Abstract
Congenital anomalies cause ~7% of all neonatal deaths, many of which have no identified pathophysiological cause. Because accurate and robust laboratory tests are unavailable for most birth defects, physicians rely on imaging such as ultrasound and MRI. Biomarkers from human body fluids are considered a powerful diagnostic tool to assess human disease and health as it mirrors an individual’s condition. Minimally invasive ‘liquid biopsies’ from blood samples are highly valuable for diagnosis, prognosis, risk assessment, and treatment of many conditions. Recent large-scale analysis (‘omics’) have enabled researchers to identify novel biomarkers in different areas. To accurately facilitate the early detection of congenital anomalies, the identification of biomarkers from maternal plasma should be promoted. This approach will uncover new opportunities in prenatal diagnosing and likely lead to a better understanding of the pathogenesis of congenital anomalies.
Similar content being viewed by others
Introduction
Congenital malformations are a major challenge for modern health care involving gynecologists, obstetricians, geneticists, neonatologists, pediatricians and pediatric surgeons. These malformations also impose an emotional burden on the parents of these children. A recent study showed that an estimated 0.5 million children aged 0–59 months died from congenital anomalies in 2015; more than from malaria.1 In the US alone, congenital anomalies were among the leading causes of infant mortality in 2013—accounting for 20% of infant deaths.2 The most common of these birth defects are congenital heart defects (CHD) and neural tube defects (NTD).3
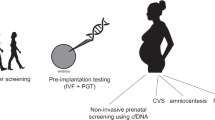
Early prenatal detection is essential to determine the prognosis and outcome of birth defects. Current diagnostic approaches predominantly rely on imaging methods, such as ultrasound (US) or fetal Magnetic Resonance Imaging (MRI). Despite improvements in prenatal imaging in regard to sensitivity and specificity, prenatal imaging is limited since the earliest possible timepoint for detection of fetal abnormalities with US is after the abnormalities are established in an irreversible state. Moreover, the sensitivity of prenatal ultrasound for some congenital malformations, like congenital diaphragmatic hernia (70%) or tracheoesophageal fistula (26%), is still unreliable.4,5 Other disadvantages of prenatal imaging are dependency on operator experience, or difficulty due to maternal obesity, or fetal position. Complementary invasive tests such as amniocentesis and chorionic villus sampling (CVS) are highly sensitive and specific for the diagnosis of chromosomal or genetic disorders of the fetus, but associated risks of miscarriage, infection, or amniotic fluid leakage are still of concern.6 Diagnostic measures using ‘liquid biopsies’ from blood, saliva, or urine to detect biomarkers are an emerging field in numerous areas, such as cardiovascular and cancer research.7,8
The most elegant approach to detect and predict outcomes of fetal malformations is to diagnose a fetal disease from maternal body fluids (Fig. 1). In contrast to imaging studies, the earliest possible timepoint for detection of e.g. circulating placental mRNA is around 4 weeks of gestation, before the embryo is fully developed.9 Such circulating molecules can be used as biomarkers for prenatal diagnosis at various stages of fetal development. They are defined as a “[…] characteristic that is measured as an indicator of normal biological processes, pathogenic processes, or a response to an exposure or intervention” (FDA-NIH: Biomarker-Working-Group, 2016).
A typical workflow for discovery and validation of biomarkers is exemplified in Fig. 2. First, a small sample size of cases and controls (n = 5) is used to profile all detectable markers from a class. A profile of up to 10,000 of molecules can be obtained from this analysis. As a second step, bioinformatics can identify the most important or significant molecules and their biological functions. Finally, during the third stage, several targets of interest have to be validated within a larger cohort (n = 50–100). Statistical analysis can narrow the number of targets to one or a small panel of sensitive and specific biomarkers that have to be further validated in large clinical studies.
The investigation of biomarkers for fetal anomalies from maternal plasma during pregnancy involves special challenges. As many of the fetal components are able to cross the placenta or be released directly from the placenta, it is imperative that the fetal contributions can be isolated and distinguished from the maternal biofluids. Furthermore, the biomarker profile is expected to change at each developmental stage with the maturation of different organ systems or the exposure of the fetus to environmental factors. As such, testing standardized timepoints during overlap of organ development should be considered to maximize return-of-investment.
Several classes of biomarkers are of interest: plasma proteins, circulating RNAs (microRNAs, circular RNAs, long non-coding RNAs) and circulating cell-free fetal DNA (Table 1). The discovery of novel biomarkers as a disease fingerprint can also provide insights into pathophysiological mechanisms of congenital malformations. This will aid translating knowledge from animal models to the human situation. Moreover, a cost-effective and simple detection method of congenital malformations using biomarkers can improve outcomes of congenital malformations in developing countries and those impacted by geography. Future technologies will enable point of care diagnostic devices to reliably and easily detect biomarkers from small samples of human body fluids. The aim of this review is to emphasize the importance of this approach and to give an overview of the pertinent molecules as a reflection of perinatal health as it relates to congenital malformations.
Proteins
Overview
The plasma proteome of humans is an invaluable resource to assess health and disease, as it mirrors an individual’s physiological status. Proteins represent a large portion of the human plasma and in clinical practice, the assessment of plasma proteins is routinely performed. The plasma concentration of CRP as an acute phase protein or the liver function tests including ALT and AST are good examples.10 Today, ~42% of all laboratory tests in clinical care are performed based on protein analysis.11 Novel proteomic approaches like liquid chromatography coupled with mass spectrometry are antibody independent, highly accurate and suitable for high throughput analysis of the human plasma proteome.12 This allows for dynamic testing from only 1 µL of plasma. In research, MS-based proteomics is revolutionary, allowing for the detection and quantification of thousands of proteins of interest within one sample.13 Geyer et al.11 proposed that >200 biomarkers can still be uncovered within human plasma proteins using this method.
Protein biomarkers in the context of congenital malformations
Proteins as biomarkers in prenatal diagnosis of congenital malformations are not sufficiently investigated and understood. One standard test is the measurement of alpha-fetoprotein (AFP) for the prenatal diagnosis of neural tube defects (NTD). Dashe et. al showed the importance of improving AFP analysis in pregnancies with NTDs as their sensitivity was 65% in their study cohort.14 In combination with standard US, detection rates can be improved to 86%. An et al.15 showed that PCSK9 could serve as a novel prenatal biomarker for NTDs from maternal serum. The authors report that PSCK9 is markedly decreased in the sera of NTD pregnancies. Additionally, the expression of PSCK9 expression of spinal cords in an animal model for NTDs was decreased. Another study presented a diagnostic model to prenatally diagnose NTDs with a panel of proteins via mass spectrometry with comparable accuracy to traditional clinical tests (sensitivity: 96%; specificity 90%).16
Differentially expressed maternal serum proteins can serve as biomarkers for different types of fetal and neonatal conditions. In one study, the sera of 370 women were analyzed with a proteomic approach and 4 cytoskeletal proteins were able to differentiate congenital heart defect cases from healthy fetuses with an area under the receiver operator characteristics curve (AUC) of 0.938.17 Others have identified PSG-5 and PSG-9 as potential biomarkers for early detection of pre-eclampsia from maternal plasma samples.18 The authors studied 3182 pregnant women at 15 weeks of gestation, of which 5.6% developed pre-eclampsia later during pregnancy. Menon et al.19 suggested salivary proteinase activity as a potential biomarker for preterm premature rupture of the membranes. They showed that pregnant women with premature rupture of the membranes had the highest activity of matrix metalloproteinase-9 compared to different control groups (non-pregnant, second trimester, active labor at term and postpartum). Patients at high risk for premature rupture of the membranes can benefit from the predictive potential of this method.
Only a few protein biomarkers for the prenatal detection of birth defects are described. The most prominent in clinical applications is the analysis of AFP for NTDs. The analysis of maternal plasma proteins holds great potential for the discovery of novel biomarkers in congenital malformations.
MicroRNAs
Overview
MicroRNAs (miR) are short RNA sequences approximately 22 nucleotides in length that regulate gene expression.20,21,22 MiRs bind messenger RNA and silence gene expression.20 Thus far, >1000 miRs capable of regulating the expression of hundreds of genes have been identified.23 This newly reported regulatory mechanism of gene expression influences the phenotype without altering the DNA sequence.23,24,25 The altered expression of specific miR signatures or microRNAomes has been linked to fetal development and the pathogenesis of congenital defects such as biliary atresia (miR-4429 and 4689), congenital diaphragmatic hernia (miR-200b), and congenital heart disease (miR-1 and 133a/b).23,26,27,28,29,30,31
From gametogenesis and implantation to embryonic development, the role of miRs has been established as molecular cues in the dynamics of pregnancy and fetal gestation.23,24,26,32,33,34 In murine models of embryo fertilization, Tan et al. showed that miR-199 was consistently downregulated as a result of in vitro fertilization compared to natural conception.35 This might explain the lower developmental potential, higher glycolic rates in blastocysts, and increased fetal losses associated with in vitro fertilization.35,36,37 On the other hand, changes in human miRs throughout development are also possible.26,27 Rodosthenous et al. demonstrated that elevated levels of miR-20b, -942, -324, -223, and -127 in maternal blood were associated with a lower likelihood of having smaller than average infants. Additionally, elevated levels of miR-661, -212, and -197 corresponded to larger than average infants.27 In silico pathway analyses of mir-197 demonstrated an association with various cancers, tumorigenesis, and neoplasms, however no fetal anomalies were reported. Carreras-Badosa et al. correlated the dysregulated profile of certain metabolic-associated placental miRs (miR-100, -1285, -296, and -487) found in pregnant obese women to low birth weight and increased postnatal weight gain in their fetuses.38
MicroRNAs in the context of congenital malformations
Similarly, a unique microRNA profile may also be used as biomarkers of congenital malformations. Congenital diseases such as congenital diaphragmatic hernia (CDH) and congenital heart disease (CHD) can be attributed to dysregulated miR expression.28,34,39,40 When Lim et al. compared 11 miR profiles in blood of non-pregnant women to women pregnant with normal fetuses and fetuses with trisomy 21, they found an elevated abundance of miR-3196 and 1973 to be associated with trisomy 21, or Down’s Syndrome.41 A constructed biological signaling network of 203 genes targeted by both miR-1973 and 3196 reported 72 genes associated with neuron projection, nervous system development, and sequence-specific DNA binding with a high confidence score of 0.7. These same miRs are associated with other congenital anomalies, mental disorders, and nervous system diseases (e.g. temporal lobe epilepsy and amyotrophic lateral sclerosis).41,42,43 We discovered that the development of pulmonary hypoplasia and pulmonary hypertension in CDH was linked to the downregulation of miR-200b in the nitrofen model of CDH corroborating with human patient data.30,31,44 More recently, Herrera-Rivero et al.45 found evidence that downregulated miR-let-7b/c, miR-1307, -185, -8084, -331, and -210 may be detrimental to lung and pulmonary vasculature development.
The cases presented identify that miR expression profiling reflects distinct changes influencing fetal development and the formation of congenital defects.32,33,46 The miR profile, obtained in the maternal blood, offers a minimally invasive method to assess the fetal condition. This unique signature is even capable of reflecting environmental exposures such as alcohol and toxic chemicals.47,48 Thus, in addition to being a diagnostic tool, the miR profile provides valuable insight into genes and molecular pathways involved in the disease pathogenesis. A miR profile can support the discovery of novel fetal therapeutics options to rescue abnormal fetal development.
Long non-coding RNAs
Overview
Long non-coding RNAs (lncRNAs) are DNA transcripts longer than ~200 nucleotides, which to date have no described protein-coding functions.49 It is recognized that lncRNAs are critical for signaling pathways and the modulation of gene expression. Hence, their dysregulation can contribute to disease phenotypes. High throughput RNA sequencing revealed that cell type specific expression patterns of lncRNAs control proliferation, differentiation, and cell death.50,51 Different roles in the regulation of gene expression are described for lncRNAs. They can modulate gene expression via binding to transcription factors, binding miRNAs, modulating splicing or effecting mRNA molecules directly.52 Interestingly, lncRNAs are differentially expressed during different stages of development and cell differentiation.53 Furthermore, studies have demonstrated that lncRNAs are stable in body fluids. They also provide a tissue specific expression pattern, which emphasizes their potential role as biomarkers.
LncRNAs are differentially expressed in human cancers and can promote metastasis and thus their role as biomarkers in several cancers has been described.54,55,56 The lncRNA HOTAIR, shows increased expression in primary breast tumors and metastasis. In contrast, suppression of HOTAIR can inhibit the invasiveness and metastasis of the same cancer.57 LncRNAs are also important in developmental processes and disease mechanisms. In this context, Szafranski et al.58 discussed that certain lung specific lncRNAs can interact with the promoter of FOXF1, a gene crucial for lung development. Disturbance in these molecular mechanisms can lead to Alveolar Capillary Dysplasia—an inherited developmental disease of the pulmonary vasculature.
Long non-coding RNAs in the context of congenital malformations
Gu et al.59 identified five lncRNAs (ENST00000436681, ENST00000422826, AA584040, AA709223, and BX478947) from maternal plasma, as potential novel biomarkers for the prenatal detection of fetal congenital heart defects. The authors analyzed the maternal plasma from 62 case-control pairs. After microarray analysis and validation via RT-qPCR, receiver operating characteristics (ROC) curve analysis showed an area under the curve (AUC) ranging from 0.755 to 0.892 for the target lncRNAs and confirmed their diagnostic value to identify congenital heart defects.59
LncRNAs have great potential as prenatal biomarkers for birth defects, but more studies are required to investigate lncRNAs in this context.
Circular RNAs
Overview
The investigation of circular RNAs (circRNA) as potential biomarkers is an emerging field in biomedical research.60 After their discovery 20 years ago, they were thought to be transcriptional ‘accidents’ not warranting further investigation. In the last decade, they have been studied extensively and associated with numerous diseases. CircRNAs are circularly shaped RNA particles generated by ligation of the ends of the pre-mRNA (‘back-splicing’).61 Their circular structure gives them an unusually high stability in biological matrices like plasma and saliva. Due to their lack of a poly-A tail, they are protected from digestion by ribonucleases. Their abundance is highest in the brain followed by plasma, where they have been shown to exist as free circulating circRNAs or encapsulated in exosomes.62,63 With up to hundreds of miRNA binding-sites, they can sequestrate miRNAs—acting as miRNA sponges.64 Interestingly, 10% of human circRNAs are tissue- and age-specific.65
CircRNAs have been associated with cancer, cardiovascular disease, osteoarthritis and diabetes mellitus.66,67,68,69 Circulating circRNA particles in plasma have been shown to be promising for biomarker discovery. Two of their major advantages for that purpose are their high stability and relative abundance in human plasma.60 Remarkably, circRNA expression patterns have been shown to be spatially and temporarily regulated and that they are involved in the development of organs and tissues.70,71 Conn et al.72 reported how circRNAs are involved and regulated during biological processes, such as epithelial-to-mesenchymal transition (EMT) an important process during organ development. They showed that the protein Quaking regulates circRNA formation during EMT. The relevance of circRNAs in abnormal fetal development has yet to be uncovered.
Circular RNAs in the context of congenital malformations
Recently, Peng et al.73 showed that circ-ZNF609 is downregulated in tissue samples from patients with Hirschsprung's disease (HD).73 Their ROC analysis showed an AUC of 0.86 to differentiate HD tissues from controls. Moreover, they show that circ-ZNF609 can sponge miR-150-5p and hence alter the expression of AKT3. They conclude that circ-ZNF609 can be involved in the onset of HD and lead to dysregulation of cell function and proliferation. Another group has recently found that embryonic heart tissues with a ventricular septal defect exhibit dysregulated circRNA profiles.74 The authors used 3 heart tissues from VSD patients and 3 control tissues without cardiac abnormality and performed a microarray analysis. They validated their findings using RT-qPCR in 12 case-control pairs of fetal heart tissue samples. This study emphasizes how circular RNA profiling in congenital heart defects can potentially lead to novel biomarker discovery.
Due to the inherent circRNAs stability and its role in cell differentiation, they qualify as potential biomarkers of congenital anomalies.
Circulating fetal DNA
Overview
Fetal cell-free DNA (cfDNA) or circulating fetal DNA was discovered 20 years ago when the Y chromosome was first isolated from the plasma of pregnant women carrying male fetuses.75 Released from the placenta into the maternal circulation during pregnancy, cfDNA originates from cytotrophoblast and syncytiotrophoblast fusion/apoptosis under physiologic conditions.76 Quantifiable as early as the 10th week of gestation, the ratio of placental to total (maternal and placental) cfDNA increases as pregnancy advances.77,78
Fetal cfDNA sequencing as a biomarker in maternal sera can provide clinical insight into the status of the fetus; aiding in disease management and counseling.79 Standardized protocols for aneuploidy confirmation: chorionic villus sampling, amniocentesis, ultrasonography, and maternal serum screening, carry low risk but possess technical limitations. Chorionic villus sampling, performed as early as 10 weeks of pregnancy, is able to detect aneuploidy from placental tissue. However, this technique has a longer learning curve and poses concerns about fetal damage and fetal infections.80 Amniocentesis can be performed at 9–11 weeks (early gestation) and 15 weeks or later (2nd trimester) is standard. Early sampling carries the risk of technical culture failure from a low extracted amniotic fluid volume, whereas sampling during the 2nd trimester may cause rupture of the membranes.80 Ultrasonography alone to survey increased nuchal translucency—the increased fluid accumulation in the fetal neck—and physical malformations does not constitute an accurate diagnosis of aneuploidy.81 Quadruple screening of maternal serum levels for: alpha fetoprotein, estriol, human chorionic gonadotropin, and inhibin are standardized to 15–22 weeks of gestation as values vary during gestation. Sequencing of fetal cfDNA offers an earlier timepoint assessment of the fetus with reduced risk and is being adopted for the most common trisomies13,18,21 and sex chromosome aneuploidies.79,82
Circulating fetal DNA in the context of congenital malformations
Non-invasive prenatal testing (NIPT) by sequencing cfDNA has garnered attention as of late 2017, with 4–6 million screens in pregnant women.78 Two-basic sequencing approaches are used to analyze circulating cfDNA. The whole-genome sequencing method randomly samples, sequences, and maps maternal and fetal cfDNA to specific chromosomes.77,78 The number of DNA molecules belonging to different chromosomes is counted and compared to a reference data set from pregnant women carrying euploid fetuses. The proportion of cfDNA molecules derived from the aneuploidic chromosome will be higher. The targeted method assesses if single-nucleotide polymorphisms occur on the chromosomes of interest. Ratios between heterozygous polymorphisms are compared with those of other targeted chromosomes. Skewed ratios corresponding to the aneuploidic chromosome is observed. A meta-analysis study by Taylor-Phillips et al. compared the standard ‘multiple marker’ prenatal screening consisting of serum biochemical assays and sonographic measurement of nuchal translucency to cfDNA sequencing.81,82 False positive rates associated with cfDNA for trisomies 21, 18, and 13 was <1/10 as high as multiple-marker screening and positive predictive values were significantly higher.82
The results of the fetal cfDNA sequencing are open to interpretation, as the fetal source is derived from the placenta.76 Placental mosaicism, maternal chromosomal abnormalities, and fetal demise may cause discordant NIPT results; leading to false positives.79,82,83 NIPT is a more robust and efficient method of global aneuploidic risk to concurrently determine uncommon fetal aneuploidies (e.x: sex chromosome aneuploidy in addition to trisomy) and maternal malignancies (e.x. malignant and benign tumors).79 CfDNA as a biomarker lends strength to make informed medical decisions without increased risk of miscarriage to pursue additional diagnostics, treatment planning, management strategies, and counseling.
Point-of-care devices
Point-of-care diagnostics (POCD) or point-of-care testing encompasses a variety of up-and-coming biosensors such as microfluidics or lab-on-a-chip to rapidly screen pertinent analytes.84,85 Applied within the context of fetal and perinatal development, this empowers physicians for quick decision making for intervention or disease management without the need for a clinical laboratory.84,86 The development of POCD is a joint venture between scientists, engineers, and clinicians leveraging the power of bioinformatics, machine learning, and biomedical engineering.87,88
First, a specific omics profile must be identified for each disease under study—matching genotype to phenotype. Large sets of patient omics (genomics, proteomics, etc.) data must be assessed using bioinformatics and machine learning, an application of artificial intelligence, to isolate disease specific biomarkers.87,88,89
Once a set of unique biomarkers has been identified, POCDs can be developed to identify the biomarkers in question within a biological sample. Biosensors such as microfluidics and lab-on-a chip provide high specificity analysis from small sample volumes with short turnaround times.84,85,90 Biosensors employing colorimetric, fluorescence, chemiluminescence, electrochemical, and label-free methods can measure the biomarkers in question.85,91 Furthermore, these technologies can be multiplexed, allowing for the simultaneous detection of multiple analytes in one sample run.91 Lafleur et al.90 provide a comprehensive review of biosensor applications and their limits of detection. Though the technology is still in its infancy, it has great potential and the possibility to be adapted for congenital and fetal diagnostics. As our knowledge of congenital biomarkers grows, it is probable for a microarray-like biosensor to be developed; allowing for the concurrent screening of various potential congenital anomalies. This could reverse the paradigm of diagnostic imaging followed by clinical confirmation to a POCD centered standard with subsequent diagnostic imaging.
Discussion
The ultimate goal of prenatal diagnosis of congenital malformations is to find accurate and minimally invasive methods to identify fetal anomalies during early pregnancy. Exciting novel applications, such as ‘omics’ approaches are likely to revolutionise pediatric medical practice and research. E.g. for bronchopulmonary dysplasia a combination of different ‘omics’ approaches are leveraged to uncover disease mechanisms and identify new biomarkers.92 Recently, Ngo et al.93 presented a method to predict gestational age and preterm birth using fetal cell-free RNA transcripts from maternal plasma and Liang et al.94 showed how artificial intelligence can diagnose a subset of pediatric diseases with equal accuracy compared to pediatricians. It will be interesting to see how novel biomarker discovery combined with deep machine learning can shape the future assessment of congenital malformations.
The search for additional biomarkers will no doubt increase the reliability of diagnostics, thus enabling more personalized prenatal therapeutic interventions in the future. Several obstacles have to be considered: heterogeneity between cases, identification of robust markers from a sample containing both fetal and maternal analytes, overlap between different diseases or the low prevalence of cases and thereby small sample sizes.87 From a prenatal fetal intervention perspective there are several criteria to assess: is the biomarker profile robust enough to distinguish one disease from another?; can we remedy the congenital anomaly?; and most importantly, will the fetus benefit from potential interventions?
We cannot assume any one specific biomarker or series of biomarkers within a category (e.g. microRNA or protein) is representative of a disease. As all biological processes are intricate and interwoven, specific biomarkers spanning several or all the biomarker categories may be required to categorize a congenital anomaly. As prenatal care standards are checkpoints for fetal development, they are not predictive of congenital anomalies. New predictive models and insights into biomarkers discovery will have to be tested in large and independent clinical cohorts to identify the unique disease biomarker fingerprint. Given a large enough cohort, biomarker screening can provide a predictive model for the incidence of congenital anomalies. Furthermore, it may be possible to determine and classify the severity of the disease according to the variations in the unique biomarker fingerprint. This would provide a basis for risk-benefit analyses for perinatal intervention where possible or management of postnatal care.
As an estimated 94% of all congenital anomalies occur in low and middle-income countries, there is a need for more economical diagnostic options.1 The discovery of biomarkers will facilitate improved prenatal diagnosing practices to alleviate financial strain and reduce expensive and invasive measures like fetal MRI, amniocentesis, and chorionic villus sampling. Better prenatal diagnosis can improve counselling with families due to more accurate risk assessments and better prediction of survival, especially for those restricted geographically.
The current technological advancement remains immature to adopt biomarkers universally—partially due to cost and limitations in computational systems biology.88 This is reflected in the cost to analyze a cohort large enough for significant conclusions and inferences. The biomarker fingerprint of a disease phenotype is intricate—regulated at multiple levels beyond the central dogma of DNA to mRNA to protein. Regulatory mechanisms such as microRNAs, circRNAs, lncRNAs, etc. add levels of complexity we have yet to understand completely. In anticipation of new technologies to improve analysis of biofluids (plasma, urine or saliva), the establishment of high-quality biobanks containing maternal biofluid specimens from cases and matched controls is crucial.95 Recruitment of cohorts large enough to infer statistical significance will require international collaborations to bring biomarkers to the forefront of medicine. This is especially true for rare congenital malformations.
The prospects of robust biomarkers combined with novel technologies will also enable the translation of science to the clinic. Machine learning is currently being implemented to determine trends within a disease cohort to assess severity.89,96 Point-of-care devices such as microarrays, lab-on-a-chip, and microfluidics may open new pathways for rapid screening in a clinical setting—providing shorter turn-around times.84,91
In this review, we emphasized the value and great potential of maternal biofluids for the early diagnosis of congenital defects and its potential to change our approach to fetal care. As our understanding of unique disease biomarker signatures deepens, more powerful and diverse biosensors can be widely adopted for point-of-care fetal screenings in clinics internationally, especially in rural settings. It is vital the molecular and clinical data generated be shared in a confidential and anonymous manner to improve the statistical interpretation and to refine severity classification. Such an initiative requires international collaborations to facilitate data accumulation. Ultimately, biomarker screening should be incorporated as standardized prenatal practice for congenital disease identification and management.
References
Wang, H. et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1459–1544 (2016).
Matthews, T. J., MacDorman, M. F. & Thoma, M. E. National vital statistics reports infant mortality statistics from the 2013 period linked birth / infant death data set. Natl Vital-. Stat. Rep. 64, 2000–2013 (2015).
Boyle, B. et al. Estimating Global Burden of Disease due to congenital anomaly: an analysis of European data. Arch. Dis. Child. Fetal Neonatal Ed. 103, 2016–311845 (2017).
Burgos C. M., et al. Prenatally versus postnatally diagnosed congenital diaphragmatic hernia – Side, stage, and outcome. J. Pediatr. Surg. 54, 651–655 (2018).
Bradshaw, C. J. et al. Accuracy of prenatal detection of tracheoesophageal fistula and oesophageal atresia. J. Pediatr. Surg. 51, 1268–1272 (2016).
Beta, J., Lesmes-HereDia, C., Bedetti, C. & Akolekar, R. Risk of miscarriage following amniocentesis and chorionic villus sampling: A systematic review of the literature. Minerva Ginecol. 70, 215–219 (2018).
Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017).
Kroh, E. M., Parkin, R. K., Mitchell, P. S. & Tewari, M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50, 298–301 (2010).
Chiu R. W. K., et al. Time profile of appearance and disappearance of cir-culating placenta-derived mRNA in maternal plasma. Clin. Chem. 1–19 (2006). https://doi.org/10.1373/clinchem.2005.059774
Anderson, N. L. et al. The human plasma proteome. Mol. Cell. Proteom. 3, 311–326 (2004).
Geyer, P. E., Holdt, L. M., Teupser, D. & Mann, M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 12, 1–14 (2016).
Geyer, P. E. et al. Plasma proteome profiling to assess human health and disease. Cell Syst. 2, 185–195 (2016).
Aebersold, R. & Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016).
Dashe, J. S., Twickler, D. M., Santos-Ramos, R., McIntire, D. D. & Ramus, R. M. Alpha-fetoprotein detection of neural tube defects and the impact of standard ultrasound. Am. J. Obstet. Gynecol. 195, 1623–1628 (2006).
An, D. et al. Identification of PCSK9 as a novel serum biomarker for the prenatal diagnosis of neural tube defects using iTRAQ quantitative proteomics. Sci. Rep. 5, 17559 (2015).
Shen, G., He, P., Du, Y. & Zhang, S. Identification of biomarkers by proteomics for prenatal screening for neural tube defects. Tohoku J. Exp. Med. 238, 123–129 (2016).
Chen, L. et al. Comprehensive maternal serum proteomics identifies the cytoskeletal proteins as non-invasive biomarkers in prenatal diagnosis of congenital heart defects. Sci. Rep. 6, 19248 (2016).
Blankley, R. T. et al. A label-free selected reaction monitoring workflow identifies a subset of pregnancy specific glycoproteins as potential predictive markers of early-onset pre-eclampsia. Mol. Cell Proteom. 12, 3148–3159 (2013).
Menon, R., McIntyre, J. O., Matrisian, L. M. & Fortunato, S. J. Salivary proteinase activity: a potential biomarker for preterm premature rupture of the membranes. Am. J. Obstet. Gynecol. 194, 1609–1615 (2006).
Bartel, D. P. MicroRNA Target Recognition and Regulatory Functions. Cell 136, 215–233 (2009).
Sayed D., Abdellatif M. Micrornas in development and disease. Physio. Rev. 91, 827–887 (2011).
Khoshgoo, N., Kholdebarin, R., Iwasiow, B. M. & Keijzer, R. MicroRNAs and lung development. Pediatr. Pulmonol. 48, 317–323 (2013).
Sadovsky, Y., Mouillet, J. F., Ouyang, Y., Bayer, A. & Coyne, C. B. The function of trophomirs and other micrornas in the human placenta. Cold Spring Harb. Perspect. Med. 5, 1–16 (2015).
Gross, N., Kropp, J. & Khatib, H. MicroRNA signaling in embryo development. Biology 6, 34 (2017).
Gebert L. F. R., MacRae I. J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 20, 21–37 (2018).
Floris, I., Kraft, J. D. & Altosaar, I. Roles of microRNA across prenatal and postnatal periods. Int. J. Mol. Sci. 17, 1–12 (2016).
Rodosthenous, R. S. et al. Second trimester extracellular microRNAs in maternal blood and fetal growth: An exploratory study. Epigenetics 12, 804–810 (2017).
Smith, T., Rajakaruna, C., Caputo, M. & Emanueli, C. MicroRNAs in congenital heart disease. Micro. Congenit. Heart Dis. 3, 1–10 (2015).
Dong R., Shen Z., Zheng C., Chen G., Zheng S. Serum microRNA microarray analysis identifies miR-4429 and miR-4689 are potential diagnostic biomarkers for biliary atresia. Sci. Rep. 1–11 (2016). https://doi.org/10.1038/srep21084
Pereira-Terra, P. et al. Unique tracheal fluid MicroRNA signature predicts response to feto in patients with congenital diaphragmatic hernia. Ann. Surg. 262, 1130–1140 (2015).
Khoshgoo N., et al. Prenatal microRNA miR-200b therapy improves nitrofen-induced pulmonary hypoplasia associated with congenital diaphragmatic hernia. Ann. Surg. 1 (2017). https://doi.org/10.1097/SLA.0000000000002595
Tsochandaridis, M., Nasca, L., Toga, C. & Levy-Mozziconacci, A. Circulating MicroRNAs as clinical biomarkers in the predictions of pregnancy complications. Biomed. Res. Int. 2015, 294954 (2014).
Winger, E. E., Reed, J. L. & Ji, X. First-trimester maternal cell microRNA is a superior pregnancy marker to immunological testing for predicting adverse pregnancy outcome. J. Reprod. Immunol. 110, 22–35 (2015).
Xie, W. Q., Zhou, L., Chen, Y. & Bin, N. Circulating microRNAs as potential biomarkers for diagnosis of congenital heart defects. World J. Emerg. Med. 7, 85–89 (2017).
Tan, K. et al. Downregulation of miR-199a-5p disrupts the developmental potential of in vitro-fertilized mouse blastocysts. Biol. Reprod. 95, 54–54 (2016).
Lee, Y., Thouas, G. & Gardner, D. Developmental kinetics of cleavage stage mouse embryos are related to their subsequent carbohydrate and amino acid utilization at the blastocyst stage. Hum. Reprod. 30, 543–552 (2015).
Bay B., Lyngsø J., Hohwü L., Kesmodel U. S. Childhood growth of singletons conceived following in vitro fertilisation or intracytoplasmic sperm injection: a systematic review and meta-analysis. BJOG 126, 158–166 (2019).
Carreras-Badosa, G. et al. Dysregulation of placental miRNA in maternal obesity is associated with pre-and postnatal growth. J. Clin. Endocrinol. Metab. 102, 2584–2594 (2017).
Li, X. & Zhao, Z. MicroRNA biomarkers for early detection of embryonic malformations in pregnancy. J. Biomol. Res. Ther. 03, 10–12 (2014).
Song, Y. et al. Clinical significance of circulating microRNAs as markers in detecting and predicting congenital heart defects in children. J. Transl. Med. 16, 1–11 (2018).
Lim, J. H. et al. MicroRNAs as potential biomarkers for noninvasive detection of fetal trisomy 21. J. Assist. Reprod. Genet. 32, 827–837 (2015).
Kan, A. A. et al. Genome-wide microRNA profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cell. Mol. Life Sci. 69, 3127–3145 (2012).
Freischmidt, A. et al. Serum microRNAs in patients with genetic amyotrophic lateral sclerosis and pre-manifest mutation carriers. Brain 137, 2938–2950 (2014).
Khoshgoo, N. et al. MicroRNA-200b regulates distal airway development by maintaining epithelial integrity. Sci. Rep. 7, 1–12 (2017).
Herrera-Rivero, M. et al. Circulating microRNAs are associated with pulmonary hypertension and development of chronic lung disease in congenital diaphragmatic hernia OPEN. Sci. Rep. 8, 10735 (2018).
Kotlabova, K., Doucha, J. & Hromadnikova, I. Placental-specific microRNA in maternal circulation - identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J. Reprod. Immunol. 89, 185–191 (2011).
Kappil, M. & Chen, J. Environmental exposures in utero and microRNA. Curr. Opin. Pediatr. 26, 243–251 (2014).
Balaraman, S. et al. Maternal and neonatal plasma MicroRNA biomarkers for fetal alcohol exposure in an ovine model. Alcohol Clin. Exp. Res. 28, 1390–1400 (2014).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Geisler, S. & Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 14, 699–712 (2013).
Fatica, A. & Bozzoni, I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21 (2013).
Wang, K. C. & Chang, H. Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011).
Batista, P. J. & Chang, H. Y. Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307 (2013).
Gutschner, T., Hämmerle, M. & Diederichs, S. MALAT1 - A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 91, 791–801 (2013).
Yamada, A. et al. A RNA-Sequencing approach for the identification of novel long non-coding RNA biomarkers in colorectal cancer. Sci. Rep. 8, 2–11 (2018).
Tang, Q. et al. Three circulating long non-coding RNAs act as biomarkers for predicting NSCLC. Cell. Physiol. Biochem. 37, 1002–1009 (2015).
Gupta, R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010).
Szafranski, P. et al. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 23, 23–33 (2013).
Gu, M. et al. Circulating LncRNAs as novel, non-invasive biomarkers for prenatal detection of fetal congenital heart defects. Cell. Physiol. Biochem. 38, 1459–1471 (2016).
Abu, N. & Jamal, R. Circular RNAs as promising biomarkers: a mini-review. Front. Physiol. 7, 355 (2016).
Lasda, E. & Parker, R. Circular RNAs: diversity of form and function. RNA 20, 1829–1842 (2014).
Memczak S., Papavasileiou P., Peters O., Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE 10, 1–13 (2015).
Lasda E., Parker R. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circrna clearance. PLoS ONE 11, 1–11 (2016).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Xia, S. et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief. Bioinform. 18, 984–992 (2017).
Liu, Q. et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation. Sci. Rep. 6, 22572 (2016).
Li, P. et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin. Chim. Acta 444, 132–136 (2015).
Kristensen L. S., Hansen T. B., Venø M. T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37, 555–565 (2018).
Viereck, J. & Thum, T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ. Res. 120, 381–399 (2017).
Barrett, S. P. & Salzman, J. Circular RNAs: analysis, expression and potential functions. Development 143, 1838–1847 (2016).
Venø, M. T. et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 16, 245 (2015).
Conn, S. J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134 (2015).
Peng, L. et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprungs disease. Oncotarget 8, 808–818 (2017).
Liu, H. et al. Differential expression of CircRNAs in embryonic heart tissue associated with ventricular septal defect. Int. J. Med. Sci. 15, 703–712 (2018).
Lo, Y. et al. Presence of fetal DNA in maternal plasma and serum. Lancet 350, 485–487 (1997).
Taglauer, E. S., Bianchi, D. W. & Street, W. Review: Cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta 35, 1–13 (2014).
Fan, H. C. et al. Non-invasive prenatal measurement of the fetal genome. Nature 487, 320–324 (2012).
Bianchi, D. W. & Chiu, R. W. K. Sequencing of Circulating Cell-free DNA during Pregnancy. N. Engl. J. Med. 379, 464–473 (2018).
Snyder H. L., Curnow K. J., Bhatt S., Bianchi D. W. Follow-up of multiple aneuploidies and single monosomies detected by noninvasive prenatal testing: implications for management and counseling. Prenat. Diagn. 36, 203–209 (2016).
Polin R. A., Fox W. W., Abman S. H. Fetal and Neonatal Physiology (Elsevier, 2011).
Malone, F. D. et al. First-trimester or second-trimester screening, or both, for Down’s Syndrome Fergal. N. Engl. J. Med. 353, 2001–2011 (2005).
Taylor-Phillips S., et al. Accuracy of non-invasive prenatal testing using cell-free DNA for detection of Down, Edwards and Patau syndromes: a systematic review and meta-analysis. Obstet. Gynecol. 6, 1–12 (2016).
McCullough R. M., et al. Non-invasive prenatal chromosomal aneuploidy testing - clinical experience: 100, 000 clinical samples. PLoS ONE 9, (2014). https://doi.org/10.1371/journal.pone.0109173.
Florkowski, C. et al. Critical Reviews in Clinical Laboratory Sciences Point-of-care testing (POCT) and evidence-based laboratory medicine (EBLM) – does it leverage any advantage in clinical decision making? Crit. Rev. Clin. Lab. Sci. 54, 471–494 (2017).
Vashist, S. K. Point-of-care diagnostics: recent advances and trends. Biosensors 7, 10–13 (2017).
National Institutes of Health. Point-of-Care Diagnostic Testing. 1–2 (2010). https://report.nih.gov/NIHfactsheets/ViewFactSheet.aspx?csid=112.
Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 18, 1–15 (2017).
Huang, S., Chaudhary, K. & Garmire, L. X. More is better: recent progress in multi-omics data integration methods. Front Genet. 8, 1–12 (2017).
Lin E., Lane H. Machine learning and systems genomics approaches for multi-omics data. Biomark. Res. 1–6 (2017). https://doi.org/10.1186/s40364-017-0082-y
Lafleur, J. P., Jönsson, A., Senkbeil, S. & Kutter, J. P. Recent advances in lab-on-a-chip for biosensing applications. Biosens. Bioelectron. 76, 213–233 (2016).
Dincer, C., Bruch, R., Kling, A., Dittrich, S. & Urban, G. A. Multiplexed point-of-care testing – xPOCT. Trends Biotechnol. 35, 728–742 (2017).
Lal, C. V., Bhandari, V. & Ambalavanan, N. Genomics, microbiomics, proteomics, and metabolomics in bronchopulmonary dysplasia. Semin. Perinatol. 42, 425–431 (2018).
Ngo, T. T. M. et al. Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science 360, 1133–1136 (2018).
Liang H., et al. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat. Med. (2019). https://doi.org/10.1038/s41591-018-0335-9
Choolani, M., Narasimhan, K., Kolla, V. & Hahn, S. Proteomic technologies for prenatal diagnostics: advances and challenges ahead. Expert Rev. Proteom. 6, 87–101 (2009).
Zeng, I. S. L. & Lumley, T. Review of statistical learning methods in integrated omics studies (An Integrated Information Science). Bioinform. Biol. Insights 12, 1–6 (2018).
Acknowledgements
We thank Clara Moy Tam for her help with the figures. R.K. holds the Thorlakson Chair in Surgical Research that supported this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wagner, R., Tse, W., Gosemann, JH. et al. Prenatal maternal biomarkers for the early diagnosis of congenital malformations: A review. Pediatr Res 86, 560–566 (2019). https://doi.org/10.1038/s41390-019-0429-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-019-0429-1
This article is cited by
-
Nanostructures in non-invasive prenatal genetic screening
Biomedical Engineering Letters (2022)