Abstract
Background
Hirschsprung’s disease (HSCR) is the most common congenital cause of intestinal obstruction in children. Sotos syndrome (SoS) is an overgrowth disorder with constipation and sometimes accompanied by HSCR. NSD1 gene mutation is the main cause of SoS. We aimed to investigate association of NSD1 common single nucleotide polymorphisms (SNPs) with HSCR susceptibility in Chinese Han population.
Method
We genotyped 15 SNPs encompassing NSD1 gene region in 420 HSCR patients and 1665 controls on Fludigm EP1 platform. Association analysis was performed between cases and controls.
Result
Rs244709 was the most associated SNP with HSCR susceptibility of the sample set (PAllelic = 9.69 × 10−5, OR = 1.37, 95% CI: 1.17–1.61). Gender stratification analysis revealed that NSD1 SNPs were associated with HSCR in males, but not in females. The nonsynonymous coding SNP rs28932178 in NSD1 exon 5 represented the most significant signal in males (PAllelic = 6.43 × 10−5, OR = 1.42, 95% CI: 1.20–1.69). The associated SNPs were expression quantitative trait loci (eQTLs) of nearby genes in multiple tissues. NSD1 expression levels were higher in aganglionic colon tissues than ganglionic tissues (P = 3.00 × 10−6).
Conclusion
NSD1 variation conferred risk to HSCR in males, indicating SoS and HSCR may share common genetic factors.
Impact
-
This is the first study to reveal that NSD1 variation conferred risk to Hirschsprung’s disease susceptibility in males of Chinese Han population, indicating Sotos syndrome and Hirschsprung’s disease may share some common genetic background.
-
This study indicates more attention should be paid to the symptom of constipation in patients with Sotos syndrome.
-
Our results raise questions about the role of NSD1 in the development of enteric nervous system and the pathogenesis of Hirschsprung’s disease.
Similar content being viewed by others
Introduction
Hirschsprung’s disease (HSCR) is a heterogeneous disorder characterized by the absence of ganglion cells along the intestine resulting in functional bowel obstruction. The incidence of HSCR is estimated to be approximately 1 in 5000 live births in Europeans and 1 in 2800 in Asians.1 This neurodevelopmental birth defect has sex difference in incidence with the male:female ratio of 4:1.2,3 Clinically, according to the length of the aganglionic segment, HSCR can be mainly classified as short-segment HSCR (S-HSCR), long-segment HSCR (L-HSCR), and total colonic aganglionosis (TCA), which count for approximately 80%, 15% and 5% of all patients, respectively.1 The underlying mechanism of HSCR pathogenesis is the failure of normal development of the enteric nervous system (ENS) in the progress of neural crest cell migration, proliferation, differentiation and survival.4 HSCR commonly occurs as an isolated phenotype, which was considered as a multifactorial disease. Common variants in several genes were found be involved in the pathogenesis of HSCR in recent studies, such as RET, NRG1 and SEMA3C-D.2,3
Sotos syndrome (SoS) is a congenital overgrowth disorder with an incidence of approximately 1 in 14,000 live births.5 It is characterized by a distinctive facial appearance, learning disability and overgrowth. SoS is believed to be an autosomal dominant disorder, and NSD1 mutations are detected in approximately 80–90% of SoS patients.5 NSD1 encodes the nuclear receptor binding SET domain protein 1, which contains multiple functional domains and functions as a bifunctional transcriptional regulator that can activate or repress transcription in response to ligand binding. Sio et al.6 reported a case of SoS patient associated with HSCR, and identified a novel NSD1 mutation responsible for this clinical syndrome. Considering that constipation is commonly seen in SoS and HSCR could be accompanied by SoS,7,8 it is reasonable to speculate that HSCR and SoS might share common genes. In this study, we therefore aimed to investigate association of common NSD1 variation with risk to HSCR in Chinese Han population.
Materials and methods
Subjects
A total of 420 sporadic HSCR patients were recruited from 2008 to 2018 who have received treatments in Xinhua Hospital affiliated to Shanghai Jiao Tong University School of Medicine. All the cases were diagnosed with HSCR by examination of biopsy specimen for the absence of the enteric ganglia. Patients were composed of 323 S-HSCR, 58 L-HSCR, and 39 TCA. Among 420 patients, 322 were males and 98 were females (the male:female ratio of 3.29:1). A group of 1665 healthy, unrelated subjects visiting Xinhua Hospital for routine health check were randomly selected as controls including 1281 males and 384 females (the male:female ratio of 3.34:1). This study was approved by the institution review board of Xinhua Hospital. Written informed consent was obtained from all participants or their parents after the procedure had been fully explained. All data were recorded anonymously, but the data were destroyed if the participants asked to withdraw their file.
SNP selecting and genotyping
We investigated single nucleotide polymorphisms (SNPs) located in the genic region and 20 kb upstream and downstream flanking regions of NSD1. TagSNPs were selected using the Genome Variation Server (http://gvs.gs.washington.edu/GVS150/) with MAF (minor allele frequency) ≥ 0.01 and r2 ≥ 0.8 based on the HapMap HCB (Han Chinese in Beijing) data. Among a cluster of interchangeable tag SNPs, SNPs with predicted deleterious effect were preferred including nonsynonymous SNPs, expression quantitative trait loci (eQTL) SNPs and SNPs occurring within transcription factor binding sites or in a regulatory region. To avoid missing information caused by failure in assay designing and genotyping, nonsynonymous SNPs and 5′UTR SNPs in high LD were all selected. Together 15 SNPs were selected, namely rs244730, rs244731, rs71601339, rs244709, rs202177679, rs3733875, rs28932178, rs79600412, rs6886255, rs35848863, rs34165241, rs78247455, rs4631, rs3088050 and rs10051822.
The selected 15 SNPs were genotyped using Fluidigm 96.96 Dynamic Array IFCs on Fludigm EP1 platform. Of the 15 SNPs genotyped, three SNPs (rs4631, rs3088050 and rs10051822) with call rate < 98% was removed for further association analysis. The data completion rate of the remaining 12 SNPs was 99.9%. The average interval between the 12 SNPs was 16.3 kb.
Functional annotation
Functional consequence of coding variants was predicted using PolyPhen 2, SIFT and PROVEAN. We visited GTEx Analysis Release v.8 (http://www.gtexportal.org/) to examine the eQTL for 12 selected SNPs of NSD1.9 We also investigated the functional consequences of the associated SNP by checking HaploRegv4.1 database. The gene expression profiles of associated genes were also acquired from GTEx database. We explored protein−protein interaction using STRING database (http://string-db.org/).10 NSD1 and its neighbor genes MXD3 and RGS14 were looked up in STRING database to explore the interaction with HSCR underlying pathway genes (RET, EDNRB, NRG1, SEMA3, SOX10 and PHOX2B).
Quantitative real-time PCR analysis
Aganglionic colon tissues and adjacent ganglionic tissues were collected from 28 HSCR patients who underwent surgery in Xinhua Hospital affiliated to Shanghai Jiao Tong University School of Medicine. Total RNA of aganglionic colon tissues and ganglionic tissues was isolated using RNeasy Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. cDNAs were synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific™, MA USA). Quantitative real-time PCR (RT-PCR) was performed with SYBR Green Kit (TaKaRa, Otsu, Japan) on a QuantStudio Dx Real-Time PCR (RT-PCR) Instrument (Applied Biosystems, Foster City, CA, USA). RT-PCR of each sample was performed in triplicate with GAPDH as an internal control. Primers are listed in Supplementary Table S1. The expression levels between aganglionic colon tissues and ganglionic tissues were compared using a paired Student’s t test.
Statistical analysis
Genetic analysis was performed using PLINK 1.09. The genotype distribution of each SNP was tested for Hardy−Weinberg equilibrium in both case and control population.
Four genetic models, including the allelic, additive, dominant and recessive models, together with a genotypic association test (2df test) were used to analyze the association for each SNP. Both allele and genotype frequencies were assessed by χ2-test between the cases and the controls. Conditional logistic analysis was performed to find additional markers with independent effect by adding the top associated markers as covariates in logistic regression. The linkage disequilibrium (LD) structure of 12 SNPs was examined by program Haploview 4.2. A study-level P value < 0.0042 (0.05/12) was considered statistically significant.
Results
Associations of NSD1 SNPs with HSCR
The genotype distribution of all 12 SNPs genotyped conformed to the Hardy−Weinberg equilibrium (P > 0.05). The allelic and genotypic distribution between cases and controls are shown in Tables 1 and 2, respectively. After multiple testing, we found that six SNPs reached the study-level significance.
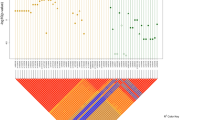
Of the 12 SNPs genotyped, rs244709 is the most associated SNP located in intron 2 of NSD1. The minor allele T of rs244709 was associated with HSCR risk (PAllelic = 9.69 × 10−5, OR = 1.37, 95% CI: 1.17–1.61; Table 1). The frequency of rs244709 allele T was 0.49 in cases and 0.42 in controls. The second association signal was two nonsynonymous SNPs in the fifth exon of NSD1, namely rs3733875 and rs28932178. These two SNPs were in perfect LD (r2 = 1) and were in high LD with rs244709 (r2 = 0.83; Fig. 1). The major allele T of rs28932178 was associated with HSCR risk (PAllelic = 7.68 × 10−4, OR = 1.30, 95% CI: 1.11–1.51; Table 1).
The plots were constructed with the program Haploview27, and r2 values (×100) were depicted in the diamonds. r2 values were estimated from the data of 420 HSCR patients and 1663 controls enrolled in the current study. r2 values of 1 represent complete LD, r2 values greater than 0.8 represent strong evidence of LD, r2 values of 0.2–0.8 represent inconclusive LD, and r2 less than 0.2 represent negligible evidence of LD. Blocks were determined using the default confidence intervals.
HSCR is more common in males than in females; we therefore performed association analysis in males and females, respectively. None of the 12 SNPs showed association with HSCR in females (P > 0.05; Table 3). However, a significant increase in risk was observed when comparing male group with the combined samples. The nonsynonymous coding SNP rs28932178 represented the most significant signal (PAllelic = 6.43 × 10−5, OR = 1.42, 95% CI: 1.20–1.69; Table 3). rs71601339 located 5′ upstream of NSD1 was in high LD with the above two missense SNPs (r2 = 0.98) and also showed significant association with HSCR risk (PAllelic = 1.63 × 10−4, OR = 1.43, 95% CI: 1.19–1.73, Table 3). Since six SNPs showed association with disease risk in males, we performed conditional logistic analysis to identify independent association signal. When conditioning on the most associated SNPs rs28932178, no other SNPs show significant association (P > 0.05).
Genotype−phenotype association study
HSCR is classified as S-HSCR, L-HSCR and TCA. S-HSCR is the most common form, which is characterized by multifactorial inheritance.3,11 L-HSCR and TCA is often characterized by autosomal dominant inheritance. However, rs2435357 risk allele at RET, the major HSCR gene, was observed to be more common in S-HSCR/L-HSCR when compared with TCA HSCR.12 Consequently, we investigated whether all six significant SNPs were restricted to specific markers of subgroups by comparing frequencies of risk alleles within cases classified by segment length of aganglionosis. Firstly, we grouped S-HSCR/L-HSCR together and compared them with TCA HSCR cases. Secondly, we grouped L-HSCR/TCA together and compared them with S-HSCR cases. Our results found none of the six associated SNPs showed significant genotype–phenotype associations as shown in Table 4.
Functional annotation of associated SNPs
SNP rs28932178 is a nucleotide transition (T > C) in exon 5 of NSD1 that causes the amino acid substitution P726S. The functional prediction results of rs28932178 from PolyPhen 2, SIFT and PROVEAN were incongruent. PolyPhen 2 predicts a probably damaging variant with a score of 0.998, PROVEAN predicts a neutral variant with a score of 0.99 (cutoff = −2.5), SIFT predicts a damaging variant with a score of 0.013 (cutoff = 0.05). Unexpectedly, the risk allele T of rs28932178 was the ancestral one. The other nonsynonymous SNP rs3733875 in the fifth exon of NSD1 was predicted to a neutral variant in three databases.
NSD1 was widely expressed and the highest level was found in cerebellar hemisphere and cerebellum from the Genotype-Tissue Expression (GTEx) portal (Supplementary Fig. S1). A higher expression level was also found in gastrointestinal tract tissues (Supplementary Fig. S1). The expression profiles of NSD1 also indicated that it might play a critical role in the development of central nerve system and enteric nerve system.
None of the six associated SNPs was identified as eQTL of NSD1, while four of the six SNPs (rs71601339, rs3733875, rs28932178, rs6886255) were found to be correlated with MXD3 expression in multiple gastrointestinal tissues (Table 5). rs244709 was associated with the expression of RGS14 as well (Table 5). Both MXD3 and RGS14 are neighboring genes of NSD1. Protein−protein interaction analysis found no interaction between NSD1 MXD3 and RGS14 with HSCR underlying pathway genes (RET, EDNRB, NRG1, SEMA3, SOX10, PHOX2B).
Gene expression analysis
We further compared the RNA expression level of NSD1, MXD13 and RGS14 in aganglionic and ganglionic colon tissues from 28 HSCR patients. The expression level of NSD1 was higher in aganglionic tissues than ganglionic tissues, with the P value of 3.00 × 10−6 (Fig. 2a), while the expression levels of RGS14 (P = 0.42) and MXD3 (P = 0.48) showed no difference (Fig. 2b, c), indicating NSD1 may participate in the pathogenesis of HSCR.
Discussion
SoS was first described by Juan Sotos in 1964 in five patients with prenatal and postnatal overgrowth, characteristic facial appearance, advanced bone age and developmental delay.13 The diagnosis of SoS is mainly based on the disease history and physical examination, which includes the cardinal features of typical facial dysmorphisms, increased velocity of growth and weight with advanced bone age, and some form of developmental delay such as in talking, walking, or other developmental milestone.6 Genetic analysis revealed that SoS was caused by mutation of NSD1.8 SoS is often accompanied by other symptoms, such as congenital heart disease, neurologic abnormalities and genitourinary problems.13 Notably, some SoS patients’ complaint was gastrointestinal symptoms, especially constipation.7,14 Sio et al.6 recently reported an SoS patient with HSCR and identified two novel rare disease-causing mutations of NSD1, c.2465C4A (p.Ser822Tyr) and c.4347T4A (p.Cys1449). In this study, we found that six common SNPs in NSD1 were associated with isolated HSCR, indicating SoS and HSCR may share overlapping genetic background. Stratification analysis revealed that NSD1 SNPs were associated with HSCR in males, but not in females.
Out of the six associated markers, rs3733875 and rs28932178 are two missense variants in the exon 5 of NSD1. rs3733875 is a G > T transversion, changing a valine residue to a leucine residue; while rs28932178 is a T > C transversion, changing a serine residue to a proline residue. The amino acid substitutions caused by missense variants are more likely to have serious consequences for the function or structural stability of NSD1, especially when the interval between these two SNPs is only 336 bps in location.15,16 The remaining four associated SNPs (rs71601339, rs3733875, rs28932178 and rs6886255) are located in the untranslated region of NSD1. We searched all the four positive SNPs in HaploReg to further explore their functions.17 Interestingly, we found rs28932178 and rs6886255 affected a PAX-family binding site (Supplementary Table S2), while PAX3 has been reported to play an important role in ENS development and in the pathogenesis of colonic aganglionosis.18 Therefore, these four noncoding SNPs might also have some functional effect involved in the pathogenesis of HSCR.
Considering the clinical manifestations, surgical options, and post-operation complications of L-HSCR and TCA patients differ significantly from S-HSCR patients, we performed an association analysis stratified by clinical subtype and did not find SNPs associated with specific subtype. In a gender-stratified analysis, we found common SNPs in NSD1 were specially associated with HSCR in males. The male-specific association might be explained by that NSD1 transcription was regulated by androgen; however, experimental evidence is still needed. Another explanation might be that the small sample size of females (98 female patients vs. 384 female controls) had not enough power to detect the moderate association of common SNPs with this disease.
NSD1 contains 23 exons and encodes a histone H3 lysine 36 methyltransferase, which is important for multiple aspects of normal embryonic development.19 NSD1 binds to various promoter elements to regulate gene transcriptions.20 NSD1 was recently recognized as an indicative factor of transcriptionally permissive chromatin state, which regulates development progress through the epigenetic mechanisms and plays a critical role in ENS development.21,22,23,24 It was also reported that NSD1 mutations altered genome-wide DNA methylation in patients with SoS.20 Our expression analysis showed that NSD1 had a higher expression level in aganglionic colon tissues than in ganglionic tissues. It is possible that NDS1 regulates expression level of the major HSCR genes such as RET, NRG1 and SEMA3C-D,2,3 and therefore contributes to the pathogenesis of HSCR. As NSD1 SNPs showed male-specific association with HSCR, it could be hypothesized that the methyltransferase function of NSD1 might be regulated by androgen. The underlying molecular basis need to be elucidated in future study.
In addition, NSD1 protein interacts with retinoic acid receptors.25 Retinoic acid is required for the efficient migration of enteric neural crest-derived cells (ENCDCs) by reducing the levels of phosphatase and tensin homolog (PTEN) protein, a critical negative regulator of ENCDC migration and proliferation.26 Whether disease-associated variation influences expression of NSD1 and thus alter RA signal was expected to be further explored. Furthermore, depletion of NSD1 reduced the expression of bone morphogenetic protein gene 4 (BMP4).27 BMP4 is known to play an important role in the formation of ENS by enhancing neuronal aggregation and controlling the clustering of ENCDCs into definitive ganglia.28 Since both RA and BMP4 influence the migration of ENCDCs, NSD1 might be involved in related signaling pathway, and plays a role in the pathogenesis of HSCR.
Of note, eQTL data from GTEx database revealed the associated SNPs in NSD1 region were eQTLs of the adjacent genes MXD3 and RGS14, but not associated with the NSD1 expression level (Table 5). Since gene expression regulation is spatio-temporal progress, the eQTL data from adult tissues could not present the expression level in embryonic stages, when the ENS developed. Therefore, whether the associated SNPs influence the expression of NSD1 in the stage of ENS development still needs further investigation. MXD3 is a transcription factor involving in the MYC/MAX/MXD transcriptional network. Knocking down of MXD3 led to a reduction in cell numbers, and overexpression of MXD3 promoted cell proliferation in mouse cerebellar.29 Interestingly, it was demonstrated that acute activation of MXD3 resulted in a transient increase in cell proliferation, while persistent activation of MXD3 resulted in an overall decrease in cell numbers, indicating MXD3 had a complicated role in the regulation of cell proliferation.30 RGS14 is a multifunctional scaffolding protein that integrates G protein and MAPK signaling pathway to regulate synaptic plasticity.31 Since these three neighboring genes all have roles in regulating the neural proliferation, it is also possible that they may work as complex to influence the development of ENS.
Our results revealed NSD1 as a susceptibility locus to HSCR in males and indicated new pathway underlying pathogenesis of HSCR. Further replication studies in other independent sample set were needed to validate this finding and functional experiments were also required to investigate the mechanisms of NSD1 influence on the development of ENS and the pathogenesis of HSCR.
Conclusion
Common variations of NSD1 conferred risk to HSCR susceptibility in Chinese Han population, indicating SoS and HSCR may share partial overlapping genetic background.
References
Haricharan, R. N. & Georgeson, K. E. Hirschsprung disease. Semin. Pediatr. Surg. 17, 266–275 (2008).
Garcia-Barcelo, M.-M. et al. Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung’s disease. Proc. Natl Acad. Sci. USA 106, 2694–2699 (2009).
Jiang, Q. et al. Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to Hirschsprung disease liability. Am. J. Hum. Genet. 96, 581–596 (2015).
Burkardt, D. D., Graham, J. M., Short, S. S. & Frykman, P. K. Advances in Hirschsprung disease genetics and treatment strategies: an update for the primary care pediatrician. Clin. Pediatr. 53, 71−81 (2014).
Lane, C., Milne, E. & Freeth, M. Cognition and behaviour in Sotos syndrome: a systematic review. PLoS ONE 11, e0149189 (2016).
Sio, C. A. et al. Sotos syndrome associated with Hirschsprung’s disease: a new case and exome-sequencing analysis. Pediatr. Res. 82, 87–92 (2017).
Faravelli, F. NSD1 mutations in Sotos syndrome. Am. J. Med. Genet. Part C: Semin. Med. Genet. 137C, 24–31 (2005).
Kurotaki, N. et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 30, 365–366 (2002).
Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein−protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 (2017).
Tam, P. K. Hirschsprung’s disease: a bridge for science and surgery. J. Pediatr. Surg. 51, 18–22 (2016).
Emison, E. S. et al. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am. J. Hum. Genet. 87, 60–74 (2010).
Leventopoulos, G. et al. A clinical study of Sotos Syndrome patients with review of the literature. Pediatr. Neurol. 40, 357–64 (2009).
Baujat, G. & Cormier-Daire, V. Sotos syndrome. Orphanet J. Rare Dis. 2, 36 (2007).
Hirschhorn, J. N. & Daly, M. J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108 (2005).
Orr, N. & Chanock, S. Common genetic variation and human disease. Adv. Genet. 62, 1–32 (2008).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Barlow, A. J., Dixon, J., Dixon, M. & Trainor, P. A. Tcof1 acts as a modifier of Pax3 during enteric nervous system development and in the pathogenesis of colonic aganglionosis. Hum. Mol. Genet. 22, 1206–17 (2013).
Portela, A. & Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 28, 1057–68 (2010).
Choufani, S. et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 6, 10207 (2015).
Hu, N., Strobl-Mazzulla, P. H. & Bronner, M. E. Epigenetic regulation in neural crest development. Dev. Biol. 396, 159–68 (2014).
Torroglosa, A. et al. Epigenetics in ENS development and Hirschsprung disease. Dev. Biol. 417, 209–216 (2016).
Strobl-Mazzulla, P. H., Sauka-Spengler, T. & Bronner-Fraser, M. Histone demethylase JmjD2A regulates neural crest specification. Dev. Cell 19, 460–468 (2010).
Tan, H., Wu, S., Wang, J. & Zhao, Z. K. The JMJD2 members of histone demethylase revisited. Mol. Biol. Rep. 35, 551–556 (2008).
Huang, N. et al. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 17, 3398–3412 (1998).
Sato, Y. & Heuckeroth, R. O. Retinoic acid regulates murine enteric nervous system precursor proliferation, enhances neuronal precursor differentiation, and reduces neurite growth in vitro. Dev. Biol. 320, 185–98 (2008).
Lucio-Eterovic, A. K. et al. Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc. Natl Acad. Sci. USA 107, 16952–16957 (2010).
Goldstein, A. M., Brewer, K. C., Doyle, A. M., Nagy, N. & Roberts, D. J. BMP signaling is necessary for neural crest cell migration and ganglion formation in the enteric nervous system. Mech. Dev. 122, 821–33 (2005).
Ngo, T. et al. Alternative splicing of MXD3 and its regulation of MXD3 levels in glioblastoma. Front. Mol. Biosci. 6, 5 (2019).
Ngo, T., Barisone, G. A., Lam, K. S. & Dίaz, E. MXD3 regulation of DAOY cell proliferation dictated by time course of activation. BMC Cell Biol. 15, 30 (2014).
Evans, P. R., Lee, S. E., Smith, Y. & Hepler, J. R. Postnatal developmental expression of regulator of G protein signaling 14 (RGS14) in the mouse brain. J. Comp. Neurol. 522, 186–203 (2014).
Acknowledgements
We thank all the participants of this research. This work was supported by National Natural Science Foundation of China (31671317, 31471190 and Key Program 81630039), Foundation of Shanghai Municipal Health Commission (201840014) and Shanghai Key Laboratory of Pediatric Gastroenterology and Nutrition (17DZ2272000).
Author information
Authors and Affiliations
Contributions
All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work; W.C. and X.C. designed the research and revised the article; X.-X.Y. drafted the article; X.C. and X.-X.Y. analyzed the data; X.-X.Y., W.-J.W., H.-L.S., Z.-L.W., M.-R.B., Y.J.L., and Y.-M.G. performed the experiment and collected the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent statement
In the present study, written informed consent was obtained from all participants or their parents.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yu, XX., Chu, X., Wu, WJ. et al. Common variation of the NSD1 gene is associated with susceptibility to Hirschsprung’s disease in Chinese Han population. Pediatr Res 89, 694–700 (2021). https://doi.org/10.1038/s41390-020-0933-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-020-0933-3