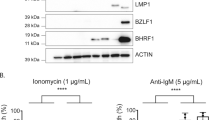
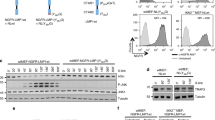
Abstract
MYC is an oncogenic transcription factor dysregulated in about half of total human tumors. While transcriptomic studies reveal more than 1000 genes regulated by MYC, a much smaller fraction of genes is directly transactivated by MYC. Virtually all Burkitt lymphoma (BL) carry chromosomal translocations involving MYC oncogene. Most endemic BL and a fraction of sporadic BL are associated with Epstein–Barr virus (EBV) infection. The currently accepted mechanism is that EBV is the BL-causing agent inducing MYC translocation. Herein we show that the EBV receptor, CR2 (also called CD21), is a direct MYC target gene. This is based on several pieces of evidence: MYC induces CR2 expression in both proliferating and arrested cells and in the absence of protein synthesis, binds the CR2 promoter and transactivates CR2 in an E-box-dependent manner. Moreover, using mice with conditional MYC ablation we show that MYC induces CR2 in primary B cells. Importantly, modulation of MYC levels directly correlates with EBV’s ability of infection in BL cells. Altogether, in contrast to the widely accepted hypothesis for the correlation between EBV and BL, we propose an alternative hypothesis in which MYC dysregulation could be the first event leading to the subsequent EBV infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The authors declare that all data that support the findings of this study are available within the paper and supplementary files.
References
Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35.
Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med. 2014;4:1–24.
Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–16.
Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA. 2006;103:17834–9.
Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–92.
Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79.
Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67.
Kress TR, Sabò A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015;15:593–607.
Kalkat M, De Melo J, Hickman KA, Lourenco C, Redel C, Resetca D, et al. MYC deregulation in primary human cancers. Genes (Basel). 2017;8:151.
Schaub FX, Dhankani V, Berger AC, Trivedi M, Richardson AB, Shaw R, et al. Pan-cancer alterations of the MYC oncogene and its proximal network across the Cancer Genome Atlas. Cell Syst. 2018;6:282–300.e2.
Delgado MD, Leon J. Myc roles in hematopoiesis and leukemia. Genes Cancer. 2010;1:605–16.
Ott G, Rosenwald A, Campo E. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Blood. 2013;122:3884–91.
Burkitt D. A sarcoma involving the jaws in African children. Br J Surg. 1958;46:218–23.
Molyneux EM, Rochford R, Griffin B, Newton R, Jackson G, Menon G, et al. Burkitt’s lymphoma. Lancet. 2012;379:1234–44.
Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–20.
Bisso A, Sabò A, Amati B. MYC in Germinal Center-derived lymphomas: mechanisms and therapeutic opportunities. Immunol Rev. 2019;288:178–97.
Methot SP, Di Noia JM. Molecular mechanisms of somatic hypermutation and class switch recombination. Adv Immunol. 2017;133:37–87.
Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–8.
Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–38.
Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61.
Lopez C, Kleinheinz K, Aukema SM, Rohde M, Bernhart SH, Hubschmann D, et al. Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma. Nat Commun. 2019;10:1459.
Farrell AS, Sears RC. MYC degradation. Cold Spring Harb Perspect Med. 2014;4:a014365.
Thorley-Lawson DA, Allday MJ. The curious case of the tumour virus: 50 years of Burkitt’s lymphoma. Nat Rev Microbiol. 2008;6:913–24.
Al-Khreisat MJ, Ismail NH, Tabnjh A, Hussain FA, Mohamed Yusoff AA, Johan MF, et al. Worldwide prevalence of Epstein-Barr virus in patients with Burkitt lymphoma: a systematic review and meta-analysis. Diagnostics (Basel). 2023;13:2068.
Dunmire SK, Verghese PS, Balfour HH Jr. Primary Epstein-Barr virus infection. J Clin Virol. 2018;102:84–92.
Young LS, Yap LF, Murray PG. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer. 2016;16:789–802.
Soldan SS, Lieberman PM. Epstein-Barr virus and multiple sclerosis. Nat Rev Microbiol. 2023;21:51–64.
Rowe M, Kelly GL, Bell AI, Rickinson AB. Burkitt’s lymphoma: the Rosetta Stone deciphering Epstein-Barr virus biology. Semin Cancer Biol. 2009;19:377–88.
Mancao C, Hammerschmidt W. Epstein-Barr virus latent membrane protein 2A is a B-cell receptor mimic and essential for B-cell survival. Blood. 2007;110:3715–21.
Kelly GL, Milner AE, Tierney RJ, Croom-Carter DS, Altmann M, Hammerschmidt W, et al. Epstein-Barr virus nuclear antigen 2 (EBNA2) gene deletion is consistently linked with EBNA3A, -3B, and -3C expression in Burkitt’s lymphoma cells and with increased resistance to apoptosis. J Virol. 2005;79:10709–17.
Wang LW, Shen H, Nobre L, Ersing I, Paulo JA, Trudeau S, et al. Epstein-Barr-virus-induced one-carbon metabolism drives B cell transformation. Cell Metab. 2019;30:539–55.e11.
Geser A, de The G, Lenoir G, Day NE, Williams EH. Final case reporting from the Ugandan prospective study of the relationship between EBV and Burkitt’s lymphoma. Int J Cancer. 1982;29:397–400.
IARC. Biological Agentes. A review of human carcinogenesis. Epstein-Barr virus. IARC Monographs. 2012;100B:49–92.
Sugden B. Epstein-Barr virus: the path from association to causality for a ubiquitous human pathogen. PLoS Biol. 2014;12:e1001939.
López C, Burkhardt B, Chan JKC, Leoncini L, Mbulaiteye SM, Ogwang MD, et al. Burkitt lymphoma. Nat Rev Dis Primers. 2022;8:78.
Allday MJ. How does Epstein-Barr virus (EBV) complement the activation of Myc in the pathogenesis of Burkitt’s lymphoma? Semin Cancer Biol. 2009;19:366–76.
Nemerow GR, Wolfert R, McNaughton ME, Cooper NR. Identification and characterization of the Epstein-Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2). J Virol. 1985;55:347–51.
Fearon DT, Carter RH. The CD19/CR2/TAPA-1 complex of B lymphocytes: linking natural to acquired immunity. Annu Rev Immunol. 1995;13:127–49.
Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–74.
Guo R, Jiang C, Zhang Y, Govande A, Trudeau SJ, Chen F, et al. MYC controls the Epstein-Barr virus lytic switch. Mol Cell. 2020;78:653–69.e8.
Gargouri B, Van Pelt J, El Feki Ael F, Attia H, Lassoued S. Induction of Epstein-Barr virus (EBV) lytic cycle in vitro causes oxidative stress in lymphoblastoid B cell lines. Mol Cell Biochem. 2009;324:55–63.
Acosta JC, Ferrandiz N, Bretones G, Torrano V, Blanco R, Richard C, et al. Myc inhibits p27-induced erythroid differentiation of leukemia cells by repressing erythroid master genes without reversing p27-mediated cell cycle arrest. Mol Cell Biol. 2008;28:7286–95.
Penn LJ, Brooks MW, Laufer EM, Land H. Negative autoregulation of c-myc transcription. Embo J. 1990;9:1113–21.
Delgado MD, Lerga A, Canelles M, Gomez-Casares MT, Leon J. Differential regulation of Max and role of c-Myc during erythroid and myelomonocytic differentiation of K562 cells. Oncogene. 1995;10:1659–65.
Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–63.
Schweighoffer E, Tybulewicz VL. Signalling for B cell survival. Curr Opin Cell Biol. 2018;51:8–14.
Ahearn JM, Fischer MB, Croix D, Goerg S, Ma M, Xia J, et al. Disruption of the Cr2 locus results in a reduction in B-1a cells and in an impaired B cell response to T-dependent antigen. Immunity. 1996;4:251–62.
Cherukuri A, Cheng PC, Pierce SK. The role of the CD19/CD21 complex in B cell processing and presentation of complement-tagged antigens. J Immunol. 2001;167:163–72.
Perez-Olivares M, Trento A, Rodriguez-Acebes S, Gonzalez-Acosta D, Fernandez-Antoran D, Roman-Garcia S, et al. Functional interplay between c-Myc and Max in B lymphocyte differentiation. EMBO Rep. 2018;19:e45770.
Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–29.
Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci USA. 2003;100:8164–9.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Vereshchagina LA, Tolnay M, Tsokos GC. Multiple transcription factors regulate the inducible expression of the human complement receptor 2 promoter. J Immunol. 2001;166:6156–63.
Ng HL, Taylor RL, Cheng J, Abraham LJ, Quail E, Cruickshank MN, et al. Notch signaling induces a transcriptionally permissive state at the Complement C3d Receptor 2 (CR2) promoter in a pre-B cell model. Mol Immunol. 2020;128:150–64.
Ulgiati D, Pham C, Holers VM. Functional analysis of the human complement receptor 2 (CR2/CD21) promoter: characterization of basal transcriptional mechanisms. J Immunol. 2002;168:6279–85.
Tolnay M, Vereshchagina LA, Tsokos GC. NF-kappaB regulates the expression of the human complement receptor 2 gene. J Immunol. 2002;169:6236–43.
Kanda K, Hu HM, Zhang L, Grandchamps J, Boxer LM. NF-kappa B activity is required for the deregulation of c-myc expression by the immunoglobulin heavy chain enhancer. J Biol Chem. 2000;275:32338–46.
Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–109.
Abate F, Ambrosio MR, Mundo L, Laginestra MA, Fuligni F, Rossi M, et al. Distinct viral and mutational spectrum of endemic Burkitt lymphoma. PLoS Pathog. 2015;11:e1005158.
Kaymaz Y, Oduor CI, Yu H, Otieno JA, Ong’echa JM, Moormann AM, et al. Comprehensive transcriptome and mutational profiling of endemic Burkitt lymphoma reveals EBV type-specific differences. Mol Cancer Res. 2017;15:563–76.
Grande BM, Gerhard DS, Jiang A, Griner NB, Abramson JS, Alexander TB, et al. Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood. 2019;133:1313–24.
Lin DC, Meng X, Hazawa M, Nagata Y, Varela AM, Xu L, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46:866–71.
Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–82.
Piris MA, Medeiros LJ, Chang KC. Hodgkin lymphoma: a review of pathological features and recent advances in pathogenesis. Pathology. 2020;52:154–65.
Price AM, Messinger JE, Luftig MA. c-Myc represses transcription of the Epstein-Barr virus latent membrane protein 1 early after primary B cell infection. J Virol. 2018;92:e01178-17.
Jochner N, Eick D, Zimber-Strobl U, Pawlita M, Bornkamm GW, Kempkes B. Epstein-Barr virus nuclear antigen 2 is a transcriptional suppressor of the immunoglobulin mu gene: implications for the expression of the translocated c-myc gene in Burkitt’s lymphoma cells. Embo J. 1996;15:375–82.
Kaiser C, Laux G, Eick D, Jochner N, Bornkamm GW, Kempkes B. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol. 1999;73:4481–4.
Bajaj R, Xu F, Xiang B, Wilcox K, Diadamo AJ, Kumar R, et al. Evidence-based genomic diagnosis characterized chromosomal and cryptic imbalances in 30 elderly patients with myelodysplastic syndrome and acute myeloid leukemia. Mol Cytogenet. 2011;4:3.
Kalchschmidt JS, Bashford-Rogers R, Paschos K, Gillman AC, Styles CT, Kellam P, et al. Epstein-Barr virus nuclear protein EBNA3C directly induces expression of AID and somatic mutations in B cells. J Exp Med. 2016;213:921–8.
Garcia-Gutierrez L, Bretones G, Molina E, Arechaga I, Symonds C, Acosta JC, et al. Myc stimulates cell cycle progression through the activation of Cdk1 and phosphorylation of p27. Sci Rep. 2019;9:18693.
Barreto V, Reina-San-Martin B, Ramiro AR, McBride KM, Nussenzweig MC. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol Cell. 2003;12:501–8.
Acknowledgements
The work was supported by grants PID2020-115903GB-100 to JL and MDD, RTI2018-095673-B-I00 to JRR, PID2019-107551RB-I00 to VG-Y, PID2020-119567RB-I00 to RM, PID2020-117539GB-I00 to IV and PID2019-106773RB-I00 to ARR, all funded by MCIN/AEI/10.13039/501100011033/, Spanish Government, and by “FEDER, Una manera de hacer Europa”, European Union, and by La Caixa HR17-0244 grant to AR. LQ and VJ were recipients of F.P.U. program and Universidad de Cantabria fellowships, respectively. LG-G was a fellow of the Maria Zambrano program, Spanish Government. We are grateful to Maria Aramburu and Patricia Arribas for technical help, Santiago Montes for useful comments and Victor Campa por assistance in microscopy and image processing.
Author information
Authors and Affiliations
Contributions
EM, LG-G: formal analysis, investigation, writing. VJ, MP-O, VG-Y, RB, AVM, JCA and LQ: formal analysis, investigation, resources. DU, RM, JRR, and IV: formal analysis and resources. IMA, AR: formal analysis, resources, editing. MDD: formal analysis, funding. JL: experimental design, formal analysis, writing, funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Molina, E., García-Gutiérrez, L., Junco, V. et al. MYC directly transactivates CR2/CD21, the receptor of the Epstein–Barr virus, enhancing the viral infection of Burkitt lymphoma cells. Oncogene 42, 3358–3370 (2023). https://doi.org/10.1038/s41388-023-02846-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02846-9