Abstract
Background
Patients with Down syndrome (DS) are at increased risk for infections and autoimmune disorders. Although several immunological abnormalities were previously found, differences in T cell receptor repertoire have never been shown. Thus we compared the T cell receptor gamma (TRG) repertoire in DS and non-syndromic pediatric patients by next-generation sequencing, in addition to other immunological markers.
Methods
Genomic DNA was extracted from thymuses of pediatric patients who underwent heart surgery, where six were with DS and six were non-syndromic patients. Peripheral blood counts, T cell subpopulations, thymus TCR excision circles (TRECs), spectratyping, and next-generation sequencing for TRG were analyzed.
Results
The mean age of the patients was 7 months and the mean lymphocyte count was slightly lower in patients with DS, whereas thymus TREC results were similar to non-syndromic patients (p = 0.197). The TRG repertoire analysis showed that patients with DS had a significantly larger number of unique TRG sequences, together with decreased clonal expansion. Lastly, the V and J gene usages in the thymus were similar in DS and non-syndromic patients.
Conclusions
Patients with DS showed increased TRG repertoire diversity with decreased clonal expansion compared to non-syndromic patients.
Impact
-
Alterations in T cell receptor gamma repertoire were found in patients with Down syndrome using next-generation sequencing (NGS) technique. Patients showed increased repertoire diversity and decreased clonal expansion compared to controls.
-
These findings add to previous reports on abnormalities of other immune system components in patients with Down syndrome. NGS technique may point out differences not seen by previous methods.
-
Repertoire abnormalities may contribute to those patients’ predisposition to infections and autoimmune diseases.
Similar content being viewed by others
Introduction
Down syndrome (DS; trisomy 21) is one of the most common genetic disorders, affecting approximately 1:700 live births.1,2 The typical features of patients with DS are the characteristic physique and mental retardation. In addition, affected patients have an increased risk to acquire infections and autoimmune disorders, such as hypothyroidism, celiac disease, alopecia areata, diabetes mellitus, and more.3,4 The factors for the association of autoimmune disorders with DS are still largely unknown. Previous reports of immune characterization of patients with DS have demonstrated decreased function of natural killer cells,5,6 diminished expansion of B lymphocytes, and abnormal humoral response.7,8,9 Numerous studies show defects in T cells from thymus morphology10,11,12 and gene expression13 to quantity and quality aspects of T cells.7,8,12,14 Patients with DS are reported to have a decreased population of cells expressing high levels of T cell receptor (TCR) αβ and a markedly higher proportion, approximately 10% of cells, expressing TCR γδ.3,8,15 In addition, AIRE (autoimmune regulator) protein, whose gene is located on 21q22.3 and was found to be downregulated in DS patients,13,16 is known to play a critical step in preventing autoimmunity by regulating the apoptosis of cells expressing TCR against self-antigens presented by the major histocompatibility complexes.
Interestingly, children with DS were found to have a significantly lower number of TCR excision circles (TRECs) in their peripheral blood cells.17 TREC is a small circle of non-replicating DNA formed as a by-product during the normal process of TCR production. Its level reflects thymic TCR gene recombination and number of recent thymic emigrants; therefore, it is a highly sensitive and specific tool to estimate T cell immunity in any medical conditions where T cells are known to be affected. The recent implementation of TREC-based assays to screen for T cell immunodeficiency early in life in every newborn enabled identification of some but not all infants with DS due to their low TREC levels.18
TCR repertoire, which is critical for induction of immunological response to an unlimited number of foreign antigens, as well as for the development of tolerance and prevention of autoimmunity, was less investigated in patients with DS. Furthermore, abnormalities in TCR repertoire determined by next-generation sequencing (NGS) have not yet been reported in DS. TCR repertoire evaluation by NGS is a technology that enables analysis of immune repertoires to a depth that was unreached before. This technique has been effective in detecting skewed TCR repertoire in multiple primary immunodeficiency syndromes, such as Omenn syndrome,19 Wiskott Aldrich syndrome,20 and agammaglobulinemia.21 It was also successfully used to determine immune balance and autoimmune activity in the affected organs, including in patients with juvenile rheumatoid arthritis,22 systemic lupus erythematosus,23 and inflammatory bowel diseases.24 Thus information on the immune repertoire of patients with DS is of great importance in comprehensively understanding the immunological component in DS.
In this work, in addition to other immunological markers, we described TCR repertoire in thymus samples obtained from patients with DS during cardiac surgery and compared to TCR repertoire of thymuses from non-syndromic patients.
Methods
Patients
Thymuses were removed as part of the standard surgical procedure from pediatric patients who underwent heart surgery due to congenital heart defects in the “Edmond and Lily Safra” Children’s Hospital, Sheba Medical Center, Tel Hashomer, Israel during the years 2009–2010. Blood tests for immunologic evaluation were taken during the hospitalization. The patients’ medical records were reviewed for clinical and demographic data, which includes age, weight, heart defect and surgical procedure, diagnosis of DS, infectious diseases, and concurrent medical conditions. Patients with suspected primary immunodeficiency were excluded. Genomic DNA from samples was extracted using the G-DEXTM IIC Kit (Intron Biotechnology, Korea).
The study was approved by the institution’s ethics committee (7730–10-SMC) and all patients’ parents signed consent forms accordingly.
TREC quantification
The amount of TRECs were quantified by real-time quantitative polymerase chain reaction (RQ-PCR) using specific fluorescent probe and primers in TaqMan assays, as previously described.25,26 Specifically, the reactions were performed using 500 ng of genomic DNA extracted from thymus cells. PCR reactions contained TaqMan universal PCR master mix (Applied Biosystems, Waltham, MA, USA), specific primers (900 nM), and FAM-TAMRA probes (250 nM). RQ-PCR was carried out in an ABI PRISM 7900 Sequence Detector System (Applied Biosystems, Waltham, MA, USA). The number of TRECs in each sample was determined by comparing cycle (Ct) value of a patient’s sample to the standard curve. The standard curve was constructed by using serial tenfold dilutions (103–106 copies) of a known TREC plasmid, kindly provided by Dr. Daniel Douek (Vaccine Research Center, National Institute of Allergy and Infectious Diseases, Bethesda, MD, USA). Amplification of RNaseP (TaqMan assay, Applied Biosystems, Waltham, MA, USA) served as a quality control to verify similar amounts of genomic DNA that were used in the assays.
Spectratyping
TCR rearrangements were amplified by PCR according to the standardized BIOMED-2 protocol with the following two sets of Vγ primers designed against different variable region genes using four fluorescently labeled sequences (either FAM or HEX fluorochrome):
Variable region | Sequence | |
|---|---|---|
Set 1 | Vγ 9/2 | [5′ ACCTGGTGAAGTCATACAGTTC 3′ FAM] |
Vγ 11 | [5′ CTTCCACTTCCACTTTGAAA 3′ HEX] | |
Set 2 | Vγ f1: Vγ 2, Vγ 3, Vγ 4, Vγ 5, Vγ 8 | [5′ ACTGGTACCTACACCAGGAGG 3′ FAM] |
Vγ 10/2 | [5′ AGCATGGGTAAGACAAGCAA 5′ HEX] |
Specifically, for each sample, two PCR reactions were performed using either set 1 or set 2 primers and a mixture of the following reverse primers: Jγ1/2 TACCTGTGACAACCAGTGTTG, JγP ACTTACCTGTAATGATAAGCTTT, and JγP1/2 TTACCAGGCGAAGTTACTATG. The PCR amplification products were visualized using GeneScan 500TM (Applied Biosystems, Waltham, MA, USA), and the analysis was carried out using the GeneMapper software (Applied Biosystems, Waltham, MA, USA).
Next-generation sequencing
TCR libraries from genomic DNA were produced using primers for various V and J genes in the TRG loci, according to the manufacturer’s protocol (LymphoTrack, Invivoscribe, San Diego, CA, USA). Same amount of 150 ng genomic DNA was taken for each sample. Quantified libraries were pooled and sequenced using Mi-Seq Illumina technology. All the samples were run in the same sequencing run. The sequences were initially subjected to bioinformatic analyses (carried out by Invivoscribe Technologies and Adaptive Biotechnologies) from fastq files for all the libraries, which were further analyzed by submitting to the IMGT HighV-QUEST webserver (http://www.imgt.org). Normalized Shannon \({\hat{H}}\) index for diversity and Dominance D for inequality or unevenness were calculated using PAST program (http://folk.uio.no/ohammer/past/) and were based on the following formulas:
where R = number of unique rearrangements and pi = proportion of the total rearrangements belonging to the “i”th unique rearrangements.
For the calculation of the diversity indices, percentile bootstrapping (with N = 9999 bootstrap replicates) was used to compute lower and upper limits for 95% confidence intervals. Analyses included only productive sequences. Tree maps of the TRG repertoire determined by NGS were generated using the TreeMap program (https://www.treemap.com/, by Macrofocus, GmbH, Switzerland). The fastq files are available in the SRA database of the National Center for Biotechnology Information (NCBI), with SRA accession number PRJNA57758. Specifically, the accession numbers are as follows: SAMN13030286, SAMN13030287, SAMN13030288, and SAMN13030289 for C1, C2, C3, and C4, respectively; SAMN13030290, SAMN13030291, SAMN13030292, and SAMN13030293 for P1, P2, P3, and P4 respectively.
Statistics
Data were analyzed using PRISM version 5 (Graph Pad) using Mann–Whitney test, where one-tailed p value < 0.05 was considered significant.
Results
Clinical and laboratory characteristics
Six DS (mean age 8.2 months) and six control (mean age 5.8 months; hereafter referred as controls) patients were included in the study. Thymuses were obtained during their cardiac surgeries. The clinical characteristics of the 12 individuals are detailed in Table 1. The prevalent surgery in patients with DS was atrioventricular canal (AV canal) repair, with or without tetralogy of Fallot (TOF) (4/6 patients), while in the control group TOF repair was the most common procedure (3/6). Patients 4 and 5 suffered from multiple infections during their hospitalizations.
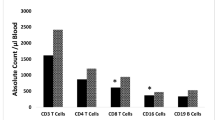
White blood cell (WBC) counts and T cell subsets of the patients and controls are described in Table 2. The mean lymphocyte count of Down patients was slightly lower than controls (4.680 cells/mm3 × 10−3 vs 6.970 cells/mm3 × 10−3, p = n.s.) and 4/6 patients demonstrated lymphopenia, as was reported in previous studies.8,27,28 T cell subpopulation analysis by flow cytometry (fluorescence-activated cell sorter) was available for 3/6 Down patients and showed low CD4 cells’ percentage in 2 patients. Unlike previous cohorts,4,29 no increase in the percentage of CD8 cells was observed in our patients with DS.
TREC levels in thymuses, representing thymic production of new T cells, were determined (Table 2) and showed no significant differences in the mean value between patients and controls (p = 0.197). Patient 4, who was also lymphopenic, had a significantly lower thymic TREC value (discussed later), whereas patient 3 had a remarkably high value (Table 2). The TREC level in peripheral blood were available in 4/6 patients. There were no significant differences in the mean blood TREC value between patients and controls (p = 0.0857). Two patients (P2 and P4) had values markedly lower than controls, albeit in the normal range (Table 2). PCR analysis of the TRG gene rearrangement (spectratyping) was done to evaluate the clonality of TRG repertoire (Fig. 1), which showed no notable clonality in either groups.
TCR gamma repertoire by NGS
Comprehensive TRG repertoire of four patients with DS and four controls was determined using NGS. One patient with DS was omitted owing to exceptional clinical course (patient 4, discussed later). In order to demonstrate the richness of our sampled sequences, we plotted rarefaction curves for the sequencing data from each of the patients and controls (Fig. 2). The rarefaction curves reach the plateau as the depth of the sequencing increases until most of the sequences in the sample have been sequenced and only rare ones remain to be sampled. Although the three patients (P1–P3) showed a higher curve compared to the controls, all the curves had reached the plateau, thus further sequencing would not reveal significantly large number of new unique sequences. Furthermore, the rarefaction curves distinctly divide the patient and control groups. The numbers of unique and total TRG sequences are shown in Fig. 3. Noticeably, while the mean number of total sequences was lower in patients compared to controls, the number of unique sequences was significantly higher in the patients with DS compared to controls (Fig. 3a, b). The overall diversity of the repertoire is represented by the calculations of the normalized Shannon’s \(\hat{H}\) diversity index and the unevenness of the repertoire is demonstrated by calculating the Dominance D. The normalized Shannon’s \(\hat{H}\) showed a tendency for broader repertoire in the patients compared to the controls (Fig. 3c). Patients also showed significantly less unevenness of the clonal distribution in the TRG repertoire, as measured by Dominance D, when compared to the controls (Fig. 3d). Furthermore, patients with DS demonstrated a significantly lower clonal expansion compared to the controls, depicted by lower frequencies of the top 100 most abundant TRG sequences, as well as the sum of the top 100 most abundant sequences (Fig. 3e).
a Number of unique sequences of TRG in Down syndrome patients (mean = 27,575) vs controls (mean = 15,651), b number of total sequences of TRG in Down syndrome patients (mean = 74,034) vs controls (mean = 99,886), c normalized Shannon index \(\hat{H}\) was higher in patients (0.9521) vs controls (0.9387), representing a trend toward higher diversity in patients. d Unevenness of the repertoire, demonstrated by calculating the Dominance D, was lower in patients (0.0001487) vs controls (0.0002317), representing a more even distribution of clonal expansion. e The relative frequencies of the most abundant 100 sequences out of the total number of sequences.
Lastly, the TRG repertoire of each patient is graphically presented as Tree maps, to visually demonstrate the overall diversity of the repertoire (Fig. 4). The Tree maps of patients with DS graphically demonstrated the increased number of sequences (depicted by increased number of squares), with less clonal expansions (depicted by the smaller size of the squares compared to controls; Fig. 4). In addition, the usages of V and J gene segments were determined in both unique and total TRG sequences, where no notable differences were observed in the patients’ TRG sequences compared to the controls (Fig. 5). Thus VJ recombination can be eliminated as a leading explanation for the repertoire alterations.
a Percent usage of specific V gene segments in the total sequences, b percent usage of specific V gene segments in the unique sequences, c percent usage of specific J gene segments in the total sequences, d percent usage of specific J gene segments in the unique sequences. The whiskers designate ±2 standard deviations.
Patient 4
This patient is discussed separately due to her remarkable clinical presentation. This was a female infant, born in Cyprus at 36 weeks’ gestation, diagnosed with DS and AV canal. She was first admitted to our hospital at the age of 4 months with multi-organ failure and underwent pulmonary artery banding and AV valvuloplasty. At the age of 14 months, she was admitted for Glenn operation, and during her hospitalization, she suffered from two episodes of bacteremia, with positive cultures of Serratia and Proteus, and from candidemia.
The laboratory studies showed that the patient was lymphopenic and had an extremely low thymus TREC number (Table 2). The TRG repertoire by NGS analysis revealed a markedly reduced repertoire, with only 5255 unique sequences, lower than the number of unique sequences in both patients and controls, as seen in Fig. 3a. The normalized Shannon’s \(\hat{H}\) diversity index was low (0.873). However, the total number of sequences (79,271 sequences) was similar to other patients. Overall, this unique pattern of the repertoire diversity was also seen in the patient’s Tree map (Fig. 6). Furthermore, the patient demonstrated a skewed usage of TRGV and TRGJ gene segments compared to the TRG repertoire of the other patients (Fig. 6).
a Tree map (left), characterized by smaller number of squares (sequences) and larger number of copies of each square compared to controls (control 4, right). b TRGV and TRGJ gene usage in the total sequences, compared to the means of Down syndrome patients and controls. The whiskers designate ±2 standard deviations.
Discussion
We aimed to characterize the TRG repertoire in order to understand the possible association of infections and autoimmunity with DS. To our knowledge, our current study is the first attempt in using the NGS technology to characterize the thymic TCR repertoire in patients with DS.
In the basic immunologic analyses, thymus TREC values of the majority of DS patients were similar to those of controls, and only one patient, who had severe infections during her hospitalizations, had a markedly lower value. Recently, Marcovecchio et al. reported thymic TREC values among patients with DS to be comparable to controls.12 Additionally, they reported reduced TREC numbers in the peripheral blood of DS patients,12 confirming previously reported studies of low peripheral TREC values determined for DS patients.17,30 Although our previous reports demonstrated a good correlation between peripheral blood and thymus TREC values,31 it is possible that this correlation does not apply in patients with DS due to the major abnormalities of their thymus.
As seen in previous studies on various immunodeficiencies, the NGS technology for studying immune repertoire allowed for higher level of sensitivity and, in certain cases, allows to determine skewed repertoire where no significant differences were found in TREC numbers or spectratyping.19,20,21 In addition, in patient 4, the data from the TRG repertoire determined by NGS supported other findings of immunodeficiency. The top 100 analysis of the NGS results reveals impaired clonal expansion that may contribute to aberrant immune response to pathogens. The TRG repertoire suggests that patients with DS show a trend for more diverse repertoire, as can be seen by the increased unique sequence numbers and higher normalized Shannon’s \(\hat{H}\) diversity index, which may contribute to the development of autoimmunity. The increased diversity may be due to defective central tolerance as a result of decreased expression of AIRE in DS,13 further leading to survival of auto-reactive cells, explaining the susceptibility to autoimmunity. Analysis of biomarkers of self-reactive T cells is needed for further investigation of this hypothesis.
Our study has a few limitations; first, the cohort was small, and since the expression of DS is heterogeneous, larger numbers of patients could further strengthen the differences between the groups. Second, we did not have the genetic–chromosomal data of our patients, since they were admitted to our tertiary hospital for heart surgery and regular follow-up was done elsewhere. Third, our patients were a few months old; however, sampling older patients may show more robust differences since premature aging was suggested as a possible mechanism for immune alterations. Fourth, we focused on TCRγ repertoire in the thymus, and NGS of TCRs α, β, and δ in the thymus and blood could further expand on the comprehensive understanding of the TCR repertoire in DS. Fifth, spectratyping and DNA analysis may represent sequences that would not be ultimately translated into proteins, albeit we included only productive rearrangements in the NGS analysis.
In summary, the analysis of TCR gamma repertoire in the thymus determined by NGS reveals that patients with DS have increased richness of the repertoire and diminished clonal expansion, although no significant differences in TRECs and spectratyping profile were observed. Furthermore, abnormalities in the adaptive immunity in the patients with DS are unlikely to be linked to VJ recombination process. Further investigations of the immune system in patients with DS, possibly with the technique of NGS for repertoire determination, may find further molecular alterations in their immunity.
References
Hobson-Rohrer, W. L. & Samson-Fang, L. Down syndrome. Pediatr. Rev. 34, 573–574 (2013).
Bishara, N. & Clericuzio, C. L. Common dysmorphic syndromes in the NICU. Neoreviews 9 e29–e38 (2007).
Murphy, M., Lempert, M. J. & Epstein, L. B. Decreased level of T cell receptor expression by Down syndrome (trisomy 21) thymocytes. Am. J. Med. Genet. 37, 234–237 (1990).
Nespoli, L., Burgio, G. R., Ugazio, A. G. & Maccario, R. Immunological features of Down’s syndrome: a review. J. Intellect. Disabil. Res. 37(Pt 6), 543–551 (1993).
Ugazio, A. G., Maccario, R., Notarangelo, L. D. & Burgio, G. R. Immunology of Down syndrome: a review. Am. J. Med. Genet. Suppl. 7, 204–212 (1990).
Cossarizza, A. et al. Precocious aging of the immune system in Down syndrome: alteration of B lymphocytes, T-lymphocyte subsets, and cells with natural killer markers. Am. J. Med. Genet. 37, 213–218 (2005).
Philip, R. et al. Abnormalities of the in vitro cellular and humoral responses to tetanus and influenza antigens with concomitant numerical alterations in lymphocyte subsets in Down syndrome (trisomy 21). J. Immunol. 136, 1661–1667 (1986).
de Hingh, Y. C. M. et al. Intrinsic abnormalities of lymphocyte counts in children with Down syndrome. J. Pediatr. 147, 744–747 (2005).
Li Volti, S. et al. Safety and effectiveness of an acellular pertussis vaccine in subjects with Down’s syndrome. Childs Nerv. Syst. 12, 100–102 (1996).
Levin, S. et al. Thymic deficiency in Down’s syndrome. Pediatrics 63, 80–87 (1979).
Larocca, L. M. et al. Morphological and immunohistochemical study of Down syndrome thymus. Am. J. Med. Genet. Suppl. 7, 225–230 (1990).
Marcovecchio, G. E. et al. Thymic epithelium abnormalities in DiGeorge and Down syndrome patients contribute to dysregulation in T cell development. Front. Immunol. 10, 447 (2019).
Lima, F. A. et al. Decreased AIRE expression and global thymic hypofunction in Down syndrome. J. Immunol. 187, 3422–3430 (2011).
Kusters, M. A. A., Verstegen, R. H. J., Gemen, E. F. A. & De Vries, E. Intrinsic defect of the immune system in children with Down syndrome: a review. Clin. Exp. Immunol. 156, 189–193 (2009).
Musiani, P. et al. Intrathymic deficient expansion of T cell precursors in Down syndrome. Am. J. Med. Genet. 37, 219–224 (1990).
Nikolich-Žugich, J., Slifka, M. K. & Messaoudi, I. The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol. 4, 123–132 (2004).
Prada, N. et al. Direct analysis of thymic function in children with Down’s syndrome. Immun. Ageing 2, 4 (2005).
Verstegen, R. H. J. et al. Impact of Down syndrome on the performance of neonatal screening assays for severe primary immunodeficiency diseases. J. Allergy Clin. Immunol. 133, 1208–1211 (2014).
Yu, X. et al. Human syndromes of immunodeficiency and dysregulation are characterized by distinct defects in T-cell receptor repertoire development. J. Allergy Clin. Immunol. 133, 1109–1115 (2014).
O’Connell, A. E. et al. Next generation sequencing reveals skewing of the T and B cell receptor repertoires in patients with Wiskott-Aldrich syndrome. Front. Immunol. 5, 340 (2014).
Ramesh, M., Simchoni, N., Hamm, D. & Cunningham-Rundles, C. High-throughput sequencing reveals an altered T cell repertoire in X-linked agammaglobulinemia. Clin. Immunol. 161, 190–196 (2015).
Henderson, L. A. et al. Next-generation sequencing reveals restriction and clonotypic expansion of Treg cells in juvenile idiopathic arthritis. Arthritis Rheumatol. 68, 1758–1768 (2016).
Thapa, D. R. et al. Longitudinal analysis of peripheral blood T cell receptor diversity in patients with systemic lupus erythematosus by next-generation sequencing. Arthritis Res. Ther. 17, 132 (2015).
Chapman, C. G. et al. Characterization of T-cell receptor repertoire in inflamed tissues of patients with Crohnʼs disease through deep sequencing. Inflamm. Bowel Dis. 22, 1275–1285 (2016).
Lev, A. et al. The kinetics of early T and B cell immune recovery after bone marrow transplantation in RAG-2-deficient SCID patients. PLoS ONE 7, e30494 (2012).
Amariglio, N. et al. Molecular assessment of thymus capabilities in the evaluation of T-cell immunodeficiency. Pediatr. Res. 67, 211–216 (2010).
Lockitch, G. et al. Age-related changes in humoral and cell- mediated immunity in Down syndrome children living at home. Pediatr. Res. 22, 536–540 (1987).
Noble, R. L. & Warren, R. P. Altered T-cell subsets and defective T-cell function in young children with Down syndrome (trisomy-21). Immunol. Invest. 16, 371–382 (1987).
Cuadrado, E. & Barrena, M. J. Immune dysfunction in Down’s syndrome: primary immune deficiency or early senescence of the immune system? Clin. Immunol. Immunopathol. 78, 209–214 (1996).
Kwan, A. et al. Newborn screening for severe combined immunodeficiency in 11 screening programs in the United States. JAMA 312, 729–738 (2014).
Machnes-Maayan, D. et al. Insight into normal thymic activity by assessment of peripheral blood samples. Immunol. Res. 61, 198–205 (2015).
Shearer, W. T. et al. Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J. Allergy Clin. Immunol. 112, 973–980 (2003).
Author information
Authors and Affiliations
Contributions
S.R.—laboratory work, acquisition of data, analysis and interpretation of data, and drafting the manuscript. A.L.—laboratory work, acquisition of data, and analysis and interpretation of data. Y.-N.L.—laboratory work, acquisition of data, analysis and interpretation of data, and revising the manuscript. D.M.-M.—conception and design, acquisition of data, analysis and interpretation of data. U.K., A.V., and D.M.—clinical input and revising the manuscript. R.S.—conception and design, analysis of data, and revising the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rabinowicz, S., Lev, A., Lee, Y.N. et al. Immune and TRG repertoire signature of the thymus in Down syndrome patients. Pediatr Res 89, 102–109 (2021). https://doi.org/10.1038/s41390-020-0857-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-020-0857-y