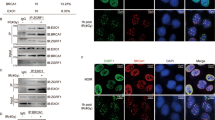
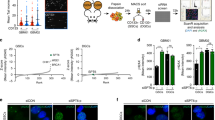
Abstract
HJURP is overexpressed in several cancer types and strongly correlates with patient survival. However, the mechanistic basis underlying the association of HJURP with cancer aggressiveness is not well understood. HJURP promotes the loading of the histone H3 variant, CENP-A, at the centromeric chromatin, epigenetically defining the centromeres and supporting proper chromosome segregation. In addition, HJURP is associated with DNA repair but its function in this process is still scarcely explored. Here, we demonstrate that HJURP is recruited to DSBs through a mechanism requiring chromatin PARylation and promotes epigenetic alterations that favor the execution of DNA repair. Incorporation of HJURP at DSBs promotes turnover of H3K9me3 and HP1, facilitating DNA damage signaling and DSB repair. Moreover, HJURP overexpression in glioma cell lines also affected global structure of heterochromatin independently of DNA damage induction, promoting genome-wide reorganization and assisting DNA damage response. HJURP overexpression therefore extensively alters DNA damage signaling and DSB repair, and also increases radioresistance of glioma cells. Importantly, HJURP expression levels in tumors are also associated with poor response of patients to radiation. Thus, our results enlarge the understanding of HJURP involvement in DNA repair and highlight it as a promising target for the development of adjuvant therapies that sensitize tumor cells to irradiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, VV (valeria.valente@unesp.br).
Materials availability
All genetic modified cell lines generated in this study are available from the lead contact.
Code availability
The standardized datasets produced in this manuscript were deposited in the Bioproject (NCBI). Access code is PRJNA855124. The code developed for the analysis of ChIPseq data can be accessed in the link: https://github.com/arnopenzias/ChIP-seq.git.
References
Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–97.
Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–84.
Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, et al. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–43.
Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–21.
Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–92.
Muller S, Montes de Oca R, Lacoste N, Dingli F, Loew D, Almouzni G. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3(CENP-A) loading. Cell Rep. 2014;8:190–203.
Miell MD, Straight AF. Regulating the timing of CENP-A nucleosome assembly by phosphorylation. Dev Cell. 2015;32:1–2.
Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, et al. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev Cell. 2015;32:68–81.
Barnhart-Dailey MC, Trivedi P, Stukenberg PT, Foltz DR. HJURP interaction with the condensin II complex during G1 promotes CENP-A deposition. Mol Biol Cell. 2017;28:54–64.
Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–42.
Maiato H, DeLuca J, Salmon ED, Earnshaw WC. The dynamic kinetochore-microtubule interface. J Cell Sci. 2004;117:5461–77.
Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–31.
Roulland Y, Ouararhni K, Naidenov M, Ramos L, Shuaib M, Syed SH, et al. The flexible ends of CENP-A nucleosome are required for mitotic fidelity. Mol Cell. 2016;63:674–85.
Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, et al. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–53.
Valente V, Teixeira SA, Neder L, Okamoto OK, Oba-Shinjo SM, Marie SK, et al. Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. BMC Mol Biol. 2009;10:17.
Valente V, Serafim RB, de Oliveira LC, Adorni FS, Torrieri R, Tirapelli DP, et al. Modulation of HJURP (Holliday Junction-Recognizing Protein) levels is correlated with glioblastoma cells survival. PLoS ONE. 2013;8:e62200.
Hu Z, Huang G, Sadanandam A, Gu S, Lenburg ME, Pai M, et al. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010;12:R18.
Li L, Li X, Meng Q, Khan AQ, Chen X. Increased expression of Holliday Junction-Recognizing Protein (HJURP) as an independent prognostic biomarker in advanced-stage serous ovarian carcinoma. Med Sci Monit. 2018;24:3050–5.
Wang CJ, Li X, Shi P, Ding HY, Liu YP, Li T, et al. Holliday junction recognition protein promotes pancreatic cancer growth and metastasis via modulation of the MDM2/p53 signaling. Cell Death Dis. 2020;11:386.
Chen T, Zhou L, Zhou Y, Zhou W, Huang H, Yin S, et al. HJURP promotes epithelial-to-mesenchymal transition via upregulating SPHK1 in hepatocellular carcinoma. Int J Biol Sci. 2019;15:1139–47.
Wei Y, Ouyang GL, Yao WX, Zhu YJ, Li X, Huang LX, et al. Knockdown of HJURP inhibits non-small cell lung cancer cell proliferation, migration, and invasion by repressing Wnt/beta-catenin signaling. Eur Rev Med Pharm Sci. 2019;23:3847–56.
Chen T, Huang H, Zhou Y, Geng L, Shen T, Yin S, et al. HJURP promotes hepatocellular carcinoma proliferation by destabilizing p21 via the MAPK/ERK1/2 and AKT/GSK3beta signaling pathways. J Exp Clin Cancer Res. 2018;37:193.
Kang DH, Woo J, Kim H, Kim SY, Ji S, Jaygal G, et al. Prognostic relevance of HJURP expression in patients with surgically resected colorectal cancer. Int J Mol Sci. 2020;21:7928.
Mao M, Jia Y, Chen Y, Yang J, Xu L, Zhang X, et al. HJURP regulates cell proliferation and chemo-resistance via YAP1/NDRG1 transcriptional axis in triple-negative breast cancer. Cell Death Dis. 2022;13:396.
Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK, et al. Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18:1061–83.
Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–93.
Noch EK, Ramakrishna R, Magge R. Challenges in the treatment of glioblastoma: multisystem mechanisms of therapeutic resistance. World Neurosurg. 2018;116:505–17.
Haar CP, Hebbar P, Wallace GC 4th, Das A, Vandergrift WA 3rd, Smith JA, et al. Drug resistance in glioblastoma: a mini review. Neurochem Res. 2012;37:1192–200.
Osuka S, Van Meir EG. Overcoming therapeutic resistance in glioblastoma: the way forward. J Clin Invest. 2017;127:415–26.
Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl J Med. 2005;352:997–1003.
Fabian D, Guillermo Prieto Eibl MDP, Alnahhas I, Sebastian N, Giglio P, Puduvalli V, et al. Treatment of glioblastoma (GBM) with the addition of tumor-treating fields (TTF): a review. Cancers. 2019;11:174.
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73.
Dou Z, Qiu C, Zhang X, Yao S, Zhao C, Wang Z, et al. HJURP promotes malignant progression and mediates sensitivity to cisplatin and WEE1-inhibitor in serous ovarian cancer. Int J Biol Sci. 2022;18:1188–210.
Yilmaz D, Furst A, Meaburn K, Lezaja A, Wen Y, Altmeyer M, et al. Activation of homologous recombination in G1 preserves centromeric integrity. Nature. 2021;600:748–53.
Kauffmann A, Rosselli F, Lazar V, Winnepenninckx V, Mansuet-Lupo A, Dessen P, et al. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27:565–73.
O’Grady S, Finn SP, Cuffe S, Richard DJ, O’Byrne KJ, Barr MP. The role of DNA repair pathways in cisplatin resistant lung cancer. Cancer Treat Rev. 2014;40:1161–70.
Atkins RJ, Ng W, Stylli SS, Hovens CM, Kaye AH. Repair mechanisms help glioblastoma resist treatment. J Clin Neurosci. 2015;22:14–20.
King HO, Brend T, Payne HL, Wright A, Ward TA, Patel K, et al. RAD51 Is a selective DNA repair target to radiosensitize glioma stem cells. Stem Cell Rep. 2017;8:125–39.
Carruthers RD, Ahmed SU, Ramachandran S, Strathdee K, Kurian KM, Hedley A, et al. Replication stress drives constitutive activation of the DNA damage response and radioresistance in glioblastoma stem-like cells. Cancer Res. 2018;78:5060–71.
Qi H, Price BD, Day TA. Multiple roles for mono- and poly(ADP-Ribose) in regulating stress responses. Trends Genet. 2019;35:159–72.
Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, et al. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191:31–43.
Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell. 2012;48:723–33.
Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JY, Berns MW, et al. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci USA. 2009;106:15762–7.
Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, et al. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev Cell. 2012;22:749–62.
Lans H, Marteijn JA, Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin. 2012;5:4.
Cheblal A, Challa K, Seeber A, Shimada K, Yoshida H, Ferreira HC, et al. DNA damage-induced nucleosome depletion enhances homology search independently of local break movement. Mol Cell. 2020;80:311–26.e4.
Min S, Ji JH, Heo Y, Cho H. Transcriptional regulation and chromatin dynamics at DNA double-strand breaks. Exp Mol Med. 2022;54:1705–12.
Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci USA. 2014;111:9169–74.
Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, et al. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep. 2014;8:1049–62.
Lee YH, Kuo CY, Stark JM, Shih HM, Ann DK. HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response. Nucleic Acids Res. 2013;41:5784–98.
Wu W, Nishikawa H, Fukuda T, Vittal V, Asano M, Miyoshi Y, et al. Interaction of BARD1 and HP1 Is Required for BRCA1 Retention at Sites of DNA Damage. Cancer Res. 2015;75:1311–21.
Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–77.
Goodarzi AA, Noon AT, Jeggo PA. The impact of heterochromatin on DSB repair. Biochem Soc Trans. 2009;37:569–76.
Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–7.
Gonzalez-Perez A, Sabarinathan R, Lopez-Bigas N. Local determinants of the mutational landscape of the human genome. Cell. 2019;177:101–14.
Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–18.
Goodarzi AA, Jeggo PA. The heterochromatic barrier to DNA double strand break repair: how to get the entry visa. Int J Mol Sci. 2012;13:11844–60.
Karagiannis TC, Harikrishnan KN, El-Osta A. Disparity of histone deacetylase inhibition on repair of radiation-induced DNA damage on euchromatin and constitutive heterochromatin compartments. Oncogene. 2007;26:3963–71.
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77.
Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–44.
Li L, Yuan Q, Chu YM, Jiang HY, Zhao JH, Su Q, et al. Advances in holliday junction recognition protein (HJURP): Structure, molecular functions, and roles in cancer. Front Cell Dev Biol. 2023;11:1106638.
Moynahan ME. The cancer connection: BRCA1 and BRCA2 tumor suppression in mice and humans. Oncogene. 2002;21:8994–9007.
Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–82.
Silver DP, Livingston DM. Mechanisms of BRCA1 tumor suppression. Cancer Discov. 2012;2:679–84.
Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–44.
Park SH, Kim JJ, Chung JS, Lee SR, Lee GY, Kim HJ, et al. RASSF1A suppresses the activated K-Ras-induced oxidative DNA damage. Biochem Biophys Res Commun. 2011;408:149–53.
Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci USA. 2011;108:3336–41.
Lopez-Contreras AJ, Gutierrez-Martinez P, Specks J, Rodrigo-Perez S, Fernandez-Capetillo O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J Exp Med. 2012;209:455–61.
Tho LM, Libertini S, Rampling R, Sansom O, Gillespie DA. Chk1 is essential for chemical carcinogen-induced mouse skin tumorigenesis. Oncogene. 2012;31:1366–75.
Agnihotri S, Gajadhar AS, Ternamian C, Gorlia T, Diefes KL, Mischel PS, et al. Alkylpurine-DNA-N-glycosylase confers resistance to temozolomide in xenograft models of glioblastoma multiforme and is associated with poor survival in patients. J Clin Invest. 2012;122:253–66.
de Sousa JF, Torrieri R, Serafim RB, Di Cristofaro LF, Escanfella FD, Ribeiro R, et al. Expression signatures of DNA repair genes correlate with survival prognosis of astrocytoma patients. Tumour Biol. 2017;39:1010428317694552.
Kun S, Duan Q, Liu G, Lu JM. Prognostic value of DNA repair genes based on stratification of glioblastomas. Oncotarget. 2017;8:58222–30.
Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, Foltz DR, et al. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 2011;7:e1002303.
Luijsterburg MS, de Krijger I, Wiegant WW, Shah RG, Smeenk G, de Groot AJL, et al. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol Cell. 2016;61:547–62.
Xu C, Xu Y, Gursoy-Yuzugullu O, Price BD. The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1. FEBS Lett. 2012;586:3920–5.
Tallis M, Morra R, Barkauskaite E, Ahel I. Poly(ADP-ribosyl)ation in regulation of chromatin structure and the DNA damage response. Chromosoma. 2014;123:79–90.
Ciccarone F, Zampieri M, Caiafa P. PARP1 orchestrates epigenetic events setting up chromatin domains. Semin Cell Dev Biol. 2017;63:123–34.
Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–6.
Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193:81–95.
Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–86.
Mathew V, Pauleau AL, Steffen N, Bergner A, Becker PB, Erhardt S. The histone-fold protein CHRAC14 influences chromatin composition in response to DNA damage. Cell Rep. 2014;7:321–30.
Hedouin S, Grillo G, Ivkovic I, Velasco G, Francastel C. CENP-A chromatin disassembly in stressed and senescent murine cells. Sci Rep. 2017;7:42520.
Mitrentsi I, Yilmaz D, Soutoglou E. How to maintain the genome in nuclear space. Curr Opin Cell Biol. 2020;64:58–66.
Ochs F, Karemore G, Miron E, Brown J, Sedlackova H, Rask MB, et al. Stabilization of chromatin topology safeguards genome integrity. Nature. 2019;574:571–4.
Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, et al. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–8.
Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–44.
Tsouroula K, Furst A, Rogier M, Heyer V, Maglott-Roth A, Ferrand A, et al. Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol Cell. 2016;63:293–305.
Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 2009;5:e1000435.
Lemaitre C, Grabarz A, Tsouroula K, Andronov L, Furst A, Pankotai T, et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 2014;28:2450–63.
Gursoy-Yuzugullu O, Ayrapetov MK, Price BD. Histone chaperone Anp32e removes H2A.Z from DNA double-strand breaks and promotes nucleosome reorganization and DNA repair. Proc Natl Acad Sci USA. 2015;112:7507–12.
Alatwi HE, Downs JA. Removal of H2A.Z by INO80 promotes homologous recombination. EMBO Rep. 2015;16:986–94.
Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–9.
Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–9.
Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA. 2010;107:18475–80.
Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–40.
Alagoz M, Katsuki Y, Ogiwara H, Ogi T, Shibata A, Kakarougkas A, et al. SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic Acids Res. 2015;43:7931–44.
Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–82.
Adkins NL, Niu H, Sung P, Peterson CL. Nucleosome dynamics regulates DNA processing. Nat Struct Mol Biol. 2013;20:836–42.
Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol. 2007;27:1602–13.
Strzyz PDN. A repair: Histones have got to go. Nat Rev Mol Cell Biol. 2017;18:138–9.
Hauer MH, Seeber A, Singh V, Thierry R, Sack R, Amitai A, et al. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat Struct Mol Biol. 2017;24:99–107.
Clouaire T, Legube G. A snapshot on the cis chromatin response to DNA double-strand breaks. Trends Genet. 2019;35:330–45.
Cejka P. DNA end resection: nucleases team up with the right partners to initiate homologous recombination. J Biol Chem. 2015;290:22931–8.
Gnugnoli M, Casari E, Longhese MP. The chromatin remodeler Chd1 supports MRX and Exo1 functions in resection of DNA double-strand breaks. PLoS Genet. 2021;17:e1009807.
Nair N, Shoaib M, Sorensen CS. Chromatin dynamics in genome stability: roles in suppressing endogenous DNA damage and facilitating DNA repair. Int J Mol Sci. 2017;18:1486.
Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–90.
Aleksandrov R, Dotchev A, Poser I, Krastev D, Georgiev G, Panova G, et al. Protein dynamics in complex DNA lesions. Mol Cell. 2018;69:1046–61.e5.
Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–6.
Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–84.
Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol. 2011;18:831–9.
Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506.
Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:366–74.
Athwal RK, Walkiewicz MP, Baek S, Fu S, Bui M, Camps J, et al. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenetics Chromatin. 2015;8:2.
Valdivia MM, Hamdouch K, Ortiz M, Astola A. CENPA a genomic marker for centromere activity and human diseases. Curr Genomics. 2009;10:326–35.
Zhang C, Han Y, Huang H, Min L, Qu L, Shou C. Integrated analysis of expression profiling data identifies three genes in correlation with poor prognosis of triple-negative breast cancer. Int J Oncol. 2014;44:2025–33.
Habel LA, Sakoda LC, Achacoso N, Ma XJ, Erlander MG, Sgroi DC, et al. HOXB13:IL17BR and molecular grade index and risk of breast cancer death among patients with lymph node-negative invasive disease. Breast Cancer Res. 2013;15:R24.
Rajput AB, Hu N, Varma S, Chen CH, Ding K, Park PC, et al. Immunohistochemical assessment of expression of centromere protein-A (CENPA) in human invasive breast cancer. Cancers. 2011;3:4212–27.
Montes de Oca R, Gurard-Levin ZA, Berger F, Rehman H, Martel E, Corpet A, et al. The histone chaperone HJURP is a new independent prognostic marker for luminal A breast carcinoma. Mol Oncol. 2015;9:657–74.
de Tayrac M, Aubry M, Saikali S, Etcheverry A, Surbled C, Guenot F, et al. A 4-gene signature associated with clinical outcome in high-grade gliomas. Clin Cancer Res. 2011;17:317–27.
Stangeland B, Mughal AA, Grieg Z, Sandberg CJ, Joel M, Nygard S, et al. Combined expressional analysis, bioinformatics and targeted proteomics identify new potential therapeutic targets in glioblastoma stem cells. Oncotarget. 2015;6:26192–215.
Serafim RB, da Silva P, Cardoso C, Di Cristofaro LFM, Netto RP, de Almeida R, et al. Expression profiling of glioblastoma cell lines reveals novel extracellular matrix-receptor genes correlated with the responsiveness of glioma patients to ionizing radiation. Front Oncol. 2021;11:668090.
Faraoni I, Graziani G. Role of BRCA mutations in cancer treatment with poly(ADP-ribose) polymerase (PARP) inhibitors. Cancers. 2018;10:487.
Sun C, Yin J, Fang Y, Chen J, Jeong KJ, Chen X, et al. BRD4 inhibition is synthetic lethal with PARP inhibitors through the induction of homologous recombination deficiency. Cancer Cell. 2018;33:401–16.e8.
Zhu H, Wei M, Xu J, Hua J, Liang C, Meng Q, et al. PARP inhibitors in pancreatic cancer: molecular mechanisms and clinical applications. Mol Cancer. 2020;19:49.
Pierce AJ, Jasin M. Measuring recombination proficiency in mouse embryonic stem cells. Methods Mol Biol. 2005;291:373–84.
Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
Shen L, Shao NY, Liu X, Maze I, Feng J, Nestler EJ. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE. 2013;8:e65598.
Steinhauser S, Kurzawa N, Eils R, Herrmann C. A comprehensive comparison of tools for differential ChIP-seq analysis. Brief Bioinform. 2016;17:953–66.
Acknowledgements
The authors thank Silvia Regina Andrade Nascimento and Chelsea Carman for technical assistance, Juliana Ferreira de Sousa for siHJURP#2 design, and Carlos Gilberto Carlotti, Elza Tiemi Sakamoto Hojo and Michael Bobola for gently providing cell lines. We also make a special acknowledgment to Mariana Santos de Queiroz for the helpful technical assistance in cloning experiments. RBS and GN were supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), and CC by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico).
Funding
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (grants #2013/13465-1, #2018/05018-9 and #2022/09066-3 to VV; grant #2013/08135-2 to WASJ; and grants #2014/18189-5 and #2021/10032-3 to EME), Programa de Apoio ao Desenvolvimento Científico (PADC) from the Faculty of Pharmaceutical Sciences of Araraquara - UNESP, Conselho Nacional de Desenvolvimento Científico e Tecnológico (#312249/2019-5 to EME) and the National Institutes of Health (NIH) (grants CA177804 and CA93602 to BDP). This study was also financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001.
Author information
Authors and Affiliations
Contributions
Investigation, RBS, CC, CBS, PS, HQ, RP and GN; Formal Analysis, RBS; Conceptualization, RBS, BDP and VV; Writing – original draft, RBS, BDP and VV; Writing – Review & Editing; BDP and VV; Funding Acquisition, BDP and VV; Supervision, BDP and VV; Resources, JPSP, WASJ, EME, MLPL, BDP and VV.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Serafim, R.B., Cardoso, C., Storti, C.B. et al. HJURP is recruited to double-strand break sites and facilitates DNA repair by promoting chromatin reorganization. Oncogene 43, 804–820 (2024). https://doi.org/10.1038/s41388-024-02937-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-024-02937-1