Abstract
Background
Medical-imaging-based three-dimensional (3D) printed models enable improvement in skills training, surgical planning, and decision-making. This pilot study aimed to use multimodality imaging and to add and compare 3D ultrasound as a future standard to develop realistic neonatal brain models including the ventricular system.
Methods
Retrospective computed tomography (CT), magnetic resonance imaging (MRI), and 3D ultrasound-based brain imaging protocols of five neonatal patients were analyzed and subsequently segmented with the aim of developing a multimodality imaging-based 3D printed model. The ventricular anatomy was analyzed to compare the MRI and 3D ultrasound modalities.
Results
A realistic anatomical model of the neonatal brain, including the ventricular system, was created using MRI and 3D ultrasound data from one patient. T2-weighted isovoxel 3D MRI sequences were found to have better resolution and accuracy than 2D sequences. The surface area, anatomy, and volume of the lateral ventricles derived from both MRI and 3D ultrasound were comparable.
Conclusions
We created an ultrasound- and MRI-based 3D printed patient-specific neonatal brain simulation model that can be used for perioperative management. To introduce 3D ultrasound as a standard for 3D models, additional dimensional correlations between MRI and ultrasound need to be examined.
Impact
-
We studied the feasibility of implementing 3D ultrasound as a standard for 3D printed models of the neonatal brain.
-
Different imaging modalities were compared and both 3D isotropic MRI and 3D ultrasound imaging are feasible for printing neonatal brain models with good dimensional accuracy and anatomical replication.
-
Further dimensional correlations need to be defined to implement it as a standard to produce 3D printed models.
Similar content being viewed by others
Introduction
Preterm and term neonates can be born with or develop neurological morbidities requiring neurosurgery, including intraventricular hemorrhage, hydrocephalus, and neonatal stroke.1,2,3 Dedicated neonatal care and collaboration with pediatric neurosurgery as well as the use of innovative technology in the diagnosis and treatment of neurosurgical problems in neonates are of immense importance. In this regard, multimodal imaging and three-dimensional (3D) printing have the potential to break the mold of conventional diagnosis, treatment planning, and preoperative training.4,5 With the possibility of producing case-specific realistic models, medical teams benefit from being well trained in both technical and non-technical skills to provide appropriate interventions during critical situations.6,7 If these requirements cannot be met, patient safety-related problems increase in frequency, possibly leading to healthcare-associated harm to patients. As a consequence, simulation-based medical education is a common practice in different medical fields, including pediatrics. It enables medical staff to improve skills without exposing patients to risk and allows standardization and consistent replication of variable clinical conditions.8,9,10,11,12 To ensure the highest possible standards for personalized and specific training for a challenging patient population, it is necessary to improve current training modalities by introducing 3D printed models that accurately present neonatal pathologies and increases training options for neurosurgical procedures.13,14
Previous studies have suggested that incorporating additively manufactured models (commonly called 3D printed models) in different pediatric medical fields, such as cranial remodeling, cardiac surgery, tumor resection, airway management, diaphragmatic hernia repair, or coronary–pulmonary artery fistula surgery, enables improvement in skills training, surgical planning, and decision-making ultimately contributing to patient safety. Most of them are based on magnetic resonance imaging (MRI) and computed tomography (CT) images, which are time consuming and partly associated with radiation exposure.14,15,16,17,18,19 As for neonatal patients, a major complication of prematurity is the development of an intraventricular hemorrhage and consequently posthemorrhagic hydrocephalus.2,3 Subsequent clinical challenges include an increase in intracranial pressure (ICP), possibly leading to irreversible damage to the brain parenchyma and seizure activity, as well as neurodevelopmental impairment. The treatment strategy at our hospital is the placement of an external ventricular drainage. Due to the small circumference of the neonatal head, different head shapes (mostly due to application of continuous positive airway pressure caps), possible asymmetric enlargement of ventricles, and drain stiffness, major complications can occur, including bleeding, misplacement, obstruction, dislocation, and infection.20 Therefore, optimal training methods are needed to reduce the risk of complications of the procedure itself. Currently, no 3D model is available for such training that precisely represents the small size of the neonatal head and adequately provides realistic material comparable to the brain tissue. A previously described 3D replication of a 14-year-old patient with hydrocephalus has been used as a model for this project.21
This study has proposed the use of multimodality imaging as an initial platform to enable and compare 3D ultrasound as a future standard to develop realistic neonatal brain models including the ventricular system. To achieve this, the project aimed at comparative evaluation of the feasibility and validity of different medical-imaging protocols in developing a brain ventricle model of the neonatal head. This prototype model is intended to find applications in the further development of 3D printed models and additionally contribute to creating models that are solely based on 3D ultrasound, which is an easily accessible bedside imaging technology, omitting radiation.
Methods
This study was a pilot study with retrospective data analysis and prospective 3D printing. We acquired CT (if available), MRI, and 3D ultrasound protocols of five neonatal patients. The available data were analyzed for image quality and printability and subsequently segmented for 3D printing. The digital models underwent dimensional and volumetric comparisons to determine the accuracy of anatomical replication.
Image acquisition
Retrospective pseudonymized CT, MRI, and 3D ultrasound imaging data were acquired from five neonatal patients (H1, H2, H3, H4, and H5) with permission from the local Institutional Review Board at the Medical University of Vienna (1989/2019). The patients were admitted to the Division of Neonatology, Intensive Care Medicine and Neuropediatrics (Clinical Department of Pediatrics and Adolescent Medicine, Medical University of Vienna) and had MRI and 3D ultrasound on the same day, and, as in the case of patient H2, also a CT as well (not on the same day). Patients were born between 25 + 6 and 41 + 6 weeks of gestational age and presented with neurologic pathologies, had an infection with cytomegalovirus, or had a cardiopulmonary resuscitation event. MRI was performed to assess the magnitude and presence of neurologic pathologies and CT was performed to evaluate a potential shunt dysfunction.
MRI and 3D ultrasound scans
Imaging data from all patients were segmented, reconstructed, analyzed, and compared by a single operator (G.O.). However, for printing the model, we focused on one patient (study ID: H1), who was born at gestational week 33 + 5 with a birth weight of 1960 g. The MRI obtained at a gestational age of 35 + 1 showed the picture of hypoxic–ischemic encephalopathy as well as intracerebral bleeding, accentuated external ventricles, and a cephalohematoma. However, the 3D cerebral ultrasound on the same day showed a non-pathological cerebral ultrasound of the internal ventricle system. The project started with an analysis of different imaging protocols using the Mimics USL software package (Materialise®, Leuven, Belgium), which consists of Mimics Research 21.0 for radiographic contrast-based segmentation and 3-Matic Research 13.0 for model reconstruction, repair and analysis.
MRI scans
We analyzed MRI images which were derived from 1.5 Tesla (Philips Ingenia 1.5 Tesla® or Siemens Aera 1.5 Tesla®) as well as 3 Tesla (Siemens Magnetom Vida 3 Tesla®) MRI examinations. T1-weighted 2D MRI sequences (repetition time, 3200 ms; echo time, 130 ms; flip angle, 90°; field of view, 80 mm; acquisition matrix, 0/180/150/0; slice thickness, 2 mm; spacing between slices, 2 mm), and additionally, T2-weighted sequences (repetition time 2500 ms, echo time 350 ms, flip angle 90°, field of view 100 mm, acquisition matrix 0/168/167/0, slice thickness 0.9 mm, spacing between slices 0.45 mm) in three orthogonal planes, integrated into a standard neonatal imaging protocol, were studied. T2-weighted 3D sequences were used to compare the applicability of imaging data for 3D printing, in addition to standard T1-weighted 3D sequences via multiplanar reformation.
3D ultrasound scans
3D ultrasound image data were acquired using a recently introduced tomographic ultrasound system (PIUR tUS Infinity®, PIUR Imaging GmbH, Vienna, Austria), which was attached to a Philips Affiniti 70 ultrasound machine (Philips, Amsterdam, Netherlands) using a linear S12-4 ultrasound probe at 37 Hz, enabling rapid image acquisition. Patients were placed in the supine position. The cerebral ultrasound was performed according to local guidelines.
CT scans
CT scans were derived from CT examinations (Siemens Somatom Drive®, Dual-Source, Siemens, Erlangen, Germany). We could only avail a CT scan for the patient with study ID: H2.
Rendering and segmentation
The CT scan did not provide any additional information compared to the other imaging modalities since only the skull could be identified in good quality; hence, we focused on MRI and ultrasound data. Furthermore, a CT scan is not the first-line imaging modality applied in the neonatal population. The Mimics USL software package was used for MRI and ultrasound data. The software automatically segmented the MRI scans by selecting a certain gray value that included all soft tissue of the brain, followed by manual extraction of the regions of interest, which in our case were ventricles and brain. For the 3D ultrasound, a complete manual segmentation of the ventricles and part of the occipital bone was performed. The occipital bone was used as a landmark for comparing the position of the ventricles and their alignment in the MRI and 3D ultrasound-based imaging data. The final 3D printed brain model was based on the rendered T2-weighted isovoxel 3D MRI images (Fig. 1c), and for the ventricular aspect, on rendered 3D ultrasound images (Fig. 1d). For assessing and comparing the two different MRI modalities, T2-weighted isovoxel 3D MRI provided the best contrast between the ventricles and the surrounding brain parenchyma, thereby making segmentation easier and anatomically more accurate compared to the T1-weighted 2D MRI protocol.
Comparative analysis of the digital models
As we rendered and segmented 3D MRI scans consisting of the (sub)cutaneous tissue, brain, ventricular system, and part of the skull (Fig. 2a) and additionally 3D ultrasound scans consisting of the ventricles and the occipital part of the skull, it was important to validate the exactness in the ventricular size derived from both these modalities. Therefore, we segmented a small portion of the occipital bone with the ventricular system, from both 3D MRI and 3D ultrasound, of the patient with study ID H1 and overlapped it with the segmented skull from the 3D MRI of the same patient, to achieve a fixed bone-ventricular distance measuring an average of 22 mm (Fig. 2a). Further, we compared the left and right ventricles in their anatomical position using segmentation analysis in the Materialise 3-Matic software 21.0 resulting in a ventricular overlap between 3D MRI and 3D ultrasound imaging data (Fig. 2b). Owing to the slight difference in ventricular orientation in 3D MRI- and 3D ultrasound-based digital models (or Standard Tessellation Language, STL models), it was imperative to compare the right and left ventricles of these modalities individually for this patient, where both showed an individual ventricular overlap between 3D MRI and 3D ultrasound STL models (Fig. 2c).
a 3D MRI and 3D ultrasound-derived ventricular dimensional validation showing an average distance of 22 mm from the base of the skull, b combined ventricular overlap between 3D MRI (red) and 3D ultrasound (blue), c individual right and left ventricular overlap between 3D MRI (red) and 3D ultrasound (turquoise).
3D printing process and material
For 3D printing of the brain model, we used the following as base material: VeroPureWhite (rigid, white), Tango+ (flexible, 30 shore hardness A), and VeroClear (rigid, transparent). A combination of VeroPureWhite and Tango+ was used to create the bony structures and the ventricles in a less brittle material. For the brain, we used Tango+ to mimic the soft tissue of the brain. After the printing procedure, the support material was removed, which was needed to create overhangs in the model, and the parts were cleaned according to the manufacturer’s cleaning instructions. For the additive manufacturing (3D printing) process of the model, we employed the Connex3 500 MJM (Multi Jet Material) printer (Stratasys®, Minnesota), which allowed the blending of definable materials from three ultraviolet-curing-based materials during the printing process.
After printing, the model was thoroughly cleaned of the support material (SUP706) mechanically and chemically, using waterjet and a 2% sodium hydroxide solution, respectively. Model cleaning is a crucial step, as the remnants of support material can affect the patency of the hollow structures in the model.
In the third step, the assembled model was prepared to present the structure, implying that the skull and the brain were separated into two parts to realize a modular design for direct access to the reconstructed ventricles and better presentation.
Results
CT scans
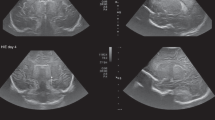
The original pixels from the images were rectangular with a resolution of 0.19 (x-axis) × 0.19 (y-axis) cm and 1 mm slice thickness. For segmentation, the pixels were automatically resliced into squares with a resolution of 0.19 (x-axis) × 0.19 (y-axis) cm with 1 mm slice thickness in the Materialise Mimics Research 21.0 software for an improved segmentation. Analysis of the CT data showed that this modality provided information about the skeletal anatomy only, and that no information could be obtained about the soft tissues in sagittal, axial, or coronal sections of the scans. The skull was portrayed in the highest resolution in the axial section as the image was acquired in this plane (Fig. 1a), while images in the other two planes were pixelated.
MRI scans
T1-weighted 2D MRI sequences provided information about both hard and soft tissues of the head including the brain and the ventricles with different signal intensities. The resolution of these images was 0.31 mm (x-axis) × 0.31 mm (y-axis) × 2.00 mm (z-axis), whereas the segmentation process of the axis that was orthogonal to the acquisition plane (mostly coronal) could not achieve the same defined results as that of segmentation within the acquisition plane (axial). As for the CT scan, the image was initially acquired in the axial plane; thus, the analysis of the sagittal and coronal sections equally resulted in pixelation (Fig. 1b).
In addition, T2-weighted sequences provided information about both hard and soft tissues of the head. The resolution of these images was 0.31 mm (x-axis) × 0.31 mm (y-axis) × 0.31 mm (z-axis) (Fig. 1c), all having a sharp resolution based on isovoxel image acquisition.
3D ultrasound
The 3D ultrasound modality provided information about the soft tissue and parts of the skull including the brain and ventricles, in different gray values (Fig. 1d). The skeletal information in 3D ultrasound images was very useful with respect to alignment and dimensional comparison of MRI and 3D ultrasound-derived ventricular models.
Segmentation analyses of digital ventricular models
The properties and dimensional aspects of the ventricles were compared between 3D ultrasound and 3D MRI-based imaging data for patients H1, H3, H4, and H5 (Fig. 3c). The MRI of patient H2 was not utilizable due to the motion of the infant during the scan; therefore, this patient was not analyzed further. Comparative parameters included the percentage of overlapping entities in various distance bands, total ventricular volume (in mm3), and surface area (in mm2) of the digital models obtained from 3D MRI and 3D ultrasound. The results from the surface entity-based segmentation analysis to examine the degree of overlap of the surface entities on the two different digital models derived from 3D MRI and 3D ultrasound showed a good overlap. The 3D ultrasound-based model showed an average of 91.50% surface entity overlap with the MRI-based digital model, while 85.25% of surface entities on the 3D MRI-derived digital model perfectly overlapped with their respective 3D ultrasound-based digital model, within the distance band of 0.00–3.00 mm (Fig. 3). The distance bands from I to VI in Fig. 3 represent the distance of deviation of the overlapping points. However, for patient H1, the results from the point-based part comparison analysis of the individual right and left ventricular models derived from 3D MRI and 3D ultrasound provided a better overlap between the two models with an average of 87.25% entities matching within a deviation range of 0.00–3.00 mm compared to the combined overlap with 84.00% match (Fig. 3). On average, 3D ultrasound-based digital models showed 5.80% lower volume and 14.90% lower surface area compared to 3D MRI-based digital models. By overlapping the occipital bone derived from the 3D ultrasound with the 3D MRI-derived skull of a neonatal patient, the anatomical structures accurately aligned, indicating that both modalities result in the same model dimensions.
a 3D ultrasound-derived digital ventricular model segmentation analysis of patient H1, b 3D MRI-derived digital ventricular model segmentation analysis of patient H1. The colors indicate the overlapping distance thresholds (I–VI). Green implies maximum surface entity overlap in threshold I, and red indicates the mismatched surface entities in threshold II and above, c Graphical representation of results from the comparative segmentation analysis of the digital model overlap between 3D ultrasound- and 3D MRI-derived models from patients H1, H3, H4, and H5. The blue series refers to analysis from ultrasound, while orange refers to MRI. The y-axis refers to the percentage of overlapping surface entities (in logarithmic scale) in the digital models and x-axis refers to the overlapping distance thresholds from I to VI in mm.
3D printing process and material
The 3D MRI digital model included the (sub)cutaneous tissue, brain, and ventricles. The additively manufactured digital model based on 3D ultrasound depicted the ventricular system and appeared to accurately fit into the 3D MRI-derived brain model.
From the latter modality, the brain and the cranial bone were taken for the printing procedure. Based on the rendered data from the different technologies of the medical-imaging systems, a set of stereolithography (STL) models representing the skull, brain, and the lateral ventricles were exported to the printing system. A modular anatomical model with multi-material parts was created (Fig. 4), with the possibility of separating and reassembling the individual parts with a small gap distance of 50.00 µm to avoid interlocking. In order to prevent printing errors resulting in undefined areas, the model was not printed in the cross-section. The printing costs were in the range of 100 euros with a printing time of 2.5 h. With a build tray size of 490 × 390 × 200 mm3, it is possible to print three neonatal brain models at a time, decreasing the overall printing time.
Discussion
Neonatal neurosurgery is a complex medical field with frequent exposure to various high-risk patients. However, the possibility of presurgical training before operating on the neonatal brain with such critical pathologies is currently inadequate. Familiarization with the procedure by repetitive and realistic training is the key to success for every clinical activity. It also provides the highest clinical standards for patients reducing patient safety-related problems. Recent research indicates that the implementation of 3D printed models in preclinical training is considered as the future of day-to-day medical care;22,23 however, current models, especially those presenting cerebral structures, need further refinement, as they show significant limitations concerning the materials used.14
This was the first attempt to implement 3D ultrasound imaging data as a standard for printing a 3D model of the neonatal brain in our department. Furthermore, we compared different imaging modalities and evaluated the feasibility of 3D ultrasound data for 3D printing. Our printed model has a high potential of being utilized as a simulation-based model for neurosurgeons to undergo realistic surgical training, for nurses to simulate the perioperative hygiene setting, and for parents to be better informed before the surgical procedure. Within this project, the neonatal lateral ventricles were realistically translated based on 3D ultrasound data with good ventricular properties and dimensions that accurately fit into the MRI-derived skull and brain, according to the ventricular system in vivo. Nevertheless, hydrocephalus is a very complex pathology, possibly causing a rise in ICP and leading to further complications, such as irreversible damage to the brain parenchyma, and seizure activity, as well as neurodevelopmental impairment. As the correlation of the change in ventricular volume with changing ICP has great clinical diagnostic value for managing neonatal hydrocephalus, the current model must be refined by testing further materials to optimize feasibility, filling the hollow ventricles with an adequate fluid and mimicking various levels of ICP. Future research is planned to assess the functional efficiency of these models for training, and to analyze its importance in decreasing the occurrence of catheter-related problems in patients. The described dimensional correlations contribute to the implementation of the 3D ultrasound system as a future standard for assessing IVH and 3D printing of hydrocephalus models. Nevertheless, the overlap between MRI and 3D ultrasound has to be increased by redefining dimensional correlations and increasing the number of studied subjects. With rising expertise on 3D printed models and their feasibility, current costs of 100 euros and printing time of 2.5 h can be reduced in future models. The long-term goal of this project is to create 3D ultrasound-based personalized 3D printed models for either preoperative use or specialized skills training for physicians and nurses to reduce complications during and after an intervention, as well as for discussion with parents to visualize their child’s pathology and the details of the planned operative procedures. With increasing usage of these models, costs can be reduced stepwise.
This study has several limitations, such as the limited number of available imaging data from neonatal patients. Furthermore, the data selection was performed by the primary author of this study based on the availability of imaging data and feasibility for a 3D printed model. Since the contrast-based segmentation procedure for all imaging data was performed by a single operator, the inter-operator discrepancy might be an interesting aspect to investigate in future models. In addition, detailed cerebral structures, such as the third ventricle, choroid plexus, blood vessels, cerebrospinal fluid, and venous sinuses, need to be translated from the imaging data into the 3D model. Finally, the acceptance of such a model among surgeons and the impact of training on patient safety need to be determined.
Conclusion
This pilot study showed the feasibility of using multimodality imaging to develop a 3D printed neonatal brain model. The 3D ultrasound-based lateral ventricles assembled in an MRI-based brain model, in an anatomically correct position, confirmed the dimensional accuracy and feasibility of deriving medical simulation models from an easily accessible bedside ultrasonographic imaging modality. This research has the potential to pave the way for the replacement of conventional training using cadavers and animal models with patient-specific 3D printed models and to introduce 3D brain ultrasound in neonatology. Further investigation in multimodality imaging, segmentation procedures, and material modifications, as well as comparison of 3D ultrasound with other imaging modalities, has the potential to further improve the application of 3D printed models.
References
Zielonka-Lamparska, E. & Wieczorek, A. P. Usefulness of 3D sonography of the central nervous system in neonates and infants in the assessment of intracranial bleeding and its consequences when examined through the anterior fontanelle. J. Ultrason. 13, 408–417 (2013).
Szpecht, D., Frydryszak, D., Miszczyk, N., Szymankiewicz, M. & Gadzinowski, J. The incidence of severe intraventricular hemorrhage based on retrospective analysis of 35939 full-term newborns-report of two cases and review of literature. Childs Nerv. Syst. 32, 2447–2451 (2016).
Miranda, P. Intraventricular hemorrhage and posthemorrhagic hydrocephalus in the preterm infant. Minerva Pediatr. 62, 79–89 (2010).
Pugliese, L. et al. The clinical use of 3D printing in surgery. Updates Surg. 70, 381–388 (2018).
Papazarkadas, X. et al. The role of 3D printing in colorectal surgery: current evidence and future perspectives. Vivo 33, 297–302 (2019).
Aggarwal, R. & Darzi, A. Technical-skills training in the 21st century. N. Engl. J. Med. 355, 2695–2696 (2006).
Cheng, A., Donoghue, A., Gilfoyle, E. & Eppich, W. Simulation-based crisis resource management training for pediatric critical care medicine: a review for instructors. Pediatr. Crit. Care Med. 13, 197–203 (2012).
Wagner, M. et al. Status quo in pediatric and neonatal simulation in four central European regions: the DACHS survey. Simul. Healthc. 13, 247–252 (2018).
Patel, E. A., Aydın, A., Desai, A., Dasgupta, P. & Ahmed, K. Current status of simulation-based training in pediatric surgery: a systematic review. J. Pediatr. Surg. 54, 1884–1893 (2019).
Everett, T. C., MacKinnon, R., de Beer, D., Taylor, M. & Bould, M. D. Ten years of simulation-based training in pediatric anesthesia: the inception, evolution, and dissemination of the Managing Emergencies in Pediatric Anesthesia (MEPA) course. Paediatr. Anaesth. 27, 984–990 (2017).
Ross, J., Rebella, G., Westergaard, M., Damewood, S. & Hess, J. Simulation training to maintain neonatal resuscitation and pediatric sedation skills for emergency medicine faculty. WMJ 115, 180–184 (2016).
Lee Chang, A. et al. Comparison between simulation-based training and lecture-based education in teaching situation awareness. A Randomized Controlled Study. Ann. Am. Thorac. Soc. 14, 529–535 (2017).
Waran, V. et al. Neurosurgical endoscopic training via a realistic 3-dimensional model with pathology. Simul. Health 10, 43–48 (2015).
Cheng, D. et al. Developing a 3D composite training model for cranial remodeling. J. Neurosurg. Pediatr 1–10 (2019).
Kiraly, L. Three-dimensional modelling and three-dimensional printing in pediatric and congenital cardiac surgery. Transl. Pediatr. 7, 129–138 (2018).
Sánchez-Sánchez, Á. et al. Three-dimensional printed model and virtual reconstruction: an extra tool for pediatric solid tumors surgery. Eur. J. Pediatr. Surg. Rep. 6, e70–e76 (2018).
Padia, R. et al. Simulation-guided tracheotomy in a patient with fibrodysplasia ossificans progressiva. Laryngoscope 129, 812–817 (2019).
Prayer, F. et al. Three-dimensional reconstruction of defects in congenital diaphragmatic hernia: a fetal MRI study. Ultrasound Obstet. Gynecol. 53, 816–826 (2019).
Misra, A., Walters, H. L. & Kobayashi, D. Utilisation of a three-dimensional printed model for the management of coronary-pulmonary artery fistula from left main coronary artery. Cardiol. Young 29, 431–434 (2019).
van Lindert, E. J., Liem, K. D., Geerlings, M. & Delye, H. Bedside placement of ventricular access devices under local anaesthesia in neonates with posthaemorrhagic hydrocephalus: preliminary experience. Childs Nerv. Syst. 35, 2307–2312 (2019).
Weinstock, P. et al. Creation of a novel simulator for minimally invasive neurosurgery: fusion of 3D printing and special effects. J. Neurosurg. Pediatr. 20, 1–9 (2017).
Tack, P., Victor, J., Gemmel, P. & Annemans, L. 3D-printing techniques in a medical setting: a systematic literature review. Biomed. Eng. Online 15, 115 (2016).
Bai, J. et al. Efficacy and safety of 3D print-assisted surgery for the treatment of pilon fractures: a meta-analysis of randomized controlled trials. J. Orthop. Surg. Res. 13, 283 (2018).
Acknowledgements
We would like to thank Editage (www.editage.com) for English language editing. This study was funded with support from the Comprehensive Center for Pediatrics Starter Grant. Part of this work has been supported by the Austrian Research Promotion Agency (FFG): “Additive Manufacturing for Medical Research, M3dRES” Project (Nr. 858060).
Author information
Authors and Affiliations
Contributions
M.W., T.W., E.U., and G.O. conceptualized and designed the study, drafted the initial manuscript, and reviewed and revised the manuscript. G.K., G.D., G.O., C.D., H.S., P.S., K.G., M.O., K.R., and A.B. helped with data collection, the analysis process, and reviewed and revised the manuscript. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Statement of consent
No patient consent was required for this study. Permission of the local Institutional Review Board at the Medical University of Vienna (1989/2019) was obtained.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wagner, M., Werther, T., Unger, E. et al. Development of a 3D printed patient-specific neonatal brain simulation model using multimodality imaging for perioperative management. Pediatr Res 91, 64–69 (2022). https://doi.org/10.1038/s41390-021-01421-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-021-01421-w
This article is cited by
-
Quality assurance of 3D-printed patient specific anatomical models: a systematic review
3D Printing in Medicine (2024)