Abstract
Colon adenocarcinoma is the third most commonly diagnosed cancer and the second deadliest one. Metabolic reprogramming, described as an emerging hallmark of malignant cells, includes the predominant use of glycolysis to produce energy. Recent studies demonstrated that mitochondrial electron transport chain inhibitor reduced colon cancer tumour growth. Accumulating evidence show that myoferlin, a member of the ferlin family, is highly expressed in several cancer types, where it acts as a tumour promoter and participates in the metabolic rewiring towards oxidative metabolism. In this study, we showed that myoferlin expression in colon cancer lesions is associated with low patient survival and is higher than in non-tumoural adjacent tissue. Human colon cancer cells silenced for myoferlin exhibit a reduced oxidative phosphorylation activity associated with mitochondrial fission leading, ROS accumulation, decreased cell growth, and increased apoptosis. We observed the triggering of a DNA damage response culminating to a cell cycle arrest in wild-type p53 cells. The use of a p53 null cell line or a compound able to restore p53 activity (Prima-1) reverted the effects induced by myoferlin silencing, confirming the involvement of p53. The recent identification of a compound interacting with a myoferlin C2 domain and bearing anticancer potency identifies, together with our demonstration, this protein as a suitable new therapeutic target in colon cancer.
Similar content being viewed by others
Introduction
Despite an encouraging drop in incidence and mortality, colon cancer is the third most common diagnosed cancer independently of gender1. The last GLOBOCAN survey revealed that 10% of new cancer cases are colon cancer2. In contrast to what is observed in older people (>50 years), incidence and death rates among younger people continue to rise1, making this malignancy the second with the highest mortality rate2, and therefore encouraging us to sustain our research efforts.
Cancer cells require catabolites to produce energy and biomass. Metabolic reprogramming of cancer cells includes the predominant use of glycolysis to produce energy (Warburg effect)3. However, oxidative phosphorylation (OXPHOS) is also an essential part of their metabolism as it was described to support growth, invasiveness and confer resistance to chemotherapy in several cancer types, including colon4,5,6. Energy metabolism reprogramming, an emerging hallmark of cancer, is necessary for tumour initiation and progression7. As such, targeting mitochondrial metabolism appears as a sound potential approach8. Accordingly, a recent study demonstrated that a mitochondrial electron transport chain inhibitor (VLX600) reduced colon cancer tumour growth9.
Energy metabolism is not only driven by intracellular enzymes but is also conditioned by the intracellular availability of nutrients uptaken through specific transporters. Their abundance at plasma membrane is controlled by several steps, including exocytosis, endocytosis, and recycling. These processes require myoferlin, a 230-kDa multiple C2-domain ferlin family member protein, mainly known for its function in myoblast membrane fusion10,11. Previously, we have described the high expression of myoferlin in several cancers12,13 and its involvement in cancer cell plasma membrane biology such as endocytosis, membrane receptor recycling, exocytosis, and exosome formation13,14,15. In a metabolic context, we have reported myoferlin as a regulator of lipid metabolism and of mitochondrial dynamics16,17. However, its mechanism of action remains poorly understood and unexplored in colon cancer.
In the continuity of our previous studies aiming at showing the importance of myoferlin in cancer aggressiveness and inspired by the lack of information regarding its expression in colon cancer, we have sought to investigate this protein in this context. We have found out that myoferlin expression is highly expressed in colon cancer and correlates with patient survival. We also have showed that myoferlin is required to maintain a high OXPHOS activity and an organised mitochondrial network. We have discovered, for the first time, that myoferlin silencing produces a DNA damage response and a p53-dependent cell cycle arrest.
Results
High myoferlin expression in colon cancer lesions is associated with low survival
Puzzled by the lack of information regarding myoferlin expression in colon cancer, we decided to mine the PrognoScan databanks18 to evaluate the consequence of myoferlin expression on colon cancer patient overall and disease-specific survival. We found a highly significant Cox P values (respectively, P < 0.01 and P < 0.001) in the GSE17536 datasets (n = 177). This result was validated using the TCGA-COAD data using the OncoLnc tool19. Kaplan–Meier analysis indicated that myoferlin expression in the malignant colon lesions was significantly associated with disease-specific survival. Patients with a high myoferlin expression have a significantly shorter survival time than in patients with low myoferlin expression (n = 440, P = 0.01—Fig. 1a). The same association was recently shown in breast16, kidney20, and pancreas17 cancers. Using TCGA-COAD data, we evaluated myoferlin gene expression according to prognostic stage (Fig. 1b) and Tumour, Node, Metastasis (TNM) categories (Supplementary figure S1). Neither the prognostic stage nor any of the TNM categories were associated with the myoferlin gene expression. In order to determine whether myoferlin was an independent prognostic factor, we performed a multivariate Cox survival analysis with clinical covariates (Supplementary Table 1). Myoferlin highest percentile was a significant (P = 0.039) factor in the prediction of the disease-specific survival with a hazard ratio of 2.618.
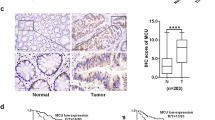
a TCGA-COAD data (http://cancergenome.nih.gov/) were analysed for disease-specific survival according to their myoferlin gene expression split into low (N = 220) and high (N = 220) according to the median. Kaplan–Meier curves were calculated for each group and a log-rank probability was calculated. b Myoferlin gene expression was analysed according to the prognostic stage. c Twenty-eight COAD sections were stained and scored for myoferlin (0—no staining, 1—faint staining, 2—medium staining, 3—strong staining). Scores were evaluated for tumour tissue and healthy adjacent
We then sought to evaluate myoferlin abundance in a collection of colon cancer specimens (n = 28). Myoferlin was strongly stained (mainly scores 2 and 3–76% of the cases) in the cytoplasm of cancer cells, whereas no or faintly detectable (mainly scores 0) in adjacent non-tumoural tissue (Fig. 1c).
Myoferlin is required to maintain a high OXPHOS activity and an organised mitochondrial network
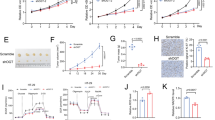
Recently, we have reported that myoferlin contributes to OXPHOS in pancreas cancer17. Accordingly, we next evaluated mitochondrial oxygen consumption rate (OCR) and mitochondrial network integrity after myoferlin silencing in colon cancer cell lines expressing highly this protein: HCT116 and SW480 (Supplementary Fig. 2A). We inhibited myoferlin translation using small interfering RNA (siRNA) and monitored the OCR after successive addition of oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), and rotenone/antimycin A mix. Myoferlin silencing significantly reduced the OCR after FCCP addition in HCT116 and SW480 cell lines (Fig. 2a). Analysis of respiration compartments revealed that maximal OCR was significantly reduced upon myoferlin silencing (Supplementary Figure S2A and S2G). In the HCT116 cell line, this reduction was already apparent in basal condition (Supplementary Figure S2B), while it was not the case in SW480 cell line (Supplementary Figure S2H). In both cell lines, myoferlin silencing led to a decrease of the metabolic potential (Supplementary Figure S2C and S2I). Because of this significant OCR reduction, we decided to evaluate the mitochondrial morphology (Fig. 2b). In HCT116 and SW480, the tetramethyl rhodamine ethyl ester (TMRE)-stained mitochondria appeared as a dense filamentous network. When myoferlin was silenced, the mitochondrial network disappeared and mitochondria turned into individual round-shape structures. Indeed, we observed a significant decrease of the number of networked mitochondria (Supplementary Figure S2D and S2J), of the mean mitochondrial network length (Supplementary Figure S2E and S2K), and of the ratio between the number of networked and individual mitochondria (Supplementary Figure S2F and S2L) when myoferlin was silenced. The mitochondrial network fragmentation induced by myoferlin silencing was concomitant to a dynamin-related protein (DRP)-1 phosphorylation on serine 616 (Fig. 2c). This phosphorylation is known to be implicated in mitochondrial fission21. These results suggest that myoferlin is needed to maintain a high and efficient OXPHOS activity in colon cancer cell lines.
a Kinetic OCR response of HCT116 and SW480 to oligomycin (O, 1 μM), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (F, 1.0 μM), rotenone, and antimycin A mix (RA, 0.5 μM each). Upon assay completion, cell number was evaluated using Hoechst incorporation (arbitrary unit, A.U.). b Tetramethyl rhodamine ethyl ester (TMRE) was used to stain mitochondria in HCT116 and SW480 living cells. At 48 h post-transfection, cells were seeded in μ-Slides 8-well at low confluence, then loaded with TMRE (1 nM). c Immunodetection of dynamin-related protein (DRP)-1 or phospho-DRP-1 in myoferlin-silenced HCT116 and SW480 cell lines. Total protein extracts (10 μg) were subjected to sodium dodecyl sulphate–polyacrylamide gel electrophoresis followed by western blot analysis with specific antibodies against myoferlin, DRP-1, or phospho-DRP-1 (ser616). HSC-70 was used as a loading control. One representative experiment out of three is illustrated. Each data point represents mean ± SD, n = 3. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05
Myoferlin silencing induces reactive oxygen species (ROS) accumulation, reduces cell growth, and induces apoptosis
We next explored the consequences of the mitochondrial alterations induced by myoferlin silencing in colon cancer cells. First, as mitochondria are the site of electron transport chain and as damaged mitochondria are considered a source of ROS, we measured ROS accumulation. In comparison to irrelevant siRNA, myoferlin silencing produced a 40% increase in mitochondrial ROS accumulation (Fig. 3a and Supplementary Figure S3). The reduced OCR and the accumulation of ROS could influence cell proliferation. Indeed, myoferlin silencing decreased sharply (~75%) the HCT116 cell growth and modestly but significantly (~30%) the SW480 one (Fig. 3b). Accordingly, myoferlin-silenced HCT116 cell produced significantly smaller tumours in a chorioallantoic in ovo model (Supplementary Figure S4). The reduced cell growth of HCT116 cell line was, at least partially, the result of an increased (more than two-fold) apoptosis rate (Fig. 3c). Intriguingly, in SW480 cell line, myoferlin silencing did not alter the percentage of apoptotic cells, remaining low (~5%) (Fig. 3c), indicating another mechanism explaining the low cell growth.
a Reactive oxygen species accumulation in HCT116 and SW480 cells after myoferlin silencing. At 48 h after myoferlin silencing, cells were harvested and loaded with CM-H2DCFDA (2 μM) for 15 min at 37 °C. Then fluorescence was measured by flow cytometry and analysed as median fluorescence intensity. b HCT116 and SW480 cell growth was assayed by Hoechst incorporation and indirect DNA quantification. Each data point represents mean ± SD, n = 3. c Percentage of apoptotic HCT116 or SW480 cells was measured by annexin V/propidium iodide flow cytometry. Insert represents the same data with a specific scale. ***P < 0.001, **P < 0.01, *P < 0.05
Myoferlin silencing activates p53 and provokes a cell cycle arrest in HCT116 cell line
Knowing the opposite p53 status of HCT116 (wild type) and of SW480 (p.R273H; p.P309S), we decided to investigate the role of p53 in the striking behaviour difference between the two cell lines upon myoferlin silencing. We first decided to evaluate the p53 abundance and its threonine 81 phosphorylation status in both cell line after myoferlin silencing (Fig. 4a). Thr81 phosphorylation was evaluated because it is known to be activated by c-Jun N-terminal kinase during ROS-induced DNA damage response22,23. In HCT116 cell line (wild-type p53), myoferlin silencing induced a two-fold increase of p53 abundance and a 2.4–4.7-fold increase of its phosphorylation on Thr81, culminating in a 2-fold increase of p21 abundance. Conversely, in SW480 cell line (mutated p53), myoferlin silencing did not alter the p53 abundance. In this cell line, p53 Thr81 phosphorylation was undetectable, as previously described24. Based on the p53 activation and p21 abundance increase, suggesting a cell cycle arrest, we decided to analyse the abundance of cyclins (Fig. 4b). In HCT116 cell line, myoferlin silencing provoked an accumulation in cyclin A (S-G2 cyclin) and B (mitotic cyclin). In SW480 cell line, no consistent cyclin modulation was observed after myoferlin depletion. In order to explore the cell cycle, HCT116 and SW480 cell lines were stained with propidium iodide and analysed by flow cytometry (Fig. 4c). It appeared that myoferlin silencing in HCT116 elicited an accumulation of cells in S phase, suggesting a blockage of the S to G2 transition.
a p53 activation by Thr81 phosphorylation and subsequent p21 abundance were evaluated in HCT116 and SW480 48 h after myoferlin silencing. b Cyclin abundance was evaluated by western blot in HCT116 and SW480 48 h after myoferlin silencing. Total protein extracts (10 μg) were subjected to sodium dodecyl sulphate–polyacrylamide gel electrophoresis followed by western blot analysis with specific antibodies. HSC-70 was used as a loading control. c Cell cycle was analysed by flow cytometry after propidium iodide incorporation in HCT116 and SW480 48 h after myoferlin silencing. Distribution of FL2 fluorescence (propidium iodide) is shown in HCT116. Proportion of cells in G1, S, or G2 is shown in HCT116 and SW480. One representative experiment out of three is illustrated. *P < 0.05
Myoferlin silencing induces a DNA damage response
The induced Thr81 phosphorylation of p53 in HCT116 upon myoferlin silencing prompted us to evaluate the cell DNA damage response. HCT116 and SW480 cell lines were immunostained for γH2Ax and observed under a confocal microscope. In both cell lines, γH2Ax foci appeared in the nucleus of myoferlin siRNA-transfected cells (Fig. 5a). Foci quantification indicated an increase of the foci number and of the foci size after myoferlin silencing in both cell lines (Fig. 5b). The accumulation of γH2Ax in transfected cells was also observed by western blot with a ten-fold accumulation in HCT116 and a two-fold accumulation in SW480 (Fig. 5c).
a HCT116 and SW480 cell lines, silenced for myoferlin during 48 h, were stained for γH2Ax and observed under a confocal microscope. b γH2Ax foci number and size were quantified using ImageJ. Number and size distributions were established (n > 210 nuclei). c γH2Ax abundance was evaluated by western blot in HCT116 and SW480 48 h after myoferlin silencing. Total protein extracts (10 μg) were subjected to sodium dodecyl sulphate–polyacrylamide gel electrophoresis followed by western blot analysis with specific antibodies. HSC-70 was used as a loading control
Myoferlin silencing induces a p53-dependent reduction of cell growth
In order to confirm the participation of p53 in the effects of myoferlin silencing, we took advantage of a HCT116 cell line deleted for p53 (HCT116 Δp53) and of SW480 cell treated with a p53-reactivating molecule (Prima-1). HCT116 Δp53 and Prima-1-treated SW480 were characterised for their myoferlin abundance and p53 and p21 status by western blot (Supplementary Figure S5). HCT116 Δp53 seemed to have slightly more myoferlin than the control HCT116 cell line. Myoferlin silencing in HCT116 Δp53 did not alter the percentage of apoptotic cells (Fig. 6a) but partly inhibited the HCT116 Δp53 cell growth (Fig. 6b). In SW480 treated with Prima-1, myoferlin silencing induced a slight but significant increase of the percentage of apoptotic cells (Fig. 6a). In these conditions, cell growth was strongly affected (Fig. 6b). These results clearly showed that p53 deletion from a p53 wild-type cell line (HCT116) or p53 chemical reactivation in a p53 mutant cell line (SW480) completely reversed the apoptotic (Fig. 7a) and growth phenotypes (Fig. 7b) observed after myoferlin silencing. Accordingly, it appeared to us that removing myoferlin should be more advantageous in p53 wild-type patients.
a Percentage of apoptotic HCT116 Δp53 cells or SW480 cells treated during 8 h with Prima-1 was measured by annexin V/propidium iodide flow cytometry 48 h after myoferlin silencing. b Cell growth of HCT116 Δp53 or SW480 treated during 8 h with Prima-1 was assayed by Hoechst incorporation and indirect DNA quantification 48 h after myoferlin silencing. Each data point represents mean ± SD, n = 3. ****P < 0.0001, **P < 0.01, *P < 0.05
a Comparison of apoptosis induction between HCT116 and HCT116 Δp53 cells and between SW480 and Prima-1 treated SW480 48 h after myoferlin silencing. Irrelevant siRNA was used as control (100%). b Comparison of 96 h cell growth between HCT116 and HCT116 Δp53 cells and between SW480 and Prima-1 treated SW480 after myoferlin silencing. Irrelevant siRNA was used as control (100%). ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05
Myoferlin expression is associated with p53 mutation status in colon cancer patients
Inspired by these results, we decided to go back to the TCGA-COAD dataset used initially and mined for an association between myoferlin expression and p53 mutation status. In the present dataset, patient survival was not associated with the p53 mutation status (Fig. 8a). However, when myoferlin expression (low vs high) was combined to the p53 mutation status (wild type vs mutated), a significant difference was found (Fig. 8b). The difference in survival remained statistically significant when myoferlin low–wild-type patients were compared to the myoferlin high–p53-mutated ones (Fig. 8c). Interestingly, myoferlin expression was significantly higher in p53 wild-type patients than in p53-mutated ones (Supplementary Figure S6).
a TCGA-COAD data were analysed for survival according to their p53 mutation status. b, c TCGA-COAD data were analysed for survival according to their p53 mutation status in combination with myoferlin expression. Kaplan–Meier curves were calculated for each group and a log-rank probability was calculated
Discussion
Most cancer cells have a glucose-oriented metabolism, preferring to use glycolysis to OXPHOS to grow. However, a successful growth requests a metabolic flexibility assured by the preservation of several functional catabolic or anaplerotic pathways. The discovery that resistance to treatment rely on OXPHOS in several cancer types5,25,26,27,28, including colon cancer5, recently drew our attention to the importance of identifying the elements rewiring metabolism towards OXPHOS. Targeting OXPHOS in cancer cells appears relevant8 as demonstrated by the use of VLX600 in colon cancer cell lines9. This compound is an iron chelator29 and inhibits respiratory complexes I, II, and IV9. Interestingly, due to its antitumour activity in human xenografts9,30, this compound was authorised in phase I in patients with refractory advanced solid tumours (clinical trials identifier NCT02222363).
Myoferlin is an emerging tumour-promoting protein described in several cancer types12,13,20,31,32,33,34,35,36, where it was often associated with a poor prognosis14,16,17,20,33,34,36. Interestingly, upon interleukin-6 stimulation myoferlin binds to the oncogenic protein signal transducer and activator of transcription factor 3 (STAT3) and participates in its nuclear translocation37. In colon cancer, STAT3 activation is associated with an increased proliferation rate, tumour growth, and a reduced survival38,39.
We have recently reported that myoferlin contributes to growth factor exocytosis14, exosome fusion to recipient cell15, lipid metabolism16, and mitochondrial fitness17. In the current study, we are investigating the involvement of this protein in colon cancer where it was only described at the exosome level40. Our results highlight for the first time an association between myoferlin expression and survival of colon cancer patients and show a consistent higher expression of this protein in tumour tissue in comparison with the non-tumoural adjacent one.
Myoferlin silencing in colon cancer cells reduces OXPHOS activity and disturbs the mitochondrial dynamics leading to mitochondrial fission. This process is mainly controlled by DRP-1 phosphorylation catalysed by several kinases, including the mitogen-activated protein kinase extracellular signal–regulated kinase 1/2 (ERK1/2)41. Interestingly, myoferlin depletion in hepatocellular carcinoma cells leads to ERK activation42 and a cleavable myoferlin is required for ERK phosphorylation in HEK293 cells43. We have previously reported this perturbation of the myoferlin-related mitochondrial dynamics in pancreas cancer17. However, unlike in pancreas, myoferlin silencing induced a low but consistent and significant accumulation of mitochondrial ROS. The cell-specific growth and the apoptosis observed after myoferlin silencing prompt us to consider their p53 mutation status.
Tumour-suppressor TP53 is the second most frequent mutated gene in colon cancer. TP53 mutations were occasionally described as being associated with low overall survival44,45. In our study, TCGA-COAD data did not associate TP53 mutation status to the patient survival confirming previously reported observation46. Despite, the lack of consistency in the relation between TP53 mutation and survival, several small molecules were designed for reactivation of p53. These promising compounds are currently tested in clinical trials for hematologic malignancies and suppresses colorectal cancer growth in xenograft mice47,48.
P53 protein is the core of a network of pathways in which ROS play critical roles. Upstream of p53, production of ROS is a fundamental mechanism of DNA damage and constitutes a trigger for p53 activation49. It appears that the wild-type p53 cells had their cell cycle stalled in S phase upon myoferlin silencing, whereas mutated p53 cells were not influenced. The reversion of the phenotype observed upon myoferlin depletion by modulating p53 activity or presence confirms the role of p53.
To our knowledge, the current study demonstrates for the first time that myoferlin silencing results in DNA damage probably due to ROS production by the fragmented mitochondria50. Interestingly, it has been previously shown that therapeutic agents generating ROS are more likely to be toxic for wild-type p53 tumour cells51. Accordingly, p53 reactivation increased the efficiency of myoferlin silencing on cell growth inhibition.
The cell growth arrest and the apoptosis generated by myoferlin silencing in p53 efficient cell line could open up new perspectives in the development of multi-modal therapies associating p53 and myoferlin targeting. In light of our results, about 75% of COAD patients exhibit a myoferlin higher expression at the protein level and, in consequence, can be considered as eligible to a myoferlin-targeted therapy. Considering a p53 mutation incidence of about 50%, half of the patients expressing myoferlin could benefit of the proposed multi-modal strategy. Currently, p53 restoration drugs are under clinical trials and C2-domain-targeting compounds are under development52. While we were preparing this manuscript, a study reported a small molecule targeting myoferlin with significant antitumour effect on breast cancer and on several other cancer cell types, including pancreas cancer, prostate cancer, and ovarian cancer53. This compound exerts promising antimetastatic activity at nanomolar concentration most probably by its interaction with the C2D domain of myoferlin. Together with our results, this discovery adds weight to the potential of myoferlin to be used as a relevant target for new anticancer therapies.
Materials and methods
Cells and chemicals
Human colon caner cells HCT116 (ATCC CCL-247) and SW480 cells (ATCC CCL-228) were purchased from ATCC. Cells were authenticated by Short-Tandem Repeat profiling (DSMZ, Braunschweig, Germany). All reagents were purchased from Sigma (Bornem, Belgium) unless mentioned otherwise. Antibodies were purchased from Sigma Life Sciences (Bornem, Belgium): myoferlin (HPA014245); Santa Cruz Biotechnology (Santa Cruz, CA): HSC70 (sc-7298), p21 (sc-6246), DRP-1 (sc-271583), Cyclin B1 (sc-752); Cell Signaling (Danvers, MA): phospho-DRP-1 (Ser616) (4494), phospho-p53 (Thr81) (2676), γH2A.X (9718), Cyclin D1 (2978); Merck-Millipore (Darmstadt, Germany): mitochondria (MAB1273), p53 clone BP53–12 (05–224); BD Biosciences: Cyclin A (611268), Cyclin E (551159).
Cell culture
HCT116 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with fetal bovine serum (10% FBS). SW480 cells were cultured in Minimum Essential Medium (MEM) supplemented with FBS (10%), L-glutamine (2 mM), sodium pyruvate (1 mM), and non-essential amino acids for MEM (Gibco #11140-085). Cells were cultured in a 37 °C, 5% CO2 incubator. Cells were recently authenticated and used between passage 1 and 10 and checked monthly for mycoplasma.
SiRNA transfection
HCT116 and SW480 cells were transfected with 40 nM siRNA, respectively, using calcium phosphate or lipofectamine as described previously17,54. All experiments were performed 48 h after transfection.
Western blotting
Cells were lysed in sodium dodecyl sulphate (SDS; 1%) in the presence of protease and phosphatase inhibitors. SDS-polyacrylamide gel electrophoresis were performed as described previously16. Band quantifications were performed with the Image Studio Lite software v5.2.5 (LI-COR Biosciences).
Extracellular flux analysis
All experiments were performed with a Seahorse XFp extracellular flux analyser (Agilent). Cells were seeded (20,000 cells/well) in XFp mini-plates (Agilent) and allowed to attach overnight. Mitochondrial OCR (pmoles/min) were measured as previously described17. Results were normalised according the cell number evaluated by Hoechst (2 µg/mL) incorporation. Results shown are representative ones out of three independent experiments.
Mitochondrial TMRE staining
TMRE was used to stain mitochondria in living cells as described previously17. Images were acquired by epifluorescence microscopy as Z-stacks with a Nikon A1R microscope equipped with ×100 Oil objective. Mitochondrial network was quantitatively assessed using image analyses with the MiNA Fiji plugin55. Results shown are representative ones out of three experiments.
Immunofluorescence
After siRNA transfection, 5 × 104 cells were seeded on sterile glass coverslips. Forty-eight hours after transfection, cells were washed, fixed, and then blocked as described17. Coverslips were incubated overnight at 4 °C with the primary antibody against γH2Ax diluted in phosphate-buffered saline–bovine serum albumin solution. Coverslips were washed and incubated with Alexafluor 546 conjugated secondary antibody for 1 h at room temperature. Sections were mounted following washes and nuclei staining with Hoechst (10 ng/mL). Z-stack images were acquired using a Nikon A1R confocal microscope equipped with a Nikon ×100 Oil objective. Results shown are representative ones out of three experiments.
Mitochondrial ROS measurement
Mitochondrial ROS production was measured by flow cytometry using Mitosox (Invitrogen) fluorescent probe according to the manufacturer’s protocol. Results shown are cumulative ones from three independent experiments.
Annexin V/propidium iodide staining
Percentage of apoptotic cells was assessed by flow cytometry using fluorescein isothiocyanate-annexin V and propidium iodide staining (BD Biosciences, Franklin Lakes, NJ) according to the manufacturer’s instructions. Results shown are cumulative ones from three independent experiments.
Cell cycle analysis
Cell were trypsinised, washed once with PBS and then fixed with ice-cold 70% ethanol for 4h at 4 °C. Fixed cells were washed once with PBS, then treated with RNAse (50µg/mL) and stained with propidium iodide (50 µg/mL) for 30 min at room temperature. Cells were analysed by flow cytometry with FACS calibur (BD Biosciences, Franklin Lakes, NJ). Results shown are cumulative ones from three independent experiments.
Cell growth
Equal number of cells were seeded in complete medium after transfection and harvested after 48 h. The cell numbers were indirectly determined using Hoechst incorporation. Results were expressed as DNA content. Results shown are representative ones out of three experiments.
Immunochemistry and staining assessment
Primary colon tumours were obtained from our institution Biobank, as formalin-fixed, paraffin-embedded tissue blocks. Sections were stained with antibodies against myoferlin. Sections were then reviewed and scored blindly by three independent investigators (G.R., M.H., and O.P.). Myoferlin scoring was performed by evaluating the intensity (ranging from 0 to 3) of each immunolabelled sample.
Chorioallantoic membrane assay (CAM)
HCT116 cells were grown on CAM for 7 days as previously mentioned14,56 and then tumour volumes have been calculated as an ellipsoid. Ten CAM were grafted in each experimental condition. Results shown are representative ones out of three experiments.
Statistics
Kaplan–Meier survival curves were established based on TCGA-COAD data. Survival curves were compared using the log-rank test. All other results are reported as means with standard deviation (SD). Two-sided statistical analyses were performed using one-way or two-way analysis of variance depending on the number of grouping factors. Unless mentioned otherwise, group means were compared by unpaired Student’s t test or Bonferroni’s post-test according to the group number. Welsch’s correction was applied when homoscedasticity was suspected. P < 0.05 was considered as statistically significant. Unlabelled differences between groups were non-significant. All experiments were performed as several independent biological replicates. Statistics were performed using R v3.457.
Change history
02 March 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41389-023-00455-5
References
Siegel, R. L. et al. Colorectal cancer statistics, 2017. CA Cancer J. Clin. 67, 177–193 (2017).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. https://doi.org/10.3322/caac.21492 (2018).
Koppenol, W. H., Bounds, P. L. & Dang, C. V. Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337 (2011).
Vellinga, T. T. et al. SIRT1/PGC1a-dependent increase in oxidative phosphorylation supports chemotherapy resistance of colon cancer. Clin. Cancer Res. 21, 2870–2879 (2015).
Denise, C. et al. 5-Fluorouracil resistant colon cancer cells are addicted to OXPHOS to survive and enhance stem-like traits. Oncotarget 6, 41706–41721 (2015).
Lin, C.-S. et al. Role of mitochondrial function in the invasiveness of human colon cancer cells. Oncol. Rep. 39, 316–330 (2018).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Weinberg, S. E. & Chandel, N. S. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 11, 9–15 (2015).
Zhang, X. et al. Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments. Nat. Commun. 5, 3295 (2014).
Doherty, K. R. et al. Normal myoblast fusion requires myoferlin. Development 132, 5565–5575 (2005).
Doherty, K. R. et al. The endocytic recycling protein EHD2 interacts with myoferlin to regulate myoblast fusion. J. Biol. Chem. 283, 20252–20260 (2008).
Turtoi, A. et al. Identification of novel accessible proteins bearing diagnostic and therapeutic potential in human pancreatic ductal adenocarcinoma. J. Proteome Res. 10, 4302–4313 (2011).
Turtoi, A. et al. Myoferlin is a key regulator of EGFR activity in breast cancer. Cancer Res. 73, 5438–5448 (2013).
Fahmy, K. et al. Myoferlin plays a key role in VEGFA secretion and impacts tumor-associated angiogenesis in human pancreas cancer. Int. J. Cancer 138, 652–663 (2016).
Blomme, A. et al. Myoferlin is a novel exosomal protein and functional regulator of cancer-derived exosomes. Oncotarget 7, 83669–83683 (2016).
Blomme, A. et al. Myoferlin regulates cellular lipid metabolism and promotes metastases in triple-negative breast cancer. Oncogene 4, 1151 (2016).
Rademaker, G. et al. Myoferlin controls mitochondrial structure and activity in pancreatic ductal adenocarcinoma, and affects tumor aggressiveness. Oncogene 66, 1–15 (2018).
Mizuno, H., Kitada, K., Nakai, K. & Sarai, A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genomics 2, 18 (2009).
Anaya, J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2, e67 (2016).
Song, D. H. et al. Prognostic role of myoferlin expression in patients with clear cell renal cell carcinoma. Oncotarget 8, 89033–89039 (2017).
Hoppins, S., Lackner, L. & Nunnari, J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76, 751–780 (2007).
Liu, Y. & Kulesz-Martin, M. p53 protein at the hub of cellular DNA damage response pathways through sequence-specific and non-sequence-specific DNA binding. Carcinogenesis 22, 851–860 (2001).
Buschmann, T. et al. Jun NH2-terminal kinase phosphorylation of p53 on Thr-81 is important for p53 stabilization and transcriptional activities in response to stress. Mol. Cell. Biol. 21, 2743–2754 (2001).
Minamoto, T. et al. Distinct pattern of p53 phosphorylation in human tumors. Oncogene 20, 3341–3347 (2001).
Ippolito, L. et al. Metabolic shift toward oxidative phosphorylation in docetaxel resistant prostate cancer cells. Oncotarget 7, 61890–61904 (2016).
Haq, R. et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell 23, 302–315 (2013).
Viale, A. et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632 (2014).
Boudreau, A. et al. Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition. Nat. Chem. Biol. 12, 779–786 (2016).
Fryknäs, M. et al. Iron chelators target both proliferating and quiescent cancer cells. Sci. Rep. 6, 38343 (2016).
Vitiello, G. A. et al. Mitochondrial inhibition augments the efficacy of imatinib by resetting the metabolic phenotype of gastrointestinal stromal tumor. Clin. Cancer Res. 24, 972–984 (2018).
Amatschek, S. et al. Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 64, 844–856 (2004).
McKinney, K. Q. et al. Discovery of putative pancreatic cancer biomarkers using subcellular proteomics. J. Proteomics 74, 79–88 (2011).
Wang, W. S. et al. ITRAQ-based quantitative proteomics reveals myoferlin as a novel prognostic predictor in pancreatic adenocarcinoma. J. Proteomics 91, 453–465 (2013).
Kumar, B. et al. High expression of myoferlin is associated with poor outcome in oropharyngeal squamous cell carcinoma patients and is inversely associated with HPV-status. Oncotarget 7, 18665–18677 (2016).
Kim, M. H. et al. Myoferlin expression and its correlation with FIGO histologic grading in early-stage endometrioid carcinoma. J. Pathol. Transl. Med. 52, 93–97 (2018).
Song, D. H. et al. Myoferlin expression in non-small cell lung cancer: Prognostic role and correlation with VEGFR-2 expression. Oncol. Lett. 11, 998–1006 (2016).
Yadav, A., Kumar, B., Lang, J. C., Teknos, T. N. & Kumar, P. A muscle-specific protein ‘myoferlin’ modulates IL-6/STAT3 signaling by chaperoning activated STAT3 to nucleus. Oncogene 36, 6374–6382 (2017).
Corvinus, F. M. et al. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia 7, 545–555 (2005).
Park, J. H. et al. Signal transduction and activator of transcription-3 (STAT3) in patients with colorectal cancer: associations with the phenotypic features of the tumor and host. Clin. Cancer Res. 23, 1698–1709 (2017).
Ji, H. et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics 13, 1672–1686 (2013).
Kashatus, J. A. et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 57, 537–551 (2015).
Hermanns, C. et al. The novel MKL target gene myoferlin modulates expansion and senescence of hepatocellular carcinoma. Oncogene 36, 3464–3476 (2017).
Piper, A.-K. et al. Enzymatic cleavage of myoferlin releases a dual C2-domain module linked to ERK signalling. Cell. Signal. 33, 30–40 (2017).
Hamelin, R. et al. Association of p53 mutations with short survival in colorectal cancer. Gastroenterology 106, 42–48 (1994).
Li, A.-J. et al. PIK3CA and TP53 mutations predict overall survival of stage II/III colorectal cancer patients. World J. Gastroenterol. 24, 631–640 (2018).
Warren, R. S. et al. Association of TP53 mutational status and gender with survival after adjuvant treatment for stage III colon cancer: results of CALGB 89803. Clin. Cancer Res. 19, 5777–5787 (2013).
Li, X.-L. et al. PRIMA-1met (APR-246) inhibits growth of colorectal cancer cells with different p53 status through distinct mechanisms. Oncotarget 6, 36689–36699 (2015).
Lu, T. et al. PRIMA-1Met suppresses colorectal cancer independent of p53 by targeting MEK. Oncotarget 7, 83017–83030 (2016).
Bykov, V. J. N., Lambert, J. M. R., Hainaut, P. & Wiman, K. G. Mutant p53 rescue and modulation of p53 redox state. Cell Cycle 8, 2509–2517 (2009).
Ježek, J., Cooper, K. F. & Strich, R. Reactive oxygen species and mitochondrial dynamics: the yin and yang of mitochondrial dysfunction and cancer progression. Antioxidants (Basel) 7, E13 (2018).
Macip, S. et al. Influence of induced reactive oxygen species in p53-mediated cell fate decisions. Mol. Cell. Biol. 23, 8576–8585 (2003).
Nicolaes, G. A. F. et al. Rational design of small molecules targeting the C2 domain of coagulation factor VIII. Blood 123, 113–120 (2014).
Zhang, T. et al. A small molecule targeting myoferlin exerts promising anti-tumor effects on breast cancer. Nat. Commun. 9, 3726 (2018).
Peixoto, P. et al. HDAC5 is required for maintenance of pericentric heterochromatin, and controls cell-cycle progression and survival of human cancer cells. Cell Death Differ. 19, 1239–1252 (2012).
Valente, A. J., Maddalena, L. A., Robb, E. L., Moradi, F. & Stuart, J. A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 119, 315–326 (2017).
Peulen, O. J. et al. The anti-tumor effect of HDAC inhibition in a human pancreas cancer model is significantly improved by the simultaneous inhibition of cyclooxygenase 2. PLoS ONE 8, e75102 (2013).
R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria (2014). http://www.R-project.org/.
Acknowledgements
The authors acknowledge Dr Chantal Humblet and Mrs Alice Marquet and Hülya Kocadag (GIGA-histology platform, ULiège) and Dr Sandra Ormenese, Dr Jean-Jacques Goval, and Mr Raafat Stephan (GIGA-imaging and flow cytometry platforms, ULiège) for their respective experimental support. We thank Mrs Caroline Ziane for proofreading and English editing. We are also thankful to the institutional Biobank of the University Hospital Liege (Dr Stéphanie Gofflot) for providing patient material. The results shown in this work are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. A.B. is Research Director, Y.H. is Senior Research Associate, and M.H. is Research Associate (FNRS). B.C. and J.B. are supported by a FNRS Télévie grant. R.P. is student-researcher supported by the ULiège Medicine Faculty. The sponsors had no other roles in the study.
Author information
Authors and Affiliations
Contributions
G.R., B.C., R.P., J.B., F.A., N.M., and O.P performed experiments and acquired data. O.P., G.R., V.C., and A.B. analysed data. O.P., G.R., and M.H. reviewed and scored immunostained sections. P.D. and Y.H. gave access to needed equipment, materials, or expertise. O.P., G.R., and V.C. conceived and designed experiments. O.P. performed statistical analysis and wrote the manuscript. All the authors proofread the manuscript, contributed to the redaction of their specific part, and approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
All human sections were used with the agreement of the Human Ethic Committee of the University of Liège and from the University Hospital (approval #2014–103). According to Belgian law, patients were informed that the residual surgical material could be used for research purpose and the consent is presumed as long as the patient does not oppose.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rademaker, G., Costanza, B., Bellier, J. et al. Human colon cancer cells highly express myoferlin to maintain a fit mitochondrial network and escape p53-driven apoptosis. Oncogenesis 8, 21 (2019). https://doi.org/10.1038/s41389-019-0130-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41389-019-0130-6
This article is cited by
-
Paladin, overexpressed in colon cancer, is required for actin polymerisation and liver metastasis dissemination
Oncogenesis (2022)
-
Prognostic value of a novel glycolysis-related gene expression signature for gastrointestinal cancer in the Asian population
Cancer Cell International (2021)
-
Lysosomal retargeting of Myoferlin mitigates membrane stress to enable pancreatic cancer growth
Nature Cell Biology (2021)
-
BRAFV600E drives dedifferentiation in small intestinal and colonic organoids and cooperates with mutant p53 and Apc loss in transformation
Oncogene (2020)