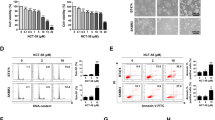
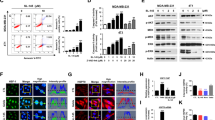
Abstract
Transmembrane glycoprotein NMB (GPNMB) is a prognostic marker of poor outcome in patients with triple-negative breast cancer (TNBC). Glembatumumab Vedotin, an antibody drug conjugate targeting GPNMB, exhibits variable efficacy against GPNMB-positive metastatic TNBC as a single agent. We show that GPNMB levels increase in response to standard-of-care and experimental therapies for multiple breast cancer subtypes. While these therapeutic stressors induce GPNMB expression through differential engagement of the MiTF family of transcription factors, not all are capable of increasing GPNMB cell-surface localization required for Glembatumumab Vedotin inhibition. Using a FACS-based genetic screen, we discovered that suppression of heat shock protein 90 (HSP90) concomitantly increases GPNMB expression and cell-surface localization. Mechanistically, HSP90 inhibition resulted in lysosomal dispersion towards the cell periphery and fusion with the plasma membrane, which delivers GPNMB to the cell surface. Finally, treatment with HSP90 inhibitors sensitizes breast cancers to Glembatumumab Vedotin in vivo, suggesting that combination of HSP90 inhibitors and Glembatumumab Vedotin may be a viable treatment strategy for patients with metastatic TNBC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–81.
Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9.
Damaskos C, Garmpi A, Nikolettos K, Vavourakis M, Diamantis E, Patsouras A, et al. Triple-negative breast cancer: the progress of targeted therapies and future tendencies. Anticancer Res. 2019;39:5285–96.
Rose AA, Grosset AA, Dong Z, Russo C, Macdonald PA, Bertos NR, et al. Glycoprotein nonmetastatic B is an independent prognostic indicator of recurrence and a novel therapeutic target in breast cancer. Clin Cancer Res. 2010;16:2147–56.
Yardley DA, Weaver R, Melisko ME, Saleh MN, Arena FP, Forero A, et al. EMERGE: a randomized phase II study of the antibody-drug conjugate glembatumumab vedotin in advanced glycoprotein NMB-expressing breast cancer. J Clin Oncol. 2015;33:1609–19.
Chen C, Okita Y, Watanabe Y, Abe F, Fikry MA, Ichikawa Y, et al. Glycoprotein nmb is exposed on the surface of dormant breast cancer cells and induces stem cell-like properties. Cancer Res. 2018;78:6424–35.
Rose AAN, Biondini M, Curiel R, Siegel PM. Targeting GPNMB with glembatumumab vedotin: current developments and future opportunities for the treatment of cancer. Pharmacol Ther. 2017;179:127–41.
Tse KF, Jeffers M, Pollack VA, McCabe DA, Shadish ML, Khramtsov NV, et al. CR011, a fully human monoclonal antibody-auristatin E conjugate, for the treatment of melanoma. Clin Cancer Res. 2006;12:1373–82.
Bendell J, Saleh M, Rose AA, Siegel PM, Hart L, Sirpal S, et al. Phase I/II study of the antibody-drug conjugate glembatumumab vedotin in patients with locally advanced or metastatic breast cancer. J Clin Oncol. 2014;32:3619–25.
Vahdat LTF-TA, Schmid P, Blackwell K, Telli ML, Melisko M, Holgado E, et al. METRIC: A randomized international phase 2b study of the antibody-drug conjugate (ADC) glembatumumab vedotin (GV) in gpNMB-overexpressing, metastatic, triple-negative breast cancer (mTNBC) (Abstract P6-20-01.). Proceedings of the 2018 San Antonio Breast Cancer Symposium; 2018; San Antonio, TX Philadelphia (PA): AACR; Cancer Res 2019;79 4 Suppl.
Ott PA, Pavlick AC, Johnson DB, Hart LL, Infante JR, Luke JJ, et al. A phase 2 study of glembatumumab vedotin, an antibody-drug conjugate targeting glycoprotein NMB, in patients with advanced melanoma. Cancer. 2019;125:1113–23.
Qian X, Mills E, Torgov M, LaRochelle WJ, Jeffers M. Pharmacologically enhanced expression of GPNMB increases the sensitivity of melanoma cells to the CR011-vcMMAE antibody-drug conjugate. Mol Oncol. 2008;2:81–93.
Rose AA, Annis MG, Frederick DT, Biondini M, Dong Z, Kwong L, et al. MAPK pathway inhibitors sensitize BRAF-mutant melanoma to an antibody-drug conjugate targeting GPNMB. Clin Cancer Res. 2016;22:6088–98.
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27.
Bergin ART, Loi S. Triple-negative breast cancer: recent treatment advances. F1000Res. 2019;8:1342.
Cheng Y, Tian H. Current development status of MEK inhibitors. Molecules. 2017;22:1551.
Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106:4519–24.
Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7.
Liu H, Murphy CJ, Karreth FA, Emdal KB, White FM, Elemento O, et al. Identifying and targeting sporadic oncogenic genetic aberrations in mouse models of triple-negative breast cancer. Cancer Discov. 2018;8:354–69.
Jing J, Greshock J, Holbrook JD, Gilmartin A, Zhang X, McNeil E, et al. Comprehensive predictive biomarker analysis for MEK inhibitor GSK1120212. Mol Cancer Ther. 2012;11:720–9.
Slade L, Pulinilkunnil T. The MiTF/TFE family of transcription factors: master regulators of organelle signaling, metabolism, and stress adaptation. Mol Cancer Res. 2017;15:1637–43.
Raben N, Puertollano R. TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu Rev Cell Dev Biol. 2016;32:255–78.
Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200:475–91.
Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014;7:ra9.
Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–14.
Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42.
Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Embo J. 2012;31:1095–108.
Hong SB, Oh H, Valera VA, Baba M, Schmidt LS, Linehan WM. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS ONE. 2010;5:e15793.
Hemesath TJ, Steingrímsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, et al. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–80.
Pogenberg V, Ogmundsdóttir MH, Bergsteinsdóttir K, Schepsky A, Phung B, Deineko V, et al. Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev. 2012;26:2647–58.
DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–20.
Le Borgne R, Planque N, Martin P, Dewitte F, Saule S, Hoflack B. The AP-3-dependent targeting of the melanosomal glycoprotein QNR-71 requires a di-leucine-based sorting signal. J Cell Sci. 2001;114:2831–41.
Starling GP, Yip YY, Sanger A, Morton PE, Eden ER, Dodding MP. Folliculin directs the formation of a Rab34-RILP complex to control the nutrient-dependent dynamic distribution of lysosomes. EMBO Rep. 2016;17:823–41.
Schmidt LS, Linehan WM. FLCN: the causative gene for Birt-Hogg-Dubé syndrome. Gene. 2018;640:28–42.
Kong T, Ahn R, Yang K, Zhu X, Fu Z, Morin G, et al. CD44 promotes PD-L1 expression and its tumor-intrinsic function in breast and lung cancers. Cancer Res. 2020;80:444–57.
Xue Y, Meehan B, Macdonald E, Venneti S, Wang XQD, Witkowski L, et al. CDK4/6 inhibitors target SMARCA4-determined cyclin D1 deficiency in hypercalcemic small cell carcinoma of the ovary. Nat Commun. 2019;10:558.
Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554.
Jolly C, Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst. 2000;92:1564–72.
Giulino-Roth L, van Besien HJ, Dalton T, Totonchy JE, Rodina A, Taldone T, et al. Inhibition of Hsp90 Suppresses PI3K/AKT/mTOR Signaling and Has Antitumor Activity in Burkitt Lymphoma. Mol Cancer Ther. 2017;16:1779–90.
Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491–9.
Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–63.
Jongsma ML, Berlin I, Wijdeven RH, Janssen L, Janssen GM, Garstka MA, et al. An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell. 2016;166:152–66.
Woodford MR, Dunn DM, Blanden AR, Capriotti D, Loiselle D, Prodromou C, et al. The FNIP co-chaperones decelerate the Hsp90 chaperone cycle and enhance drug binding. Nat Commun. 2016;7:12037.
Pick E, Kluger Y, Giltnane JM, Moeder C, Camp RL, Rimm DL, et al. High HSP90 expression is associated with decreased survival in breast cancer. Cancer Res. 2007;67:2932–7.
Boshuizen J, Peeper DS. Rational cancer treatment combinations: an urgent clinical need. Mol Cell. 2020;78:1002–18.
van der Lienden MJC, Gaspar P, Boot R, Aerts J, van Eijk M. Glycoprotein non-metastatic protein B: an emerging biomarker for lysosomal dysfunction in macrophages. Int J Mol Sci. 2018;20:1.
Noda Y, Tsuruma K, Takata M, Ishisaka M, Tanaka H, Nakano Y, et al. GPNMB induces BiP expression by enhancing splicing of BiP Pre-mRNA during the endoplasmic reticulum stress response. Sci Rep. 2017;7:12160.
Martina JA, Puertollano R. TFEB and TFE3: The art of multi-tasking under stress conditions. Transcription. 2017;8:48–54.
Tanaka M, Homme M, Yamazaki Y, Shimizu R, Takazawa Y, Nakamura T. Modeling alveolar soft part sarcoma unveils novel mechanisms of metastasis. Cancer Res. 2017;77:897–907.
Zhitomirsky B, Assaraf YG. Lysosomal sequestration of hydrophobic weak base chemotherapeutics triggers lysosomal biogenesis and lysosome-dependent cancer multidrug resistance. Oncotarget. 2015;6:1143–56.
Li B, Castano AP, Hudson TE, Nowlin BT, Lin SL, Bonventre JV, et al. The melanoma-associated transmembrane glycoprotein Gpnmb controls trafficking of cellular debris for degradation and is essential for tissue repair. Faseb J. 2010;24:4767–81.
Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838.
Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33.
Vega-Rubin-de-Celis S, Peña-Llopis S, Konda M, Brugarolas J. Multistep regulation of TFEB by MTORC1. Autophagy. 2017;13:464–72.
Puertollano R, Ferguson SM, Brugarolas J, Ballabio A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. Embo J. 2018;37:e98804.
Martina JA, Diab HI, Brady OA, Puertollano R. TFEB and TFE3 are novel components of the integrated stress response. Embo J. 2016;35:479–95.
Wada S, Neinast M, Jang C, Ibrahim YH, Lee G, Babu A, et al. The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev. 2016;30:2551–64.
Hoffmann RM, Coumbe BGT, Josephs DH, Mele S, Ilieva KM, Cheung A, et al. Antibody structure and engineering considerations for the design and function of Antibody Drug Conjugates (ADCs). Oncoimmunology. 2018;7:e1395127.
Wang H, Lu M, Yao M, Zhu W. Effects of treatment with an Hsp90 inhibitor in tumors based on 15 phase II clinical trials. Mol Clin Oncol. 2016;5:326–34.
Jansson KH, Tucker JB, Stahl LE, Simmons JK, Fuller C, Beshiri ML, et al. High-throughput screens identify HSP90 inhibitors as potent therapeutics that target inter-related growth and survival pathways in advanced prostate cancer. Sci Rep. 2018;8:17239.
Bekki H, Kohashi K, Maekawa A, Yamada Y, Yamamoto H, Harimaya K, et al. Elevated expression of HSP90 and the antitumor effect of an HSP90 inhibitor via inactivation of the Akt/mTOR pathway in undifferentiated pleomorphic sarcoma. BMC Cancer. 2015;15:804.
Butler LM, Ferraldeschi R, Armstrong HK, Centenera MM, Workman P. Maximizing the therapeutic potential of HSP90 inhibitors. Mol Cancer Res. 2015;13:1445–51.
Dai C. The heat-shock, or HSF1-mediated proteotoxic stress, response in cancer: from proteomic stability to oncogenesis. Philos Trans R Soc Lond B Biol Sci. 2018;373:20160525.
Wang Y, McAlpine SR. N-terminal and C-terminal modulation of Hsp90 produce dissimilar phenotypes. Chem Commun. 2015;51:1410–3.
Pu J, Guardia CM, Keren-Kaplan T, Bonifacino JS. Mechanisms and functions of lysosome positioning. J Cell Sci. 2016;129:4329–39.
Kallunki T, Olsen OD, Jäättelä M. Cancer-associated lysosomal changes: friends or foes? Oncogene. 2013;32:1995–2004.
Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202:1107–22.
El-Houjeiri L, Possik E, Vijayaraghavan T, Paquette M, Martina JA, Kazan JM, et al. The transcription factors TFEB and TFE3 link the FLCN-AMPK signaling axis to innate immune response and pathogen resistance. Cell Rep. 2019;26:3613–28.e6.
Lauwers E, Wang YC, Gallardo R, Van der Kant R, Michiels E, Swerts J, et al. Hsp90 mediates membrane deformation and exosome release. Mol Cell. 2018;71:689–702.e9.
Park S, Park JA, Jeon JH, Lee Y. Traditional and novel mechanisms of heat shock protein 90 (HSP90) inhibition in cancer chemotherapy including HSP90 cleavage. Biomol Ther. 2019;27:423–34.
Kopp LM, Malempati S, Krailo M, Gao Y, Buxton A, Weigel BJ, et al. Phase II trial of the glycoprotein non-metastatic B-targeted antibody-drug conjugate, glembatumumab vedotin (CDX-011), in recurrent osteosarcoma AOST1521: a report from the Children’s Oncology Group. Eur J Cancer. 2019;121:177–83.
Nytko KJ, Thumser-Henner P, Russo G, Weyland MS, Rohrer Bley C. Role of HSP70 in response to (thermo)radiotherapy: analysis of gene expression in canine osteosarcoma cells by RNA-seq. Sci Rep. 2020;10:12779.
El-Houjeiri L, Biondini M, Paquette M, Kuasne H, Pacis A, Park M, et al. Folliculin impairs breast tumor growth by repressing TFE3-dependent induction of the Warburg effect and angiogenesis. J Clin Investig. 2021;131:e144871.
McGuirk S, Audet-Delage Y, Annis MG, Xue Y, Vernier M, Zhao K, et al. Resistance to different anthracycline chemotherapeutics elicits distinct and actionable primary metabolic dependencies in breast cancer. Elife. 2021;10:e65150.
Sirois I, Aguilar-Mahecha A, Lafleur J, Fowler E, Vu V, Scriver M, et al. A unique morphological phenotype in chemoresistant triple-negative breast cancer reveals metabolic reprogramming and PLIN4 expression as a molecular vulnerability. Mol Cancer Res. 2019;17:2492–507.
Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–22.
Acknowledgements
We acknowledge technical assistance from the McGill GCRC Histology core facility for routine histological services. We acknowledge McGill Life Science Complex Advanced BioImaging Facility (ABIF) for technical assistance in acquisition/analysis of images. We acknowledge McGill Platform for Cellular Perturbation (MPCP) for providing the shRNAs libraries and technical assistance with screnning process and analysis. We acknowledge McGill Life Science Complex Cell Vision Core for technical assistance in acquisition/analysis of flow cytometry data. We acknowledge Génome Quebec and McGill University for support of the Q-CROC-03 trial. We acknowledge Dr. Alana Welm for providing us with HCI-010 PDX tumor model. We thank members of the Siegel laboratory, all collaborators and Dr. Ursini-Siegel for thoughtful discussions and critical reading of the paper. MB was the recipient of a Fonds de recherche du Québec—Santé (FRQS) post-doctoral fellowship. PMS is a William Dawson Scholar of McGill University. This work was supported by an operating grant from the Canadian Institutes of Health Research to PMS (PJT-153327).
Author information
Authors and Affiliations
Contributions
MB and PMS designed the research. MB, AK, MGA, BEH, JAM, GM, and LE performed experiments. AANR, AAM, TG, RMJ, GM, and EB, performed computational analysis, statistical analysis, biostatistics. IS, AAM, SM, JSP, UMT, MB, RP, OS, SH, TK, and AP, provided reagents and expertise. MB and PMS wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Biondini, M., Kiepas, A., El-Houjeiri, L. et al. HSP90 inhibitors induce GPNMB cell-surface expression by modulating lysosomal positioning and sensitize breast cancer cells to glembatumumab vedotin. Oncogene 41, 1701–1717 (2022). https://doi.org/10.1038/s41388-022-02206-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02206-z
This article is cited by
-
Combination therapy involving HSP90 inhibitors for combating cancer: an overview of clinical and preclinical progress
Archives of Pharmacal Research (2024)
-
CCN3/NOV promotes metastasis and tumor progression via GPNMB-induced EGFR activation in triple-negative breast cancer
Cell Death & Disease (2023)
-
GPNMB: a potent inducer of immunosuppression in cancer
Oncogene (2022)