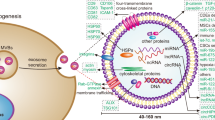
Abstract
Increasing studies have indicated that circular RNAs (circRNAs) play pivotal roles in various cancers. Here, we aimed to explore the roles of circRNAs in breast cancer. We identified a novel circRNA circKDM4B (hsa_circ_0002926) by whole-transcriptome sequencing and validated this by Real-time quantitative polymerase chain reaction (RT-qPCR) and Sanger sequencing. It was significantly decreased in breast cancer tissues compared with adjacent non-tumor tissues. Furthermore, circKDM4B, which is mainly localized in the cytoplasm, was more resistant to actinomycin D or ribonuclease R than its linear transcript KDM4B. In addition, the overexpression of circKDM4B inhibited cell migration and invasion in vitro, while knockdown of circKDM4B induced the opposite effects. In vivo, circKDM4B suppressed tumor growth and metastasis. Additionally, circKDM4B inhibited migration and tube formation of human umbilical vein endothelial cells (HUVECs) in vitro and angiogenesis in vivo. Mechanically, circKDM4B sponged miR-675 to upregulate the expression of NEDD4-like E3 ubiquitin protein ligase (NEDD4L), which catalyzes ubiquitination of PI3KCA, thereby inhibiting PI3K/AKT and VEGFA secretion. Collectively, these findings uncovered the tumor-suppressor role of circKDM4B in breast cancer, especially in angiogenesis and tumor metastasis, indicating that circKDM4B could be a potential therapeutic target for breast cancer progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438–51.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33.
Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 2020;30:226–40.
Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64.
Hanniford D, Ulloa-Morales A, Karz A, Berzoti-Coelho MG, Moubarak RS, Sanchez-Sendra B, et al. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell. 2020;37:55–70 e15.
Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 2019;30:157–73 e157.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48.
Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8:4003–15.
Xu JZ, Shao CC, Wang XJ, Zhao X, Chen JQ, Ouyang YX, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10:175.
Liang Y, Song X, Li Y, Su P, Han D, Ma T, et al. circKDM4C suppresses tumor progression and attenuates doxorubicin resistance by regulating miR-548p/PBLD axis in breast cancer. Oncogene. 2019;38:6850–66.
Karedath T, Ahmed I, Al Ameri W, Al-Dasim FM, Andrews SS, Samuel S, et al. Silencing of ANKRD12 circRNA induces molecular and functional changes associated with invasive phenotypes. BMC Cancer. 2019;19:565.
Gyparaki M-T, Basdra EK, Papavassiliou AG. MicroRNAs as regulatory elements in triple negative breast cancer. Cancer Lett. 2014;354:1–4.
Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6.
Zheng J, Zhang H, Ma R, Liu H, Gao P. Long non-coding RNA KRT19P3 suppresses proliferation and metastasis through COPS7A-mediated NF-kappaB pathway in gastric cancer. Oncogene. 2019;38:7073–88.
Luo Z, Rong Z, Zhang J, Zhu Z, Yu Z, Li T, et al. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression. Mol Cancer. 2020;19:86.
Goel P, Manning JA, Kumar S. NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene. 2015;557:1–10.
Guarnieri AL, Towers CG, Drasin DJ, Oliphant MUJ, Andrysik Z, Hotz TJ, et al. The miR-106b-25 cluster mediates breast tumor initiation through activation of NOTCH1 via direct repression of NEDD4L. Oncogene. 2018;37:3879–93.
Kovacevic Z, Chikhani S, Lui GY, Sivagurunathan S, Richardson DR. The iron-regulated metastasis suppressor NDRG1 targets NEDD4L, PTEN, and SMAD4 and inhibits the PI3K and Ras signaling pathways. Antioxid Redox Signal. 2013;18:874–87.
Wang Z, Dang T, Liu T, Chen S, Li L, Huang S, et al. NEDD4L protein catalyzes ubiquitination of PIK3CA protein and regulates PI3K-AKT signaling. J Biol Chem. 2016;291:17467–77.
Cheng J, He S, Wang M, Zhou L, Zhang Z, Feng X, et al. The Caspase-3/PKCdelta/Akt/VEGF-A signaling pathway mediates tumor repopulation during radiotherapy. Clin Cancer Res. 2019;25:3732–43.
Suhasini AN, Wang L, Holder KN, Lin AP, Bhatnagar H, Kim SW, et al. A phosphodiesterase 4B-dependent interplay between tumor cells and the microenvironment regulates angiogenesis in B-cell lymphoma. Leukemia. 2016;30:617–26.
Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273:114–27.
Viallard C, Larrivee B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20:409–26.
Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17:679–92.
Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214–27.
Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. 2018;17:140.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777.
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–4.
Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–59.
Liu G, Xiang T, Wu QF, Wang WX. Long noncoding RNA H19-derived miR-675 enhances proliferation and invasion via RUNX1 in gastric cancer cells. Oncol Res. 2016;23:99–107.
Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315–22.
Tsang WP, Ng EKO, Ng SSM, Jin H, Yu J, Sung JJY, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–8.
Guo J, Cheng J, Zheng N, Zhang X, Dai X, Zhang L, et al. Copper promotes tumorigenesis by activating the PDK1-AKT oncogenic pathway in a copper transporter 1 dependent manner. Adv Sci. 2021;8:e2004303. https://doi.org/10.1002/advs.202004303.
Protopsaltis NJ, Liang W, Nudleman E, Ferrara N. Interleukin-22 promotes tumor angiogenesis. Angiogenesis. 2019;22:311–23.
Morland C, Andersson KA, Haugen OP, Hadzic A, Kleppa L, Gille A, et al. Exercise induces cerebral VEGF and angiogenesis via the lactate receptor HCAR1. Nat Commun. 2017;8:15557.
Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–17.
He Q, Zhao L, Liu Y, Liu X, Zheng J, Yu H, et al. circ-SHKBP1 regulates the angiogenesis of U87 glioma-exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol Ther Nucleic Acids. 2018;10:331–48.
Barbagallo D, Caponnetto A, Brex D, Mirabella F, Barbagallo C, Lauretta G. et al. CircSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers. 2019;11:194. https://doi.org/10.3390/cancers11020194.
Acknowledgements
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant nos. 81872362 and 82072665), the Taishan Scholars Program of Shandong Province (Grant no. ts201511096).
Author information
Authors and Affiliations
Contributions
X-YG and PG conceived the experiment and wrote the manuscript. X-YG performed the experiments and analysed the data. T-TL, W-J, H-TL, G-HZ, L-S, R-NZ, and XC helped with the experiments and analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
All authors agree with the content of the manuscript.
Ethics
This study was approved by the Ethics Committee of Shandong University and conformed to the ethical standards of Helsinki Declaration. Informed consent was obtained from all patients included in this study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Availability of supporting data The data supporting the findings of this study are available in the article or available from the corresponding author upon reasonable request.
Rights and permissions
About this article
Cite this article
Guo, XY., Liu, TT., Zhu, WJ. et al. CircKDM4B suppresses breast cancer progression via the miR-675/NEDD4L axis. Oncogene 41, 1895–1906 (2022). https://doi.org/10.1038/s41388-022-02232-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02232-x
This article is cited by
-
Exosomal circZNF800 Derived from Glioma Stem-like Cells Regulates Glioblastoma Tumorigenicity via the PIEZO1/Akt Axis
Molecular Neurobiology (2024)
-
CircHERC1 promotes non-small cell lung cancer cell progression by sequestering FOXO1 in the cytoplasm and regulating the miR-142-3p-HMGB1 axis
Molecular Cancer (2023)
-
CircRNA MBOAT2 promotes intrahepatic cholangiocarcinoma progression and lipid metabolism reprogramming by stabilizing PTBP1 to facilitate FASN mRNA cytoplasmic export
Cell Death & Disease (2023)
-
CircSTX6 promotes pancreatic ductal adenocarcinoma progression by sponging miR-449b-5p and interacting with CUL2
Molecular Cancer (2022)
-
EIF4A3-induced circBRWD3 promotes tumorigenesis of breast cancer through miR-142-3p_miR-142-5p/RAC1/PAK1 signaling
BMC Cancer (2022)