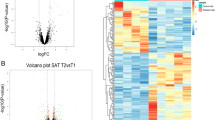
The Rat Clariom S Array (Affymetrix) performed in retroperitoneal adipose tissue analyzed 21,848 coding gene transcripts. From all of them, 546 (2.5%) were found to be DEGs between the R/C and R/R groups (│log2FC│ > 3 and FDR-adjusted p value < 0.05) (Fig. 1a). Of the 546 DEGs, 327 were upregulated and 219 were downregulated (Fig. 1b and Supplementary Table S1).
a Heatmap of the 546 DEGs detected by the gene expression profile of retroperitoneal adipose tissue. b Summary of upregulated and downregulated DEGs in R/C as compared to R/R. c Semantic similarity-based scatterplot of the relevant pathways using all DEGs performed with REViGO. Bubble color indicates the user-provided p value, and size indicates the frequency of the GO term in the underlying GO analysis database. d Table of the selected pathways and DEGs presented within each pathway. e Array analysis Log2 fold change (FC) and false rate discovery (FDR) adjusted p value of the selected genes for validation. f Diagram of the selection of candidate genes for validation.
GO analysis retrieved enrichment in 172 different biological pathways (Supplementary Table S2). After summarizing and removing the redundant GO terms with REViGO, 56 pathways were grouped according to semantic similarity (Fig. 1c and Supplementary Table S2). In order to validate genes with functional relevance, we identified the pathways that contained genes from the 20 top-ranked upregulated or downregulated DEGs. Using this combined approach, six pathways were selected: immune system process, together with regulation of leukocyte proliferation and response to cytokines, lipid metabolic process, brown fat cell differentiation, and regulation of PI3K (Fig. 1d).
Then, all genes from our DEGs from these six selected pathways were chosen to be validated in all study samples: Cd79b, Itk, Cr2, Npr3, Ccl19, Ccl21, and Serpina3n related to the immune system; Apod, Pck1, and Pnpla3 related to lipid metabolism; Adrb3, Plac8, Pparg, Slc2a4, and Ucp1 for brown fat-related pathways; and Agt, Cd28, Igf1, Sema4d, and Serpina12 for positive regulation of PI3K (Fig. 1e, f).
DEGs between R/C and R/R: validation analysis
The validation by RT-qPCR demonstrated that Ccl21 (response to cytokines) was significantly downregulated and Npr3 (regulation of leukocyte proliferation); Serpina3n (response to cytokines) Pnpla3 (lipid metabolic process), Slc2a4 (related to brown fat cell differentiation), and Serpina12 (positive regulation of PI3K) were significantly upregulated in R/C vs R/R group (Fig. 2). Differential gene expression was not confirmed for the remaining selected genes. Available C/C control group from the experimental setting was also assessed for confirmation. In all cases, the C/C control group gene expression was in between gene expression of R/R and R/C groups (Fig. 2).
Association of DEGs with body composition and metabolic parameters
Correlation analysis showed significant associations between the validated genes and several metabolic parameters, after correcting for multiple comparisons (Table 2). Ccl21 showed negative correlations with body weight at PD42, postnatal body weight gain, plasma triglycerides, and HOMA-IR (all p < 0.05). Npr3 showed positive correlations with body weight at PD42, postnatal body weight gain, amount of brown adipose tissue, plasma triglycerides, and HOMA-IR (all p < 0.01). Serpina3n and Serpina12 showed positive correlations with body weight at PD42, postnatal body weight gain, amount of white and brown adipose tissue, plasma triglycerides, and HOMA-IR (all between p < 0.0001 and ≤0.001); and Serpina3n was negatively correlated with total plasma adiponectin (Supplementary Fig. S2). Pnpla3 showed positive correlations with body weight at PD42, postnatal body weight gain, and the amount of white and brown adipose tissue and HOMA-IR (all p ≤ 0.01). Slc2a4 showed positive correlations with plasma triglycerides (all p < 0.05). Most of these associations were maintained in multivariate regression analyses, adjusting for the sex of the animals, with the exception of Slc2a4 that maintained association only with triglycerides (data not shown).
Discussion
Using the same rat model of catch-up growth that we previously validated,21 we show for the first time a gene expression profile in adipose tissue of juvenile rats with catch-up growth following intrauterine restriction and report associations of several dysregulated genes in adipose tissue with fat expansion and metabolic parameters. We have compared restricted pups with and without catch-up growth in order to study the exclusive effect of postnatal growth on metabolic alterations. Given that low birth weight children with catch-up growth can manifest early abnormalities at school age15 and that adipose tissue has been identified as one of the principal targets affected by catch-up growth,19,21 the data obtained in this rat model provide interesting information regarding the molecular mechanisms involved in the visceral fat expansion and metabolic abnormalities following catch-up growth.
The present results showed changes in the gene expression of retroperitoneal adipose tissue in R/C group as compared to R/R. Pathway enrichment analysis showed that DEGs were mainly involved in six metabolic pathways, including immune and inflammatory processes (regulation of leukocyte proliferation and response to cytokines), lipid metabolic process, brown fat cell differentiation, and upregulation of PI3K signalling. Dysregulated gene expression of the studied genes occurred in parallel to (a) fat expansion, as catch-up pups showed increased weight gain from birth, increased weight at PD42, increased retroperitoneal adipose tissue mass, and also hypertrophy of adipocytes in the retroperitoneal adipose tissue;21 and (b) lipid and glucose metabolism as catch-up pups showed increased levels of triglycerides and HOMA-IR.21
Ccl21 expression has been described in adipocytes as well as in dendritic cell/macrophage populations.27 Many articles report that high fat diet-induced obesity enhances the expression of tissue chemokines, including CCL21 and CCL19, during the course of low-grade inflammation.28 Moreover, Ccl21 has been closely related to atherosclerosis as its expression is elevated in ruptured lesions of coronary arteries and in carotid plaques.29 Then, dysregulated Ccl21 expression in retroperitoneal fat during catch-up growth may contribute to atherosclerotic events29 or impair immune system tissue responses.30
High Npr3 levels in adipose tissue of obese mice increased the clearance of lipolytic atrial natriuretic peptide (ANP), abrogating its locally lipolytic effect and thus contributing to obesity.31 The regulation of Npr3 over lipolytic ANP may only be manifested in states of low insulin action or in combination with other factors (e.g., obesity) that contribute to pre-existing insulin resistance.31 As already shown by our research group, R/C rats exhibited an insulin-resistant state;21 thus high Npr3 expressions in this context may facilitate the observed altered metabolism and fat expansion. Our results showed positive correlations of Npr3 with HOMA-IR and also with body weight gain and the amount of brown adipose tissue. The association with brown adipose tissue is in line with observations from other authors describing that Npr3 can confer thermogenic characteristics to rodent adipose tissues.32 Npr3 contribution to both obesity and enhancing thermogenesis may seem contradictory at first sight, but a plausible explanation may lie in the potential effect of Npr3 on beige adipocytes. Beige adipocytes (basically white adipocytes that present brown adipocyte characteristics) are observed during the browning of white adipose tissues through the activation of natriuretic peptides.32 Despite observing a similar expansion of both white and brown adipose tissues in the catch-up growth group, the browning of white adipose tissue may be different under catch-up growth conditions. Thus, higher Npr3 during catch-up could at the same time modulate white adipose tissue expansion and enhance its thermogenic potential through a browning effect.
We also showed that Serpina3n was upregulated in retroperitoneal adipose tissue of R/C rats and was positively correlated with postnatal body weight, weight gain, the amount of visceral adipose tissue, and plasma triglycerides. Serpina3n, a serine protease inhibitor that forms part of the SERPINA clade, is the murine ortholog of the human anti-chymotrypsin and has been identified as an inhibitor of both human and rodent granzyme B in vitro.33 Recently, Serpina3n has been identified as a serum biomarker in the development of the early stages of T2D and it is believed that the increased expression of Serpina3n might contribute to impaired insulin secretion in the pancreas in the diabetic state.34,35 The same authors pointed out the possibility that the increase in Serpina3n might reflect the obesity-related phenotype, as the weight gain was more prominent in diabetic than control rats. This is in line with the associations we observed between Serpina3n and body composition during catch-up growth in our juvenile rats. It is known that rats with intrauterine restriction and postnatal catch-up growth show increased susceptibility to diabetes mellitus in adulthood;36 therefore, increased Serpina3n expression could facilitate the appearance of T2D in these rats. Increase in Serpina3n expression was also observed in response to inflammation in the hypothalamus of rats subjected to high fat diet.37 Adiponectin, an anti-inflammatory cytokine of the adipose tissue, is related to obesity-induced chronic inflammation. In our study, total plasma adiponectin was negatively correlated with Serpina3n, supporting that overexpression of Serpina3n in R/C rats may be involved in inflammatory responses under low-grade inflammation during catch-up growth.
Pnpla3 encodes for a protein called adiponutrin or triacylglycerol lipase involved in several lipid metabolic processes, including lipid hydrolysis, and possibly also in energy balance and storage in adipocytes.38 Pnpla3 has been described to drive fatty acids excess into tissue lipid droplet storage,39 which strengthens our data on its relation to increased visceral fat depots and/or hypertrophy. Pnpla3 has also been related to liver fat deposition.40 In fact, a well-known PNPLA3 polymorphism has been identified as a childhood predictor of adulthood fatty liver.41,42 The same variant of Pnpla3 has been associated with metabolic and anthropometric T2D traits in adult patients with metabolic syndrome.43 Moreover, increased Pnpla3 mRNA expression has been previously described in adult obese rats (both in brown and white adipose tissue) and has been attributed to vesicular targeting and protein transport between both types of adipose tissue.44,45 These findings are consistent with our results showing increased expression of Pnpla3 in catch-up rats and its positive correlation with the amount of retroperitoneal (white) and brown adipose tissue.
The expression of Slc2a4 was found to be significantly increased in R/C as compared to R/R rats. Slc2a4 gene is a member of the solute carrier family and encodes the GLUT4 protein. Slc2a4/Glut4, highly expressed in adipose tissue and skeletal muscle, functions as an insulin-regulated facilitative glucose transporter and is a key regulator of whole-body glucose homeostasis.46 Several studies in animal models showed decreased Slc2a4/Glut4 expression in offspring adipose tissue after maternal undernutrition,23,47 although its expression has also been described as being downregulated in adipose tissue of obese subjects.48,49
We hypothesize that the Slc2a4 upregulation in adipose tissue of R/C vs R/R rats may have a compensatory role to improve the dysregulated insulin-resistant state observed in these catch-up rats. An excess of glucose can be up-taken by adipocytes and converted to glycerol as previously described in vitro in primary cell cultures of adipocytes.50 The upregulation of Slc2a4 could thus be a compensatory mechanism used by white adipose tissue to convert the excess of glucose input into glycerol, not only to dispose of the glucose excess but also to produce a more readily usable metabolite and redistribute this secondary product into other peripheral tissues.
In addition to being crucial for glucose uptake,51 augmented PI3K signalling may cause exacerbated lipogenesis by increasing the assortment of the required substrates from the citric acid cycle. Serpina12 also known as vaspin (visceral adipose tissue-derived serine protease inhibitor) has been associated with insulin resistance, negative regulation of gluconeogenesis, and increased carotid artery intima–media thickness in subjects with metabolic syndrome.52 Moreover, this vaspin adipokine negatively regulates lipid biosynthesis as it contributes to the metabolic regulation of triglycerides in vitro51 and in vivo.53 Human studies have also shown that mRNA expression of vaspin in the adipose tissue could be induced by an increased fat mass.54 Our results head in the same direction as previous investigations as Serpina12 positively correlates with body weight and plasma triglycerides. In our model, we observed increased Serpina12 expression in the retroperitoneal adipose tissue in R/C rats, which was positively correlated with both brown and white adipose tissue weight. We, therefore, suggest that higher Serpina12 expression could contribute to the visceral fat and brown adipose tissue expansion in R/C rats.
To summarize, we have identified several dysregulated genes (Ccl21, Npr3, Serpina3n, Pnpla3, Slc2a4, and Serpina12) in adipose tissue of juvenile rats that could be involved in catch-up induced visceral fat expansion and glucose and lipid metabolism. However, we acknowledge some limitations in our study. The study is mainly descriptive; nevertheless, the information obtained from our array and validation increases knowledge and may inspire other studies on adipose tissue during catch-up growth. Moreover, other significant DEG genes could be chosen for further studies such as angiopoietin-like proteins. The selected Agt (angiotensinogen) did not get validated from our array results; nonetheless, their implication in vascularization and/or blood pressure regulation could be of study using models assessed in later time-points until adulthood or aging. On the other hand, retroperitoneal adipose tissue availability was scarce in these animals, precluding the assessment of protein expression as well as the separation of different cell population fractions (e.g., adipocytes) from whole adipose tissue samples, in order to complement the transcript gene expression profile described in this study.
In conclusion, genes related to immune/inflammatory and metabolic processes as well as brown fat cell differentiation were upregulated in retroperitoneal adipose tissue following catch-up growth in juvenile rats and were found to be associated with fat expansion and metabolic parameters. Our results provide evidence for several dysregulated genes in white adipose tissue that could help develop novel strategies to prevent the metabolic abnormalities associated with catch-up growth.
References
Riaz, H. et al. Association between obesity and cardiovascular outcomes: a systematic review and meta-analysis of mendelian randomization studies. JAMA Netw. Open. 1, e183788 (2018).
Ong, K. K. Catch-up growth in small for gestational age babies: good or bad? Curr. Opin. Endocrinol. Diabetes Obes. 14, 30–34 (2007).
Olbertz, D. M. et al. Identification of growth patterns of preterm and small-for-gestational age children from birth to 4 years - do they catch up? J. Perinat. Med. 47, 448–454 (2019).
Hales, C. N. & Barker, D. J. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia 35, 595–601 (1992).
Hales, C. N. et al. Fetal and infant growth and impaired glucose tolerance at age 64. BMJ 303, 1019–1022 (1991).
Barker, D. J. et al. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia 36, 62–67 (1993).
Li, Y. et al. Exposure to the chinese famine in early life and the risk of metabolic syndrome in adulthood. Diabetes Care 34, 1014–1018 (2011).
Wang, N. et al. The famine exposure in early life and metabolic syndrome in adulthood. Clin. Nutr. 36, 253–259 (2017).
Xu, H., Zhang, Z., Li, L. & Liu, J. Early life exposure to China’s 1959-61 famine and midlife cognition. Int. J. Epidemiol. 47, 109–120 (2018).
Embleton, N. D. et al. Catch-up growth and metabolic outcomes in adolescents born preterm. Arch. Dis. Child. 101, 1026–1031 (2016).
Tang, A. et al. Catch-up growth, metabolic, and cardiovascular risk in post-institutionalized Romanian adolescents. Pediatr. Res. 84, 842–848 (2018).
Somm, E. et al. Early metabolic defects in dexamethasone-exposed and undernourished intrauterine growth restricted rats. PLoS ONE 7, e50131 (2012).
de Zegher, F., Garcia Beltran, C., Lopez-Bermejo, A. & Ibanez, L. Metformin for rapidly maturing girls with central adiposity: less liver fat and slower bone maturation. Horm. Res. Paediatr. 89, 136–140 (2018).
Stettler, N., Zemel, B. S., Kumanyika, S. & Stallings, V. A. Infant weight gain and childhood overweight status in a multicenter, cohort study. Pediatrics 109, 194–199 (2002).
Ibanez, L., Ong, K., Dunger, D. B. & de Zegher, F. Early development of adiposity and insulin resistance after catch-up weight gain in small-for-gestational-age children. J. Clin. Endocrinol. Metab. 91, 2153–2158 (2006).
Araujo de Franca, G. V. et al. Associations of birth weight, linear growth and relative weight gain throughout life with abdominal fat depots in adulthood: the 1982 Pelotas (Brazil) birth cohort study. Int. J. Obes. 40, 14–21 (2016).
Malpique, R. et al. Liver volume and hepatic adiposity in childhood: relations to body growth and visceral fat. Int. J. Obes. 42, 65–71 (2018).
Myrie, S. B. et al. Intrauterine growth-restricted Yucatan miniature pigs experience early catch-up growth, leading to greater adiposity and impaired lipid metabolism as young adults. Appl. Physiol. Nutr. Metab. 42, 1322–1329 (2017).
Yan, H. et al. Postnatal high-fat diet enhances ectopic fat deposition in pigs with intrauterine growth retardation. Eur. J. Nutr. 56, 483–490 (2017).
Lizarraga-Mollinedo, E. et al. Early and long-term undernutrition in female rats exacerbates the metabolic risk associated with nutritional rehabilitation. J. Biol. Chem. 290, 19353–19366 (2015).
Carreras-Badosa, G. et al. Dlk1 expression relates to visceral fat expansion and insulin resistance in male and female rats with postnatal catch-up growth. Pediatr. Res. 86, 195–201 (2019).
Guan, H. et al. Adipose tissue gene expression profiling reveals distinct molecular pathways that define visceral adiposity in offspring of maternal protein-restricted rats. Am. J. Physiol. Endocrinol. Metab. 288, E663–E673 (2005).
Bol, V. V., Delattre, A. I., Reusens, B., Raes, M. & Remacle, C. Forced catch-up growth after fetal protein restriction alters the adipose tissue gene expression program leading to obesity in adult mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 297, R291–R299 (2009).
Cacho, J., Sevillano, J., de Castro, J., Herrera, E. & Ramos, M. P. Validation of simple indexes to assess insulin sensitivity during pregnancy in Wistar and Sprague-Dawley rats. Am. J. Physiol. Endocrinol. Metab. 295, E1269–E1276 (2008).
Eden, E., Navon, R., Steinfeld, I., Lipson, D. & Yakhini, Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 10, 48 (2009).
Supek, F., Bosnjak, M., Skunca, N. & Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 (2011).
Badr, G., Ebaid, H., Mohany, M. & Abuelsaad, A. S. Modulation of immune cell proliferation and chemotaxis towards CC chemokine ligand (CCL)-21 and CXC chemokine ligand (CXCL)-12 in undenatured whey protein-treated mice. J. Nutr. Biochem. 23, 1640–1646 (2012).
Jung, J. I. et al. High-fat diet-induced obesity increases lymphangiogenesis and lymph node metastasis in the B16F10 melanoma allograft model: roles of adipocytes and M2-macrophages. Int. J. Cancer 136, 258–270 (2015).
Akhavanpoor, M. et al. CCL19 and CCL21 modulate the inflammatory milieu in atherosclerotic lesions. Drug Des. Dev. Ther. 8, 2359–2371 (2014).
Weitman, E. S. et al. Obesity impairs lymphatic fluid transport and dendritic cell migration to lymph nodes. PLoS ONE 8, e70703 (2013).
Morton, N. M. et al. A stratified transcriptomics analysis of polygenic fat and lean mouse adipose tissues identifies novel candidate obesity genes. PLoS ONE 6, e23944 (2011).
Neinast, M. D. et al. Activation of natriuretic peptides and the sympathetic nervous system following Roux-en-Y gastric bypass is associated with gonadal adipose tissues browning. Mol. Metab. 4, 427–436 (2015).
Ang, L. S. et al. Serpina3n attenuates granzyme B-mediated decorin cleavage and rupture in a murine model of aortic aneurysm. Cell Death Dis. 2, e209 (2011).
Takahashi, E. et al. Proteomic analysis of serum biomarkers for prediabetes using the Long-Evans Agouti rat, a spontaneous animal model of type 2 diabetes mellitus. J. Diabetes Investig. 8, 661–671 (2017).
Takahashi, E. et al. Differential proteome analysis of serum proteins associated with the development of type 2 diabetes mellitus in the KK-A(y) mouse model using the iTRAQ technique. J. Proteom. 84, 40–51 (2013).
Casimir, M. et al. A role for pancreatic beta-cell secretory hyperresponsiveness in catch-up growth hyperinsulinemia: Relevance to thrifty catch-up fat phenotype and risks for type 2. Diabetes Nutr. Metab. 8, 2 (2011).
Dalby, M. J. et al. Diet induced obesity is independent of metabolic endotoxemia and TLR4 signalling, but markedly increases hypothalamic expression of the acute phase protein, SerpinA3N. Sci. Rep. 8, 15648 (2018).
Umano, G. R., Martino, M. & Santoro, N. The association between pediatric NAFLD and common genetic variants. Children 4, 49 (2017).
Chamoun, Z., Vacca, F., Parton, R. G. & Gruenberg, J. PNPLA3/adiponutrin functions in lipid droplet formation. Biol. Cell 105, 219–233 (2013).
Romeo, S. et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 40, 1461–1465 (2008).
Goran, M. I. et al. Effects of PNPLA3 on liver fat and metabolic profile in Hispanic children and adolescents. Diabetes 59, 3127–3130 (2010).
Suomela, E. et al. Childhood predictors of adult fatty liver. The Cardiovascular Risk in Young Finns Study. J. Hepatol. 65, 784–790 (2016).
Wei, F. J. et al. Quantitative candidate gene association studies of metabolic traits in Han Chinese type 2 diabetes patients. Genet. Mol. Res. 14, 15471–15481 (2015).
Baulande, S., Lasnier, F., Lucas, M. & Pairault, J. Adiponutrin, a transmembrane protein corresponding to a novel dietary- and obesity-linked mRNA specifically expressed in the adipose lineage. J. Biol. Chem. 276, 33336–33344 (2001).
Caimari, A., Oliver, P. & Palou, A. Regulation of adiponutrin expression by feeding conditions in rats is altered in the obese state. Obesity 15, 591–599 (2007).
Huang, S. & Czech, M. P. The GLUT4 glucose transporter. Cell Metab. 5, 237–252 (2007).
Gardner, D. S. et al. Programming of glucose-insulin metabolism in adult sheep after maternal undernutrition. Am. J. Physiol. Regul. Integr. Comp. Physiol. 289, R947–R954 (2005).
Garvey, W. T. et al. Pretranslational suppression of a glucose transporter protein causes insulin resistance in adipocytes from patients with non-insulin-dependent diabetes mellitus and obesity. J. Clin. Invest. 87, 1072–1081 (1991).
Sinha, M. K. et al. Adipose tissue glucose transporters in NIDDM. Decreased levels of muscle/fat isoform. Diabetes 40, 472–477 (1991).
Rotondo, F. et al. Glycerol is synthesized and secreted by adipocytes to dispose of excess glucose, via glycerogenesis and increased acyl-glycerol turnover. Sci. Rep. 7, 8983 (2017).
Sheu, M. L., Ho, F. M., Chao, K. F., Kuo, M. L. & Liu, S. H. Activation of phosphoinositide 3-kinase in response to high glucose leads to regulation of reactive oxygen species-related nuclear factor-kappaB activation and cyclooxygenase-2 expression in mesangial cells. Mol. Pharmacol. 66, 187–196 (2004).
Karbek, B. et al. Relationship of vaspin and apelin levels with insulin resistance and atherosclerosis in metabolic syndrome. Minerva Endocrinol. 39, 99–105 (2014).
Gao, J. H. et al. Visceral adipose tissue-derived serine protease inhibitor accelerates cholesterol efflux by up-regulating ABCA1 expression via the NF-kappaB/miR-33a pathway in THP-1 macropahge-derived foam cells. Biochem. Biophys. Res. Commun. 500, 318–324 (2018).
Kloting, N. et al. Vaspin gene expression in human adipose tissue: association with obesity and type 2 diabetes. Biochem. Biophys. Res. Commun. 339, 430–436 (2006).
Acknowledgements
The study was supported by grant no. PI13/01257 and PI16/01335 (to A.L.-B.) and PI14/01625 (to J.B.) from the National Institute of Health Carlos III (Fund for Health Research FIS, Spain), projects co-financed by FEDER.
Author information
Authors and Affiliations
Contributions
E.L.-M. contributed to the study design, carried out experiments, analyzed data, and wrote the first draft of the manuscript. G.C.-B. contributed to the study design, carried out experiments, analyzed data, and participated in the writing of the manuscript. S.X.-T., X.R., B.M.-P., and A.P.-P. carried out experiments and reviewed the manuscript. F.d.Z. and L.I. reviewed the manuscript. A.L.-B. and J.B. designed and supervised the study and participated in the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deceased: Esther Lizarraga-Mollinedo
Supplementary information
Rights and permissions
About this article
Cite this article
Lizarraga-Mollinedo, E., Carreras-Badosa, G., Xargay-Torrent, S. et al. Catch-up growth in juvenile rats, fat expansion, and dysregulation of visceral adipose tissue. Pediatr Res 91, 107–115 (2022). https://doi.org/10.1038/s41390-021-01422-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-021-01422-9
This article is cited by
-
Local ancestry and selection in admixed Sanjiang cattle
Stress Biology (2023)