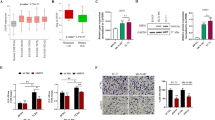
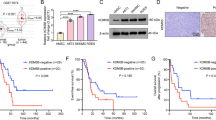
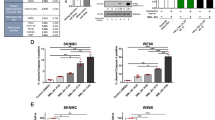
Abstract
EWS/ETS fusion transcription factors, most commonly EWSR1::FLI1, drives initiation and progression of Ewing sarcoma (EwS). Even though direct targeting EWSR1::FLI1 is a formidable challenge, epigenetic/transcriptional modulators have been proved to be promising therapeutic targets for indirectly disrupting its expression and/or function. Here, we identified structure-specific recognition protein 1 (SSRP1), a subunit of the Facilitates Chromatin Transcription (FACT) complex, to be an essential tumor-dependent gene directly induced by EWSR1::FLI1 in EwS. The FACT-targeted drug CBL0137 exhibits potent therapeutic efficacy against multiple EwS preclinical models both in vitro and in vivo. Mechanistically, SSRP1 and EWSR1::FLI1 form oncogenic positive feedback loop via mutual transcriptional regulation and activation, and cooperatively promote cell cycle/DNA replication process and IGF1R-PI3K-AKT-mTOR pathway to drive EwS oncogenesis. The FACT inhibitor drug CBL0137 effectively targets the EWSR1::FLI1-FACT circuit, resulting in transcriptional disruption of EWSR1::FLI1, SSRP1 and their downstream effector oncogenic signatures. Our study illustrates a crucial role of the FACT complex in facilitating the expression and function of EWSR1::FLI1 and demonstrates FACT inhibition as a novel and effective epigenetic/transcriptional-targeted therapeutic strategy against EwS, providing preclinical support for adding EwS to CBL0137’s future clinical trials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNAseq raw data are accessible at the NCBI Gene Expression Omnibus (GEO) accession code GSE195803 and GSE195804. ChIPseq raw data are accessible at the accession code GSE195802.
References
Grunewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Alava E, Kovar H, et al. Ewing sarcoma. Nat Rev Dis Prim. 2018;4:5.
Riggi N, Suva ML, Stamenkovic I. Ewing’s sarcoma. N. Engl J Med. 2021;384:154–64.
Gangwal K, Sankar S, Hollenhorst PC, Kinsey M, Haroldsen SC, Shah AA, et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc Natl Acad Sci USA. 2008;105:10149–54.
Boulay G, Volorio A, Iyer S, Broye LC, Stamenkovic I, Riggi N, et al. Epigenome editing of microsatellite repeats defines tumor-specific enhancer functions and dependencies. Genes Dev. 2018;32:1008–19.
Musa J, Cidre-Aranaz F, Aynaud MM, Orth MF, Knott MML, Mirabeau O, et al. Cooperation of cancer drivers with regulatory germline variants shapes clinical outcomes. Nat Commun. 2019;10:4128.
Tomazou EM, Sheffield NC, Schmidl C, Schuster M, Schonegger A, Datlinger P, et al. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep. 2015;10:1082–95.
Janknecht R. EWS-ETS oncoproteins: the linchpins of Ewing tumors. Gene 2005;363:1–14.
Tanaka K, Iwakuma T, Harimaya K, Sato H, Iwamoto Y. EWS-Fli1 antisense oligodeoxynucleotide inhibits proliferation of human Ewing’s sarcoma and primitive neuroectodermal tumor cells. J Clin Invest. 1997;99:239–47.
Ouchida M, Ohno T, Fujimura Y, Rao VN, Reddy ES. Loss of tumorigenicity of Ewing’s sarcoma cells expressing antisense RNA to EWS-fusion transcripts. Oncogene 1995;11:1049–54.
Flores G, Grohar PJ. One oncogene, several vulnerabilities: EWS/FLI targeted therapies for Ewing sarcoma. J Bone Oncol. 2021;31:100404.
Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci USA. 2009;106:5324–9.
Gollavilli PN, Pawar A, Wilder-Romans K, Natesan R, Engelke CG, Dommeti VL, et al. EWS/ETS-driven Ewing Sarcoma requires BET Bromodomain proteins. Cancer Res. 2018;78:4760–73.
Lin L, Huang M, Shi X, Mayakonda A, Hu K, Jiang YY, et al. Super-enhancer-associated MEIS1 promotes transcriptional dysregulation in Ewing sarcoma in co-operation with EWS-FLI1. Nucleic Acids Res. 2019;47:1255–67.
Sanchez-Molina S, Figuerola-Bou E, Blanco E, Sanchez-Jimenez M, Taboas P, Gomez S, et al. RING1B recruits EWSR1-FLI1 and cooperates in the remodeling of chromatin necessary for Ewing sarcoma tumorigenesis. Sci Adv. 2020;6:eaba3058.
Schmidt O, Nehls N, Prexler C, von Heyking K, Groll T, Pardon K, et al. Class I histone deacetylases (HDAC) critically contribute to Ewing sarcoma pathogenesis. J Exp Clin Cancer Res. 2021;40:322.
Pishas KI, Drenberg CD, Taslim C, Theisen ER, Johnson KM, Saund RS, et al. Therapeutic targeting of KDM1A/LSD1 in Ewing Sarcoma with SP-2509 engages the endoplasmic reticulum stress response. Mol Cancer Ther. 2018;17:1902–16.
Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 1999;400:284–8.
Formosa T, Winston F. The role of FACT in managing chromatin: disruption, assembly, or repair? Nucleic Acids Res. 2020;48:11929–41.
Kim M, Neznanov N, Wilfong CD, Fleyshman DI, Purmal AA, Haderski G, et al. Preclinical validation of a single-treatment infusion modality that can eradicate extremity melanomas. Cancer Res. 2016;76:6620–30.
Somers K, Kosciolek A, Bongers A, El-Ayoubi A, Karsa M, Mayoh C, et al. Potent antileukemic activity of curaxin CBL0137 against MLL-rearranged leukemia. Int J Cancer. 2019;146:1902–16.
Barone TA, Burkhart CA, Safina A, Haderski G, Gurova KV, Purmal AA, et al. Anticancer drug candidate CBL0137, which inhibits histone chaperone FACT, is efficacious in preclinical orthotopic models of temozolomide-responsive and -resistant glioblastoma. Neuro Oncol. 2017;19:186–96.
Carter DR, Murray J, Cheung BB, Gamble L, Koach J, Tsang J, et al. Therapeutic targeting of the MYC signal by inhibition of histone chaperone FACT in neuroblastoma. Sci Transl Med. 2015;7:312ra176.
Mo J, Liu F, Sun X, Huang H, Tan K, Zhao X, et al. Inhibition of the FACT complex targets aberrant hedgehog signaling and overcomes resistance to smoothened antagonists. Cancer Res. 2021;81:3105–20.
Wang J, Sui Y, Li Q, Zhao Y, Dong X, Yang J, et al. Effective inhibition of MYC-amplified group 3 medulloblastoma by FACT-targeted curaxin drug CBL0137. Cell Death Dis. 2020;11:1029.
Savola S, Klami A, Myllykangas S, Manara C, Scotlandi K, Picci P, et al. High expression of Complement Component 5 (C5) at tumor site associates with superior survival in Ewing’s Sarcoma family of tumour patients. ISRN Oncol. 2011;2011:168712.
Postel-Vinay S, Veron AS, Tirode F, Pierron G, Reynaud S, Kovar H, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012;44:323–7.
Scotlandi K, Remondini D, Castellani G, Manara MC, Nardi F, Cantiani L, et al. Overcoming resistance to conventional drugs in Ewing sarcoma and identification of molecular predictors of outcome. J Clin Oncol. 2009;27:2209–16.
Liu D, Sartor MA, Nader GA, Pistilli EE, Tanton L, Lilly C, et al. Microarray analysis reveals novel features of the muscle aging process in men and women. J Gerontol A Biol Sci Med Sci. 2013;68:1035–44.
Bilke S, Schwentner R, Yang F, Kauer M, Jug G, Walker RL, et al. Oncogenic ETS fusions deregulate E2F3 target genes in Ewing sarcoma and prostate cancer. Genome Res. 2013;23:1797–809.
Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suva ML, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;26:668–81.
Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–84.
Wang S, Hwang EE, Guha R, O’Neill AF, Melong N, Veinotte CJ, et al. High-throughput chemical screening identifies focal adhesion Kinase and Aurora Kinase B Inhibition as a synergistic treatment combination in Ewing Sarcoma. Clin Cancer Res. 2019;25:4552–66.
Wakahara K, Ohno T, Kimura M, Masuda T, Nozawa S, Dohjima T, et al. EWS-Fli1 up-regulates expression of the Aurora A and Aurora B kinases. Mol Cancer Res. 2008;6:1937–45.
Chang HW, Valieva ME, Safina A, Chereji RV, Wang J, Kulaeva OI, et al. Mechanism of FACT removal from transcribed genes by anticancer drugs curaxins. Sci Adv. 2018;4:eaav2131.
Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods. 2016;13:521–7.
Hancock JD, Lessnick SL. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle. 2008;7:250–6.
Adane B, Alexe G, Seong BKA, Lu D, Hwang EE, Hnisz D, et al. STAG2 loss rewires oncogenic and developmental programs to promote metastasis in Ewing sarcoma. Cancer Cell. 2021;39:827–44.e10.
Sen N, Cross AM, Lorenzi PL, Khan J, Gryder BE, Kim S, et al. EWS-FLI1 reprograms the metabolism of Ewing sarcoma cells via positive regulation of glutamine import and serine-glycine biosynthesis. Mol Carcinog. 2018;57:1342–57.
Heitzeneder S, Sotillo E, Shern JF, Sindiri S, Xu P, Jones R, et al. Pregnancy-Associated Plasma Protein-A (PAPP-A) in Ewing Sarcoma: role in tumor growth and immune evasion. J Natl Cancer Inst. 2019;111:970–82.
Fukuma M, Okita H, Hata J, Umezawa A. Upregulation of Id2, an oncogenic helix-loop-helix protein, is mediated by the chimeric EWS/ets protein in Ewing sarcoma. Oncogene 2003;22:1–9.
Kirschner A, Thiede M, Grunewald TG, Alba Rubio R, Richter GH, Kirchner T, et al. Pappalysin-1 T cell receptor transgenic allo-restricted T cells kill Ewing sarcoma in vitro and in vivo. Oncoimmunology 2017;6:e1273301.
Ohmura S, Marchetto A, Orth MF, Li J, Jabar S, Ranft A, et al. Translational evidence for RRM2 as a prognostic biomarker and therapeutic target in Ewing sarcoma. Mol Cancer. 2021;20:97.
Goss KL, Gordon DJ. Gene expression signature based screening identifies ribonucleotide reductase as a candidate therapeutic target in Ewing sarcoma. Oncotarget 2016;7:63003–19.
Toretsky JA, Thakar M, Eskenazi AE, Frantz CN. Phosphoinositide 3-hydroxide kinase blockade enhances apoptosis in the Ewing’s sarcoma family of tumors. Cancer Res. 1999;59:5745–50.
Martins AS, Mackintosh C, Martin DH, Campos M, Hernandez T, Ordonez JL, et al. Insulin-like growth factor I receptor pathway inhibition by ADW742, alone or in combination with imatinib, doxorubicin, or vincristine, is a novel therapeutic approach in Ewing tumor. Clin Cancer Res. 2006;12(11 Pt 1):3532–40.
Amin HM, Morani AC, Daw NC, Lamhamedi-Cherradi SE, Subbiah V, Menegaz BA, et al. IGF-1R/mTOR Targeted Therapy for Ewing Sarcoma: A meta-analysis of five IGF-1R-related trials matched to proteomic and radiologic predictive biomarkers. Cancers. 2020;12:1768.
Lamhamedi-Cherradi SE, Menegaz BA, Ramamoorthy V, Vishwamitra D, Wang Y, Maywald RL, et al. IGF-1R and mTOR blockade: novel resistance mechanisms and synergistic drug combinations for Ewing Sarcoma. J Natl Cancer Inst. 2016;108:djw182.
Loganathan SN, Tang N, Holler AE, Wang N, Wang J. Targeting the IGF1R/PI3K/AKT pathway sensitizes Ewing Sarcoma to BET Bromodomain inhibitors. Mol Cancer Ther. 2019;18:929–36.
Xiao L, Somers K, Murray J, Pandher R, Karsa M, Ronca E, et al. Dual targeting of chromatin stability by the Curaxin CBL0137 and Histone Deacetylase Inhibitor Panobinostat shows significant preclinical efficacy in neuroblastoma. Clin Cancer Res. 2021;27:4338–52.
Song H, Xi S, Chen Y, Pramanik S, Zeng J, Roychoudhury S, et al. Histone chaperone FACT complex inhibitor CBL0137 interferes with DNA damage repair and enhances sensitivity of medulloblastoma to chemotherapy and radiation. Cancer Lett. 2021;520:201–12.
Seong BKA, Dharia NV, Lin S, Donovan KA, Chong S, Robichaud A, et al. TRIM8 modulates the EWS/FLI oncoprotein to promote survival in Ewing sarcoma. Cancer Cell. 2021;39:1262–78.e7.
Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12:828–63.
Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554.
Liu F, Jiang W, Sui Y, Meng W, Hou L, Li T, et al. CDK7 inhibition suppresses aberrant hedgehog pathway and overcomes resistance to smoothened antagonists. Proc Natl Acad Sci USA. 2019;116:12986–95.
Chiou YY, Hu J, Sancar A, Selby CP. RNA polymerase II is released from the DNA template during transcription-coupled repair in mammalian cells. J Biol Chem. 2018;293:2476–86.
Grunewald TG, Bernard V, Gilardi-Hebenstreit P, Raynal V, Surdez D, Aynaud MM, et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet. 2015;47:1073–8.
Mo JL, Liu Q, Kou ZW, Wu KW, Yang P, Chen XH, et al. MicroRNA-365 modulates astrocyte conversion into neuron in adult rat brain after stroke by targeting Pax6. Glia 2018;66:1346–62.
Surdez D, Zaidi S, Grossetete S, Laud-Duval K, Ferre AS, Mous L, et al. STAG2 mutations alter CTCF-anchored loop extrusion, reduce cis-regulatory interactions and EWSR1-FLI1 activity in Ewing sarcoma. Cancer Cell. 2021;39:810–26.e9.
Volchenboum SL, Andrade J, Huang L, Barkauskas DA, Krailo M, Womer RB, et al. Gene expression profiling of Ewing Sarcoma tumors reveals the prognostic importance of tumor-stromal interactions: a report from the Children’s Oncology Group. J Pathol Clin Res. 2015;1:83–94.
Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, et al. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics 2006;7:67–80.
Feng J, Meyer CA, Wang Q, Liu JS, Shirley Liu X, Zhang Y. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 2012;28:2782–8.
Tang Y, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med. 2014;20:732–40.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–5.
Acknowledgements
This work was supported by Chinese Universities Scientific Fund, Innovative Research Team of High-Level Local Universities in Shanghai (SHSMU-ZDCX20212700), National Natural Science Foundation of China (81772655, 81972646 to YT, 82002978 to JM), Shanghai Frontiers Science Center of Cellular Homeostasis and Human Diseases, the Innovation Program of Shanghai Municipal Education Commission (2019-01-07-00-01-E00036, China), the Recruitment Program of Global Experts of China (YT), Postdoctoral Science Foundation of China (2019M651527 to JM), and National Research Center for Translational Medicine, Ruijin Hospital affiliated to Shanghai Jiao Tong University School of Medicine (Open research program to YT). We thank Jing Xue (Shanghai Renji Hospital) and Bing Li (Shanghai Jiao Tong University School of Medicine) for reagents and/or helpful suggestions.
Author information
Authors and Affiliations
Contributions
JM: Conceptualization, project administration, funding acquisition, methodology, investigation, writing (original draft and review and editing) and visualization. KT: Investigation, writing (original draft and review and editing) and visualization. YD: Software, writing (original draft) and visualization. WL: Investigation and visualization. FL: Investigation. YM: Investigation. WL: Investigation. HH: Investigation. KZ: Investigation. ZL: Conceptualization, funding acquisition, resources, supervision. YY: Conceptualization, software, methodology and visualization. YT: Conceptualization, formal analysis, project administration, funding acquisition, resources, supervision, writing (original draft and review and editing) and visualization.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mo, J., Tan, K., Dong, Y. et al. Therapeutic targeting the oncogenic driver EWSR1::FLI1 in Ewing sarcoma through inhibition of the FACT complex. Oncogene 42, 11–25 (2023). https://doi.org/10.1038/s41388-022-02533-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02533-1
This article is cited by
-
CRISPR-Cas9 knockout screening identifies KIAA1429 as an essential gene in Ewing sarcoma
Journal of Experimental & Clinical Cancer Research (2023)