Abstract
Castration-resistant prostate cancer (CRPC) is a highly malignant type of advanced cancer resistant to androgen deprivation therapy. One of the important mechanisms for the development of CRPC is the persistent imbalanced regulation of AR and AR splice variants (AR/AR-Vs). In this study, we reported KDM4A-AS1, a recently discovered lncRNA, as a tumor promoter that was significantly increased in CRPC cell lines and cancer tissues. Depletion of KDM4A-AS1 significantly reduced cell viability, proliferation, migration in vitro, and tumor growth in vivo. We found that by binding to the NTD domain, KDM4A-AS1 enhances the stability of USP14-AR/AR-Vs complex, and promoted AR/AR-Vs deubiquitination to protect it from MDM2-mediated ubiquitin-proteasome degradation. Moreover, KDM4A-AS1 was found to enhance CRPC drug resistance to enzalutamide by repressing AR/AR-Vs degradation; antisense oligonucleotide drugs targeting KDM4A-AS1 significantly reduced the growth of tumors with enzalutamide resistance. Taken together, our results indicated that KDM4A-AS1 played an important role in the progression of CRPC and enzalutamide resistance by regulating AR/AR-Vs deubiquitination; targeting KDM4A-AS1 has broad clinical application potential.
Similar content being viewed by others
Introduction
In recent years, prostate cancer (PCa) has become the primary malignant tumor endangering male health. According to 2020’s statistics by the American Cancer Society, PCa has raised to the first in male urinary cancer estimated new cases (cases = 191,930) and the second in estimated deaths (death cases = 33,330) [1]. According to the response to androgen deprivation therapy (ADT), PCa can be classified into two subtypes, including hormone sensitive prostate cancer (HSPC) and castration-resistant prostate cancer (CRPC). As a standard treatment, although PCa patients benefit from ADT at the beginning, most of them will relapse into CRPC within 2–3 years [2]. Several mechanisms were found to result in CRPC, including long noncoding RNAs (lncRNAs) [3, 4].
LncRNAs are key regulators of gene expression patterns and have recently received widespread attention due to its involvement in regulating disease progression including cancers [5, 6]. Studies have confirmed that lncRNA could regulate the expression of key proteins in various cancers through direct regulation of target genes or epigenetic pathways [7,8,9]. LncRNAs like HOTAIR, GAS5, and ANCR could act as post-translational regulators through degradation patterns to regulate key protein expressions in cancers [3, 10, 11]. However, mechanisms between protein regulation and lncRNAs in PCa, especially CRPC, remain unclear.
An important mechanism of CRPC development is the continuous activation of androgen receptor (AR). Studies have shown that, even under the premise of castration, CRPC itself can still synthesize androgens de novo from cholesterol, keeping AR highly active [12, 13]. Previous studies have shown that USP14, a deubiquitination enzyme, could protect AR from ubiquitin-mediated protein degradation [14, 15]. Meanwhile, the expression of AR-Vs was also reported to be regulated by ubiquitin-proteasome pathway [16, 17]. High expression of AR-Vs was closely related to drug resistance including enzalutamide [18]. In this study, we examined the important role of a newly discovered lncRNA KDM4A-AS1 and its potential mechanisms in PCa. Our results indicated that KDM4A-AS1 contributed to CRPC development and was closely related to clinical outcomes. Through binding with USP14-AR/AR-Vs complex, KDM4A-AS1 enhanced complex stability and promoted AR/AR-Vs deubiquitination, which resulted in an escape of AR/AR-Vs degradation, and finally led to CRPC progression and drug resistance to enzalutamide.
Result
KDM4A-AS1 is upregulated in patients with CRPC and related to poor clinical outcomes in PCa patients
In order to observe the expression change of lncRNA in PCa cell after castration, we processed microarray on LNCaP and LNCaP-AI cells [4]. Compared with LNCaP, totally 1520 differentially expressed lncRNAs were detected in LNCaP-AI under the threshold (|FoldChange| >1.5, P value < 0.05), including 734 upregulated and 786 downregulated. Considering the value of clinical application, we discarded the low expression, unstable expression, and unnamed lncRNAs in the data. On the other hand, we also considered the function of the mRNA near these lncRNAs in CRPC. Among the remaining lncRNAs, KDM4A-AS1 caught our mind (Fig. 1A). KDM4A, which is also known as JMJD2A, has been shown to drive prostate tumorigenesis through ETV1 [19]. As the antisense transcript of KDM4A, KDM4A-AS1 deserves an in-depth research. According to the UCSC genome browser, KDM4A-AS1 contains 1539 bp and is located near the 3’UTR of KDM4A with a CpG island around its 5’ start and a polyA tail at the 3’ end (Fig. 1B).
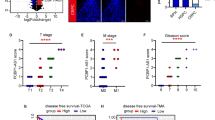
A Microarray volcano plot indicates a significant upregulation of KDM4A-AS1 in LNCaP-AI. The red dots represent lncRNAs that are significantly upregulated in LNCaP-AI, and the blue dots represent lncRNAs that are significantly downregulated in LNCaP-AI. Threshold: |logFC| >1, P < 0.05, compared with LNCaP. B Genome location of KDM4A-AS1. C Validation of KDM4A-AS1 expression in BPH, HSPC and CRPC tissue microarray using RNAScope. Objective lens magnification: 20X and 40X, eye piece magnification: 10X. D The percentage of stacked column chart was used to calculate the results of tissue microarray RNAScope, indicating that the expression of KDM4A-AS1 at different stages of the disease has significant difference. E KDM4A-AS1 RNA expression in prostate cancer cell lines using qPCR. F, G Disease-free survival of patients in microarray and TCGA database using Kaplan–Meier survival analysis.
We next detected the expression of KDM4A-AS1 using RNAScope in a tissue microarray of 4 benign prostatic hyperplasia (BPH), 28 HSPC, and 13 CRPC tissues (Fig. 1C). Result showed that the expression of KDM4A-AS1 could be determined in HSPC and CRPC, but few expression could be observed in BPH tissues (Fig. 1D and Supplementary Fig. 1A). In PCa samples, 3.57% of patients scored 3 in HSPC (no patient scored 4 in HSPC), while 38.5% patients in CRPC scored ≥3 (two scored 4 and three scored 3). Similar results were also confirmed in PCa cell lines. Compared with LNCaP cells, KDM4A-AS1 was significantly upregulated in LNCaP-AI, 22RV1, and C4-2 cells, but showed no changes in PC-3 and DU145 cells (Fig. 1E).
We then counted the patient’s clinical data, including age, smoke history, alcohol history, disease-free survival (DFS), disease status (HSPC or CRPC), and Gleason score. We divided clinical information into groups and calculate the average score of KDM4A-AS1 in different groups. Subsequently, we calculated the significance between the expression of KDM4A-AS1 and clinical information through the χ2 test. As can be seen in Table 1 and Supplementary Table 1, KDM4A-AS1 level was only linked with PCa patients’ disease status (HSPC or CRPC, P value = 0.0364), KDM4A-AS1 expression was closely related to DFS time in CRPC patients (P value = 0.0119) and was independent with patients’ age, smoke history, alcohol history, and Gleason score, indicating a specific potential role of KDM4A-AS1 in CRPC. Meanwhile, Kaplan–Meier survival analysis of these patients and TCGA database also indicated that high KDM4A-AS1 expression is related to poor clinical outcomes in PCa patients (Fig. 1F, G).
Targeting KDM4A-AS1 could reduce prostate cancer cell proliferation and migration
In order to further validate the potential function and phenotype of KDM4A-AS1 in CRPC, a knockdown shRNA sequence targeting KDM4A-AS1 was used to interfere KDM4A-AS1 expression in C4-2 and LNCaP-AI (Fig. 2A, B). KDM4A-AS1 depletion showed a significant effect on cell viability in both LNCaP-AI and C4-2 (Fig. 2C, D). Moreover, we performed Transwell and colony formation assay to test cell migration and cell proliferation ability after KDM4A-AS1 knockdown; subsequent analysis by ImageJ software showed that after knocking down KDM4A-AS1, the migration and proliferation of C4-2 and LNCaP-AI cells were significantly inhibited (Fig. 2E–H), indicating KDM4A-AS1 played a role in CRPC cells’ proliferation and migration.
A, B KDM4A-AS1 knockdown efficiency in LNCaP-AI and C4-2 cell lines. Repeated three times with three biological replicates each time. C, D Validation of cell viability change after KDM4A-AS1 depletion by MTT in LNCaP-AI and C4-2 cell lines. Repeated three times with six biological replicates each time. E, F Validation of cell migration ability after KDM4A-AS1 depletion by transwell assay in LNCaP-AI and C4-2 cell line. Barplots represent the cell count of the corresponding cell line. Repeated three times with three biological replicates each time. G, H Validation of cell proliferation ability after KDM4A-AS1 depletion by colony formation assay in LNCaP-AI and C4-2 cell line. Barplots represent the colony count of the proliferation ability of the corresponding cell lines. Repeated three times with three biological replicates each time.
KDM4A-AS1 depletion promotes AR ubiquitin-proteasome degradation in CRPC
According to results above, we hypothesized that KDM4A-AS1 might regulate CRPC progression. In order to further study the mechanism, we used the published RPI-Seq algorithm to predict the proteins that interacts with KDM4A-AS1 potentially (Table 2) [20]. The results exhibited that AR ranks first in both random forest (score = 0.8) and support vector machines classifiers (score = 0.93). To further confirm the relationship between KDM4A-AS1 and AR protein, we performed immunohistochemistry on the same series tissue array to identify AR protein expression in same patients (Fig. 3A). By comparing the staining scores, we found that KDM4A-AS1 showed a high positive correlation with AR in CRPC (R = 0.6, P value = 0.037, Fig. 3B).
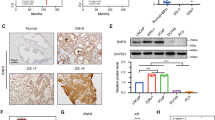
A IHC validation of AR in TMA. B Pearson correlation between KDM4A-AS1 and AR scores. C RNA expression of AR signal by qPCR. D Protein expression of AR signal by western blot, GAPDH was used as loading control. E Binding ability between AR protein and KDM4A-AS1 by RIP. F AR degradation rate detection with or without KDM4A-AS1 depletion. CHX concentration is 10 µg/ml. G Detection of AR degradation pattern regulated by KDM4A-AS1. Expression detected after 36 h. H Detection of AR ubiquitination with indicated condition. MG-132 treated for 10 h before detection. I Binding ability between USP14 protein and KDM4A-AS1 by RIP. J Binding ability between AR and indicated proteins using IP. K Binding ability between AR and indicated proteins. MG-132 treated time: 10 h. L Graph perform plasmids containing different domains of AR proteins. M, N Detection of binding regions between AR and KDM4A-AS1 using RIP-qPCR. O Detection of binding regions between AR and KDM4A-AS1 using RNA pulldown.
The results above suggested that there might be a potential regulatory relationship between KDM4A-AS1 and AR. To verify this, we performed qPCR to validate AR, KLK3, and FKBP5 mRNA expression in shKDM4A-AS1 and shScramble cells. Depletion of KDM4A-AS1 decreased KLK3 and FKBP5 expression in RNA level on AR (Fig. 3C). However, knocking down KDM4A-AS1 could significantly decrease AR, KLK3, and FKBP5 in protein level (Fig. 3D). Subsequent RNA immunoprecipitation (RIP) assay also showed that KDM4A-AS1 binds to AR in LNCaP-AI and C4-2 cells (Fig. 3E), which is consistent with previous RPI-Seq result, indicating that KDM4A-AS1 regulated AR protein through a post-translation manner.
To further clarify the potential relationship between KDM4A-AS1 and AR, we treated LNCaP-AI and C4-2 cells with cycloheximide (CHX) and found that KDM4A-AS1 depletion reduced the stability of AR protein (Fig. 3F). On the other hand, we overexpressed KDM4A-AS1 in LNCaP cells, and found that the protein expression of AR and its downstream KLK3 and FKBP5 were significantly upregulated, and the degradation rate of AR slows down significantly (Supplementary Fig. 2), which indicated the potential role of KDM4A-AS1 in AR degradation. Since lysosomal and ubiquitin-proteasome-dependent manner are two important ways to degrade proteins including AR, chloroquine and MG-132 were used to distinguish how KDM4A-AS1 affects the degradation of AR. The results in Fig. 3G demonstrated that KDM4A-AS1 depletion enhances AR degradation through ubiquitin-proteasome system instead of lysosomal system. To verify the above results, we performed immunoprecipitation using AR antibody to detect the ubiquitination of AR in LNCaP-AI and C4-2 cells. We found that knocking down KDM4A-AS1 significantly induced the ubiquitination level of AR protein (Fig. 3H). Overall, the results indicated that KDM4A-AS1 bound with AR and regulated AR degradation through ubiquitination.
KDM4A-AS1 inhibits AR degradation by stabilizing USP14- AR complex
To further explore the potential mechanism, we used biotin-labeled KDM4A-AS1 probes to perform RNA pulldown experiments and used iTRAQ to find proteins involved in AR ubiquitination. To our surprise, we did not find any meaningful ubiquitin-related proteins, instead, we identified three deubiquitin-related enzymes, USP14, USP5, and OTUB1 (Table 3). In addition, in previous RPI-Seq prediction, the binding potential of KDM4A-AS1 and USP14 ranked second, which second only to AR (Table 2). Then through RIP experiments, we confirmed that KDM4A-AS1 can bind to USP14 in LNCaP-AI and C4-2 cells (Fig. 3I). Next, we verified the binding potential of these three proteins with AR by immunoprecipitation. The results suggest that AR did not bind to USP5 and OTUB1, but bound to USP14, which is consistent with the previous report (Fig. 3J) [14, 15].
Therefore, we believed that KDM4A-AS1 might be involved in the regulation of the USP14-AR complex. To confirm this, we added 10 mM MG-132 to the cell culture medium, and compared the binding ability of AR with USP14 before and after knocking down KDM4A-AS1. Since the binding of MDM2 is one of the main mechanisms of AR ubiquitination, previous study has confirmed that MDM2 and USP14 have a competitive binding relationship with AR. We also detected the binding ability of MDM2 and AR. It was found that in the LNCaP-AI and C4-2 cell lines, the depletion of KDM4A-AS1 could significantly reduce the stability of the USP14-AR complex and enhance the binding of MDM2 with AR (Fig. 3K and Supplementary Fig. 3). This indicated that KDM4A-AS1 might enhance the deubiquitination of AR by stabilizing the form of the USP14-AR complex, and prevent AR from being degraded by the MDM2-mediated ubiquitin-proteasome pathway. In addition, we found that after knocking down KDM4A-AS1, the expression of USP14 was not affected (Supplementary Fig. 4), which suggested that KDM4A-AS1 regulation of USP14-AR complex and AR deubiquitination is not achieved by increasing the expression of USP14.
In order to further explore the specific binding regions of AR and KDM4A-AS1, we designed plasmids containing different domains of AR and plasmid expressing KDM4A-AS1. All AR plasmids contained MYC tags to ensure that our results were objective and accurate (Fig. 3L). Subsequent RIP-qPCR using MYC-tag antibody showed that compared to the control group, MYC-AR-FL, MYC-NTD, and MYC-NTD+DBD could detect significant amplification of KDM4A-AS1, while MYC-DBD+LBD had no significant difference to the control group, which suggested that KDM4A-AS1 might bind to the NTD domain of AR (Fig. 3M, N). Meanwhile, we also used the biotin-labeled KDM4A-AS1 probe for RNA pulldown. And through RNA pulldown assay, we found that only MYC-AR-FL, MYC-NTD, MYC-NTD+DBD can detect the MYC-tag signal, which was consistent with RIP-qPCR results (Fig. 3O).
In short, the results above indicated that KDM4A-AS1 bound to the NTD domain of AR and protected AR from degradation by the ubiquitin-proteasome pathway as well as stabilizing the USP14-AR complex.
Rescue of KDM4A-AS1 stabilizes USP14-AR complex and promotes CRPC progression
Based on the result above, we designed the rescue assay to determine whether we can restore the function of KDM4A-AS1 to USP14-AR complex. After rescue of KDM4A-AS1 in shKDM4A-AS1 cell line (reKDM4A-AS1, Fig. 4A, B), we found a recovery of KLK3 and FKBP5 in the RNA level (Fig. 4C, D) and a recovery of AR, KLK3, and FKBP5 in the protein level (Fig. 4E). After blocking protein synthesis with CHX, the degradation of AR was also significantly reduced with the rescue of KDM4A-AS1 (Fig. 4F). Compared with the KDM4A-AS1 knockdown cells, the binding ability of AR to USP14 was also significantly restored after KDM4A-AS1 was rescued, while the combination of MDM2 and AR was significantly repressed (Fig. 4G). Moreover, cell migration and proliferation ability were also restored in reKDM4A-AS1 cells (cell viability: Fig. 4H, I; cell migration: Fig. 4J–L; cell proliferation: Fig. 4M–O). The results above not only indicated that the regulation of AR by KDM4A-AS1 can be reproduced by restoring the expression of KDM4A-AS1, but further also confirmed that the stability of the USP14-AR complex can be regulated by KDM4A-AS1.
A, B KDM4A-AS1 knockdown and rescue efficiency in LNCaP-AI and C4-2 cell line. C, D qPCR detected the mRNA expression of AR and downstream KLK3, FKBP5 in LNCaP-AI and C4-2 cell lines before and after rescue KDM4A-AS1, GAPDH was used as loading control. E Validation of AR and downstream KLK3, FKBP5 protein expression in LNCaP-AI and C4-2 cell lines before and after rescue KDM4A-AS1 using western blot, GAPDH was used as loading control. F Detection of AR degradation rate before and after rescue KDM4A-AS1. The corresponding cells were treated with 10 ug/ml CHX according to the time indicated, and the expression of AR was detected by western blot. GAPDH was used as a loading control. G Detection of USP14 and MDM2 binding ability with AR before and after KDM4A-AS1 rescue. Corresponding cells were treated with 10 mM MG-132 for 12 h, cells were then lysate and subjected to immunoprecipitation using anti-AR antibody, followed by western blot with corresponding antibodies. GAPDH was used as loading control. H, I Validation of cell viability change after KDM4A-AS1 depletion and rescue by MTT in LNCaP-AI and C4-2 cell line. Repeated three times with eight biological replicates each time. J–L Validation of cell migration ability after KDM4A-AS1 rescue by transwell assay in LNCaP-AI and C4-2 cell line. Barplots represent the cell count of the migration ability of the corresponding cell line. Repeated three times with three biological replicates each time. M–O Validation of cell proliferation ability after KDM4A-AS1 rescue by colony formation assay in LNCaP-AI and C4-2 cell line. Barplots represent the colony count of the proliferation ability of the corresponding cell line. Repeated three times with three biological replicates each time.
Targeting KDM4A-AS1 could reverse enzalutamide resistance CRPC by degradating AR splice variants in vitro and in vivo
The abnormal expression of AR may lead to ADT resistance and cancer progression. Therefore, we hypothesized that the regulation of AR by KDM4A-AS1 may be one of the reasons for the cancer resistance to ADT drugs like enzalutamide. We generated enzalutamide-resistant C4-2 cell line (C4-2-EnzR, details in Methods) and found an upregulation of KDM4A-AS1 in C4-2-EnzR comparing normal C4-2 (Fig. 5A), while AR, AR-V7, and AR-V567es were also upregulated in the protein level (Fig. 5B). Depletion of KDM4A-AS1 significantly reduced the expression of AR, AR-V7, AR-V567es and downstream KLK3 and FKBP5 in the protein level (Fig. 5C, D). Since KDM4A-AS1 bound to the NTD of AR, it might also bind to AR-V7 and AR-V567es. Previous study has confirmed that AR-Vs were also degraded through MDM2-mediated ubiquitin-proteasome pathway [21, 22]. After repressing protein synthesis with CHX and knocking down KDM4A-AS1 in C4-2-EnzR (EnzR-shLnc), we observed an enhanced level of AR, AR-V7, and AR-V567es degradation (Fig. 5E). Moreover, the combination of USP14 with AR/AR-Vs was repressed after KDM4A-AS1 depletion in EnzR-shLnc, indicating that USP14 might take part in AR/AR-Vs ubiquitin-proteasome-mediated degradation and KDM4A-AS1 could stabilize the USP14-AR/AR-Vs complex (Fig. 5F). In order to further study the relationship between KDM4A-AS1’s regulation of AR/AR-Vs and C4-2-EnzR enzalutamide resistance, we validated the effects of different treatments indicated in Fig. 5G on cell viability through cytotoxicity experiment. After enzalutamide treatment, the cell viability of C4-2 was significantly repressed (PA-B < 0.0001), while C4-2-EnzR showed no difference (PC-D = 0.1737). Compared with DMSO treatment, enzalutamide significantly repressed cell viability of EnzR-shLnc (PE-F = 0.0006), we also observed a significant repress between C4-2-EnzR and EnzR-shLnc under the condition of enzalutamide treatment (PD-F < 0.0001), indicating that KDM4A-AS1 depletion may increase C4-2-EnzR cell sensitivity to enzalutamide.
A KDM4A-AS1 expression in C4-2 and C4-2-EnzR. B Expression of indicated proteins in C4-2 and C4-2-EnzR. C KDM4A-AS1 knockdown efficiency in C4-2-EnzR. D expression of indicated proteins before and after KDM4A-AS1 depletion in C4-2-EnzR. E Degradation rate of indicated proteins after 10 µg/ml CHX treatment. F Binding abilities of indicated proteins with USP14 with or without KDM4A-AS1 depletion. G Cell viability of indicated groups. Enzalutamide concentration: 20 µM. H Tumor volume after 4-week growth with indicated condition. I Tumor volume growth curves of nude mice in different treatment groups. The time point of enzalutamide and AAV application is also indicated. Volume unit: cm3. J KDM4A-AS1 expression in different groups with indicated treatment. K AR, AR-V7, and AR-V567es expressions in different groups. HE staining is listed on the left panel. L Detection of KDM4A-AS1 expression in different groups using qPCR. M Detection of AR signal expression using western blot. GAPDH was used as loading control.
At present, enzalutamide resistance in CRPC patients is relatively common, which is also one of the current treatment dilemmas. Whether KDM4A-AS1 has similar functions in vivo is worthy of further investigation. Therefore, we performed subcutaneous transplantation tumor using C4-2-EnzR cells in nude mice. Detail workflow is shown in Supplementary Fig. 5. We found that there was no significant difference in tumor growth between the different treatment groups after enzalutamide or DMSO (solvent) treatment. Subsequently, 4–6 days after intratumoral injection of shKDM4A-AS1 adeno-associated virus (AAV) or shScramble AAV, the tumor volume in the D: Enz+/AAV+ group gradually decreased, and the tumor growth in the B: Enz-/AAV+ group was significantly slower. Meanwhile, the tumors of the A: Enz-/AAV- and C: Enz+/AAV- groups continued to grow, and there was no significant difference between these two groups (PA-B = 0.0459, PB-C = 0.0122, PA-D = 0.0055, PC-D = 0.0008, Fig. 5I). After the mice were euthanized, tumors in each group were harvested. By comparing the tumor size, significant differences between the different groups can also be observed (Fig. 5H). The above results suggest that in in vivo experiments, knocking down KDM4A-AS1 alone can significantly inhibit tumor growth, while the combined application of enzalutamide can reduce the tumor volume and restore C4-2-EnzR sensitivity to enzalutamide. Subsequently, we divided the tumor into two parts, extracted the total RNA and protein or embedding in paraffin, and using RNAScope (Fig. 5J and Supplementary Fig. 1B), immunohistochemistry (Fig. 5K), qPCR (Fig. 5L), and western blot (Fig. 5M) to detect the expression KDM4A-AS1, AR, AR-V7, and AR-V567es in each group. The results suggest that after knocking down KDM4A-AS1, the expression of AR/AR-Vs in tumors decreased significantly, indicating that KDM4A-AS1 can regulate the protein expression of AR/AR-Vs in vivo. Taken together, in vivo experiments show that knocking down KDM4A-AS1 can significantly reduce the expression of AR/AR-Vs in subcutaneous transplantation tumors. Although inhibiting KDM4A-AS1 alone can also significantly inhibit tumor growth, the combined application with enzalutamide can restore the sensitivity of drug-resistant tumors to enzalutamide, which has important clinical significance.
Antisense oligonucleotide drugs targeting KDM4A-AS1 restore enzalutamide sensitivity in vivo and in vitro
Based on the above results, we further explore the clinical application potential of targeting KDM4A-AS1. We constructed KDM4A-AS1 antisense oligonucleotides, a new type of targeted therapy vector, and first verified the efficacy in vitro (ASO-KDM4A-AS1, Fig. 6A). It can be seen from the results that after the addition of ASO, the expression of KDM4A-AS1 in C4-2-EnzR cells decreased significantly, and subsequently, the protein expression of AR, AR-V7, and AR-V567es decreased significantly (Fig. 6B). In subsequent phenotyping experiments, the addition of ASO-KDM4A-AS1 can significantly inhibit the cell migration ability and clone formation ability of C4-2-EnzR (Fig. 6C, D), and the protein degradation rate of AR/AR-Vs is significantly increased (Fig. 6E). These data indicate that ASO-KDM4A-AS1 can significantly inhibit the activation of AR signaling pathway in vitro, and inhibit the proliferation and migration of C4-2-EnzR cells.
A ASO-KDM4A-AS1 drug depletion efficiency. B Indicated protein expression after drug treatment. C Transwell assay in C4-2-EnzR cell line with ASO-KDM4A-AS1 or PBS treatment. D Colony formation assay in C4-2-EnzR cell line with ASO-KDM4A-AS1 or PBS treatment. E Indicated protein degradation rate in C4-2Enz+/ASO- and C4-2Enz+/ASO+ cell line. F Tumor size after in vivo experiment. G Tumor growth rate of indicated groups. H Blood routine value of indicated groups. I Blood biochemistry value of indicated groups. J KDM4A-AS1 expression in indicated groups, GAPDH used as control. K HE and immunohistochemistry staining of indicated proteins and tissues.
After confirming the in vitro efficacy of ASO-KDM4A-AS1, we further carried out in vivo experiments. As shown in Fig. 6F–G, compared with the control group, intraperitoneal injection of ASO-KDM4A-AS1 (concentration: 10 mg/kg) can significantly reduce the volume of tumors in enzalutamide conditions. In order to verify its biological safety, we collected blood from nude mice and performed blood routine and blood biochemical tests (Fig. 6H, I). The results showed that there was no significant change between the two groups. On the other hand, no significant difference was observed in the HE staining results of kidney tissue and liver tissue of nude mice (Fig. 6K). The above results suggest that ASO-KDM4A-AS1 has high biological safety. We also detected KDM4A-AS1, AR, AR-V7, and AR-V567es in tumor tissues by qPCR and immunohistochemistry (Fig. 6J, K). The results showed that ASO-KDM4A-AS1 can target the expression of KDM4A-AS1, and reduce the protein expression of AR/AR-Vs. Our results show that ASO-KDM4A-AS1 has high clinical application value, and may be able to provide new ideas for the treatment of clinically refractory CRPC.
Discussion
In this study, we first revealed the critical role of KDM4A-AS1 in CRPC. By stabilizing the USP14-AR/AR-Vs complex, KDM4A-AS1 protects AR/AR-Vs from degradation by the ubiquitin-proteasome pathway, promoting the progression of CRPC and resistance to enzalutamide (Fig. 7).
The upper panel shows that in HSPC, KDM4A-AS1 is under-expressed, MDM2 antagonizes USP14 and binds to AR. AR protein is degraded through the ubiquitin-proteasome pathway after being ubiquitinated by MDM2. The lower part shows that in CRPC, KDM4A-AS1 is highly expressed and stabilizes the USP14-AR/AR-Vs complex, KDM4A-AS1 enhances the deubiquitination of AR/AR-Vs, prevents AR/AR-Vs from being degraded by MDM2, and further promotes the progress of CRPC.
LncRNA was reported to be involved in regulating gene expression in multiple aspects [23, 24]. In transcriptional level, lncRNA can participate in the regulation of chromatin structure by recruiting chromatin modifying enzymes, leading to the regulation of downstream genes [25]. For example, GAS5 prevented glucocorticoid receptor from approaching the target DNA by competitively binding to the DNA binding domain of the glucocorticoid receptor [26]. Although LncRNA plays a vital role at the transcription level, the regulation of LncRNA at the translation and post-translational level has been gradually explored. LincRNA-p21 regulated hypoxia-enhanced glycolysis by inhibiting the ubiquitination of hypoxia inducible factor 1α, which in turn led to apoptosis of cells from mice [27]. For the post-translational regulation, lncRNAs such as HOTAIR, GIAT4RA, and ANCR have been reported to be involved in the regulation of ubiquitination pathways. HOTAIR and ANCR may function through stable protein complex and enhance ubiquitin-proteasome degradation [11, 28, 29]. However, HOTAIR was also reported to reduce the ubiquitination degradation of AR by preventing its combination with MDM2, and GIAT4RA was recently reported to interfere the interaction between Uchl3 and LSH to repress ubiquitin-proteasome pathway [30], which indicate multiple roles of lncRNA in ubiquitination. In comparison, in PCa, the research on the mechanism of deubiquitination and lncRNA is not thorough enough.
Herein, for the first time, we demonstrated that KDM4A-AS1 protects AR/AR-Vs from ubiquitin-mediated degradation through enhancing the stability of USP14-AR/AR-Vs complex. Our research also revealed a new potential mechanism of enzalutamide resistant, we found that not only AR, but also AR-V7 and AR-V567es could be stabled by KDM4A-AS1. As the receptor of androgen, in addition to being related to reproduction, AR can also maintain hormone balance, stimulate protein synthesis or metabolism, and promote nitrogen deposition [31]. In prostate malignant tumors, the abnormal expression of AR can maintain and promote the malignant progression [32]. With the deepening of related research, drugs like enzalutamide targeting AR became an important part of ADT and showed a significant benefit to PCa patients. Although most patients respond initially, they become resistant to ADT, and relapse into CRPC, which is highly malignant and incurable [33]. AR mutations, AR amplification, and AR splice variants (AR-Vs) play roles in CRPC ADT resistance, mutations in AR may cause receptor activation by bicalutamide [34]. AR amplification is observed in up to 80% CRPC patients and leading to AR protein overexpression, overexpression of AR contributes to CRPC and poor clinical outcomes [35, 36]. AR splice variants, like AR-V7 and AR-V567es, are often detected in CRPC, and one of the common characteristic of these alternative spliced variants is the lack of ligand binding domain [37, 38]. According to previous research studies, AR-Vs are connected with drug resistance; inhibition of AR-Vs may restore the sensitivity of PCa cells to enzalutamide [39, 40]. Thus, research studies on therapies targeting AR and AR-Vs are urgent and important, among which, the degradation mechanism of AR/AR-Vs is the focus of related research. One of the main degradation processes of AR/AR-Vs is the ubiquitin-proteasome pathway [16, 17, 41]. Ubiquitination is the process of covalently binding ubiquitin to the substrate lysine. It is an important post-translational modification that can regulate various biological processes [42]. Protein ubiquitination has been proved to play an important role in protein degradation. Ubiquitin ligase Siah2 was found to regulate AR ubiquitin-dependent degradation and promote expression of AR target genes in lipid metabolism [43]. Wild-type SPOP was found to enhance ubiquitination and degradation of AR through a specific binding motif (ASSTT) in AR hinge region [44]. Moreover, high expression of MDM2 [45], CHIP [46], or SKP2 [47] were also reported to regulate AR ubiquitination mediated degradation.
Unlike most related studies focusing on the enhancement or inhibition of ubiquitinating enzymes, research on the regulatory mechanism of deubiquitination enzymes on AR is relatively scarce. As a deubiquitinating enzyme, USP14 has previously been reported to enhance the deubiquitination of AR and resist the binding of MDM2, but the specific mechanism is still unclear and needs further exploration. We found that KDM4A-AS1 may be one of the important stabilizing factors, and we further discovered that KDM4A-AS1 has similar regulatory effects on AR-V7 and AR-V567es. Our research confirms that KDM4A-AS1 may inhibit the ubiquitin-mediated degradation process by stabilizing the binding of USP14 to AR or AR-Vs and leading to enzalutamide resistance in CRPC cell lines. Moreover, our in vivo experiments also suggest that KDM4A-AS1 may be a good therapeutic target to CRPC.
Combination therapy committed to solve the problem of drug resistance, and combination therapy for PCa is an emerging concept. Research has proven that the combination of ADT and radiation therapy can significantly increase the survival rates of PCa patients [48]. On the other hand, combination of different drugs may also benefit patients. Anti-MDSC multi-kinase inhibitor combined with anti-PD-L1 therapy may help PCa patients overcome immune escape to increase the efficacy of immunity therapy [49]. As an important RNA that can regulate AR/AR-Vs protein expression and recover PCa cell sensitivity to enzalutamide, target KDM4A-AS1 through nucleic acid drugs may significantly benefit CRPC patients. From the results of this study, the depletion of KDM4A-AS1 alone can significantly inhibit subcutaneous xenograft tumors and cell proliferation, while the combined application of enzalutamide can reverse the resistance of C4-2-EnzR. Therefore, we believe that the combined treatment of the two may become an important treatment for enzalutamide-resistant PCa.
Although we have revealed the important role of KDM4A-AS1 in PCa for the first time here, this is still only a preliminary study. From the results, we can infer that the regulation of AR by KDM4A-AS1 may be only one of its multiple regulatory mechanisms. Whether KDM4A-AS1 can regulate the development of CRPC through AR-independent mechanisms remains to be further studied. From the perspective of clinical application, the safety and effectiveness of ASO drugs are still unclear. Whether targeting KDM4A-AS1 will cause other adverse reactions needs further exploration and verification of clinical cohorts. Although there is no commercial application in the treatment of PCa, it is undeniable that as a very potential treatment, lncRNA such as KDM4A-AS1 is worthy of in-depth exploration. In conclusion, our research highlights the importance of lncRNA KDM4A-AS1-induced stabilization in AR/AR-Vs expression through the USP14-mediated deubiquitination process in CRPC progression and enzalutamide resistance. The inhibition of KDM4A-AS1-USP14-AR/AR-Vs axis is worth a further research as a potential method for patients with enzalutamide resistance.
Materials and methods
Cell culture
Seven PCa cell lines were used in this research, including LNCaP, LNCaP-AI, C4-2, C4-2-EnzR, PC-3, DU145, and 22RV1. LNCaP, C4-2, PC-3, DU145, and 22RV1 were cultured in RPMI-1640 medium (BI, 01-100-1ACS, USA) supplemented with 10% fetal bovine serum (ExCell Bio, FND500, New Zealand). LNCaP-AI was generated in our previous research and was cultured in RPMI-1640 medium (BI, 01-100-1ACS, USA) supplemented with 10% certified charcoal stripped foetal bovine serum (BI, 04-201-1A, USA). As for C4-2-EnzR cell generation, C4-2 cells were first cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and 1 μM enzalutamide (Sigma, 532996, Germany). After the cell growth was not restricted, we increased the enzalutamide concentration gradually at 2, 5, 10, and 20 μM. When C4-2 cells can grow stably in the medium containing 20 μM enzalutamide, we maintain this concentration and pass it 50 times, and consider this cell as an enzalutamide-resistant C4-2 cell (C4-2-EnzR).
Primers, plasmids, and shRNAs
Primers used in this research were provided by Sangon Biotech (Shanghai) Co., Ltd. Lentiviral shRNA and overexpression vectors targeting human KDM4A-AS1 were purchased from Shanghai Genechem Co., Ltd (www.genechem.com; Shanghai, China). AAV targeting nude mice KDM4A-AS1 and a none-targeting control vector were also acquired from Shanghai Genechem. Plasmids overexpressing KDM4A-AS1 and truncated MYC-AR fragments (MYC-AR-FL, MYC-AR-NTD, MYC-AR-NTD+DBD, MYC-AR-DBD+LBD) were generated by cloning cDNAs encoding KDM4A-AS1 and were packaged into CMV-MCS-polyA-EF1A-zsGreen-sv40-puromycin vector (Genechem, Shanghai, China). All sequence details are listed in Supplementary Table 2.
Clinical specimens and tissue microarray
BPH (n = 4), HSPC (n = 28), and CRPC (n = 13) samples were approved by the Ethics Committee of the Second Hospital of Tianjin Medical University. All samples’ disease type were identified by three professional pathologists and clinical doctors. Tissue microarrays using these samples were designed and manufactured by Shanghai Outdo Biotech Company (Cat No. T17–1413 TMA, Lot No. T17–1413).
RNAScope
RNAScope assay was performed with RNAScope® Multiplex Fluorescent Reagent Kit (Advanced Cell Diagnostics, 323100, USA) to detect the expression levels of KDM4A-AS1 in patients and nude mice tissues according to the manual instruction(). The probe targeting KDM4A-AS1 was designed by Advanced Cell Diagnostics (KDM4A-AS1 probe catalog number: REF: 451021, Advanced Cell Diagnostics). Sample was visually scored based on the average number of dots per cell using the following criteria: no staining or <1 dot/10 cells = 0, 1–3 dots/cell = 1, 4–9 dots/cell (none or very few dot clusters) = 2, 10–15 dots/cell (<10% dots are in clusters) = 3, and >15 dots/cell (>10% dots are in clusters) = 4. KDM4A-AS1 expression of samples were scored based on the maximum score of the cell.
Immunohistochemistry
Xenograft tumors were fixed with formalin solution overnight, and embedded in paraffin. Tissue microarrays and tissue sections from xenograft tumor were first deparaffinized. The slides were immersed in 0.01 M sodium citrate buffer (Solarbio, C1010, China) and placed in a microwave oven for 5 min on high heat, and then adjusted to medium-low heat for 10 min to restore the antigen. A total of 3% H2O2 was added to remove endogenous peroxidase in tissue samples. Cover the tissue on the slide with the primary antibody, place it in a humid box, and incubate overnight at 4 °C. After rewarming at room temperature for 30 min, horseradish peroxidase-linked secondary antibody (ZSGB-BIO, PV-6000, China) was added to the specimen and incubate the slides at room temperature for 30 min. After stained with the DAB solution, the slides were immediately placed in water to stop dyeing, slides were subsequently counterstained with hematoxylin. The tissue is then dehydrated and preserved with neutral balsam (OriGene, ZLI-9555, China).
We used the immunohistochemical scoring (IHS) method to score the results by combining the proportion of positive cells and the strength of positive cell staining: A: percentage of positive cells (no positive cells = 0, positive cells accounted for 1–10% = 1, 11–50% = 2, 51–80% = 3, 81–100% = 4), B: positive cell staining strong or weak (negative = 0, weak positive = 1, moderate positive = 2, strong positive = 3). The product of A and B is the IHS of the tissue in this case. To ensure that IHS and RNAScope score ranges are as consistent as possible, we divide the result by 2 to narrow the range.
Western blot (immunoblot analysis)
RIPA protein lysis buffer (Solarbio, Cat#R0020, Beijing, China) and protease inhibitor PMSF were used to extract total cell protein. The volume ratio of RIPA and PMSF is 100 μl:1 μl. We used the Bradford to measure the protein concentration at 595 nm. As for gel prepare, we used the SDS-PAGE Gel Kit (Solarbio, P1200-50T, China) to prepare 10% gel according to its manuscription. The prepared protein sample is added to the gel channel for constant voltage electrophoresis. After the electrophoresis, the protein in the SDS-PAGE gel is transferred to the PVDF membrane. Subsequently, the PVDF membrane was placed in 5% fat-free milk and blocked for 1 h at room temperature. Then the appropriate diluted primary antibody was used to incubate the PVDF membrane at 4 °C overnight. On the second day, wash the PVDF membrane three times with TBST for 10 min each time. After washing, the blots were incubated with HRP conjugated anti rabbit or antimice IgG for 1 h. The blots were developed in ECL mixture (Vector Laboratories, Burlingame, CA) and visualized by Imager. The corresponding antibody and dilution ratio are shown in the list A.
RNA isolation and quantitative real-time PCR
Trizol reagent (Invitrogen, 15596018, USA) was used to isolated cell total RNA. Then, the RNA was used as a template strand for reverse transcription into cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific,00887496) according to the operation manual. Finally, Biosystems 7900 Real Time PCR System (Thermo Scientific) and SYBR Green PCR Master Mix (Roche) were used to analyze the cDNA following the Instruction manual. GAPDH was used as an internal control. The 2-ΔΔCt method was used to calculate the relative quantitative value. All primers used are listed in Supplementary Table 2.
MTT assay
The MTT experiment was used to detect the effect of shKDM4A-AS1 on the viability of PCa cells and the sensitivity of C4-2-EnzR to enzalutamide. Seed cells in 96-well plates. Each cell line has six repeat wells, each well was planted with 6 × 103 cells, and the cells in the 96-well plate were cultured in a cell incubator at 37 °C with corresponding medium. After 24, 48, and 72 h, 30 μl of 5 mg/ml MTT (Solarbio, M8180, China) solution was added to each well, and incubated at 37 °C for 4 h. Then 150 μl of DMSO was added to each well and shaken on a shaker for 20 min. Finally, the absorbance value of each well was detected with a microplate reader at 490 nm.
Transwell assay
Transwell was used to evaluate the migration of cells. Add 200 μl of serum-free RPMI-1640 medium to the upper chamber of the transwell chamber (Corning, 09820034, USA), and seed 2 × 104 cells in it. Add 800 μl of serum-containing medium to the lower chamber. Transwell chambers were cultured at 37 °C with 5% CO2. After 48–72 h, chambers seeded with C4-2 cells were taken out and fixed with 4% paraformaldehyde (Biosharp, BL539A, China) at 4 °C for 30 min, followed by staining with crystal violet for 30 min at room temperature. As for LNCaP-AI cells, chambers seeded with LNCap-AI cells were taken out after 96–120 h and treated in the same way. Finally, wipe off the cells in the upper chamber with a cotton swab. Graphs were taken by microscope and subsequently analyzes by ImageJ software.
Colony formation assay
The monolayer cultured cells were digested with 0.25% Trypsin-EDTA solution (Solarbio, T1320, China) and pipetted into single cells. The cells were seeded at 1 × 103 per well in a 6-well plate, and was cultured in a 37 °C cell incubator with 5% CO2. After 14 days, remove the medium in the 6-well plate and fix the cells with 4% paraformaldehyde at 4 °C for 30 min, then remove the paraformaldehyde and stain with crystal violet at room temperature for 30 min. After removing the crystal violet, dry the plate at room temperature. Graphs were taken by camera and subsequently analyzed by ImageJ software.
RNA immunoprecipitation (RIP)
Magna RIPTM Kit (Millipore, 3480215, USA) was used to detect the combination of long noncoding RNA KDM4A-AS1 with AR and USP14 antibody according to the manufacturer’s protocol. Briefly, cells (1 × 107) were lysed with ice -cold RIP Lysis Buffer. Collect and store the cell lysate at −80 °C. Prepare the magnetic bead-antibody complex with 5 μg of the above-mentioned target antibody or control IgG and 50 μl of protein A/G magnetic beads, rotating at room temperature for 30 min. Take an equal volume of cell lysate and incubate the magnetic bead-antibody complex with rotation at 4 °C overnight so that the antibody can fully contact and bind to the protein. At the same time,10 μl cell lysate was extracted and used as input. Next day, RNA was extracted and purified with the prepared proteinase K buffer. Acquired RNA was used as a template to synthesize the corresponding cDNA. Use PCR technology to amplify the obtained cDNA and the products were subjected to agarose gel electrophoresis analysis or qPCR.
RNA pulldown assay
RNA pulldown assay was performed using PierceTM Magnetic RNA-Protein Pull-Down Kit (Thermo Scientific, 20164, USA) according to the operation instruction. Before RNA pulldown. MEGAscript T7 Transcription Kit (Thermo Scientific, AM1333, USA) was used to generate KDM4A-AS1 RNA from pre-cut plasmid provided by FulenGen (Guangzhou, China). PierceTM RNA 3’ End Desthiobiotinylation Kit (Thermo Scientific, 20163, USA) was used to label the in vitro transcription product of KDM4A-AS1 with biotin. Proteins bound to KDM4A-AS1 were subjected to western bolt or iTRAQ.
Co-immunoprecipitation (Co-IP) assay
Co-IP assay was performed with Pierce Classic Magnetic IP/Co-IP Kit (Thermo Scientific, 88804, USA) following the manufacturer’s suggestions. In short, add ice-cold IP Lysis/Wash Buffer to the cell. Transfer the lysate to a 1.5 ml microcentrifuge tube and centrifuge at ~13,000 × g for 10 min to acquire supernatant. Incubate acquired protein (500 μg) with 5 μg of AR antibody or IgG overnight with rotation at 4 °C. Bind antigen/antibody complex to Protein A/G magnetic beads for 1 h at room temperature. Then, wash beads twice with IP Lysis/Wash Buffer and once with purified water. Elute the antigen/antibody complex. Western blot assay was performed to detect AR, AR-V7, AR-V567es, USP14, MDM2 with corresponding antibodies. GAPDH antibody was used as an internal control.
In vitro ubiquitination assay
C4-2, C4-2-shKDM4A-AS1, LNCaP-AI, and LNCaP-AI-shKDM4A-AS1 cells were treated with 10 μM proteasome inhibitor MG-132 for 10 h. Then, the cells were lysed with IP Lysis/Wash Buffer, followed by immunoprecipitation with AR antibody and western blotting analysis with ubiquitin or AR antibody. GAPDH antibody was used as an internal control.
In vivo animal experiments
The animal experiments involved in our research were approved by Tianjin Institute of Urology, Tianjin, China. Six-week-old male nude mice were purchased from Beijing HFK Bioscience Co. Ltd. (Beijing, China). Male nude mice were divided into two groups: one group was subcutaneously implanted with C4-2 cells (5 × 106), and the other group was injected with C4-2-EnzR cells (5 × 106). Enzalutamide was administered once a day via oral gavage at 10 mg/kg in 1% carboxymethyl cellulose, 0.1% Tween-80, and 5% DMSO. Two weeks later, the C4-2 nude mice model group (n = 10) and the C4-2-EnzR nude mice model group (n = 10) were randomly divided into two groups, and were injected with AAV carrying control shRNA or AAV carrying KDM4A-AS1 shRNA, respectively. The tumor size was monitored every 2 days for 2 weeks with formula V = 1/2 × length × width2. After a week, tumor samples were harvested and subjected to western blot, qRT-PCR, RNAScope, hematoxylin-eosin staining, and immunohistochemistry analysis (Supplementary Fig. 1).
As for ASO in vivo experiments, 6-week-old male nude mice were subcutaneously implanted with C4-2-EnzR cells (5 × 106). Enzalutamide was treated as above at day 7, and mice were divided into two groups at day 14: one was injected with PBS solution, the other group was intraperitoneal injected with ASO-KDM4A-AS1 at the concentration of 10 mg/kg. The tumor size was monitored every 2 days for 2 weeks with formula V = 1/2 × length × width2. After 2 weeks, tumor samples were harvested and subjected to western blot, qRT-PCR, hematoxylin-eosin staining, and immunohistochemistry analysis. The ASO-KDM4A-AS1 was conducted by Guangzhou Ribobio Co., Ltd.
Antibodies, sequences, and primers
Antibody, sequence, and primer detail information can be found in Supplementary Table 2.
Kaplan–Meier survival analysis
Kaplan–Meier survival analysis was performed to compare the difference between high KDM4A-AS1 expression patients and low KDM4A-AS1 expression patients. Survival analysis of TCGA patients was done by GEPIA website (http://gepia.cancer-pku.cn/). TCGA patients were divided into two groups according to the median expression of KDM4A-AS1. The end point was patients’ DFS, which is given by TCGA clinical information. As for the patients in tissue microarray, patients with score >2 were grouped as high risk and patients with score ≤2 were grouped as low risk. The end point of DFS was defined by the value of PSA and the follow-up results. When the patient’s PSA rises by 0.2 ng/ml for two consecutive times, the disease is considered to have recurred. The patient’s survival status is confirmed by the follow-up results. As for data calculation and graph drawing, R package “survminer” and “survival” were used. “survdiff()” and “survfit()” codes were used for analysis.
Statistical analysis
All statistical significance were calculated by R-4.0.0. “edgeR” package was used for counting differentially expressed genes. “Survminer” and “survival” package were used for Kaplan–Meier survival analysis. GraphPad-prism 8, “ggplot2” and “ggsci” package were used for image drawing and output.
Student’s t-test was used for paired comparisons. ANOVA is used to compare multiple groups. Log-rank P value was used in survival analysis. “Mean ± SD” was used for experiments with data points ≥5, “mean ± SEM” was used for experiments with data points <5.
Significant status, na: P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Experimental and biological replicates
All qPCR experiments were done three times, with three biological replicates each time. All western blots were done at least three times, among which, Co-IPs and RIPs were repeated at least three times to ensure the accuracy, two replicates were used each time. MTT assays were repeated three times, with six biological replicates each time. Transwell assays were repeated three times, with three replicates each time. Colony formation assays were repeated three times, with three biological replicates each time. For Transwell assays, we took six unique photos for each biological replicate and counted them using ImageJ software.
Code availability
All R codes used in this research are available, please contact BZ (zhangboya_0722@163.com).
Change history
31 July 2023
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1038/s41388-023-02790-8
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol. 2009;6:76–85.
Zhang A, Zhao JC, Kim J, Fong KW, Yang YA, Chakravarti D, et al. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 2015;13:209–21.
Shang Z, Yu J, Sun L, Tian J, Zhu S, Zhang B, et al. LncRNA PCAT1 activates AKT and NF-kappaB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKalpha complex. Nucleic Acids Res. 2019;47:4211–25.
Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407.
Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–7.
Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43.
Grillone K, Riillo C, Scionti F, Rocca R, Tradigo G, Guzzi PH, et al. Non-coding RNAs in cancer: platforms and strategies for investigating the genomic “dark matter”. J Exp Clin Cancer Res. 2020;39:117.
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18:143.
Li Z, Hou P, Fan D, Dong M, Ma M, Li H, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24:59–71.
Montgomery RB, Mostaghel EA, Vessella R, Hess DL, Kalhorn TF, Higano CS, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68:4447–54.
Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308.
Liao Y, Xia X, Liu N, Cai J, Guo Z, Li Y, et al. Growth arrest and apoptosis induction in androgen receptor-positive human breast cancer cells by inhibition of USP14-mediated androgen receptor deubiquitination. Oncogene. 2018;37:1896–910.
Liao Y, Liu N, Hua X, Cai J, Xia X, Wang X, et al. Proteasome-associated deubiquitinase ubiquitin-specific protease 14 regulates prostate cancer proliferation by deubiquitinating and stabilizing androgen receptor. Cell Death Dis. 2017;8:e2585.
Liu C, Yang JC, Armstrong CM, Lou W, Liu L, Qiu X, et al. AKR1C3 promotes AR-V7 protein stabilization and confers resistance to AR-targeted therapies in advanced prostate cancer. Mol Cancer Ther. 2019;18:1875–86.
Li Y, Xie N, Gleave ME, Rennie PS, Dong X. AR-v7 protein expression is regulated by protein kinase and phosphatase. Oncotarget. 2015;6:33743–54.
Yin Y, Li R, Xu K, Ding S, Li J, Baek G, et al. Androgen receptor variants mediate DNA repair after prostate cancer irradiation. Cancer Res. 2017;77:4745–54.
Kim TD, Jin F, Shin S, Oh S, Lightfoot SA, Grande JP, et al. Histone demethylase JMJD2A drives prostate tumorigenesis through transcription factor ETV1. J Clin Invest. 2016;126:706–20.
Muppirala UK, Honavar VG, Dobbs D. Predicting RNA-protein interactions using only sequence information. BMC Bioinforma. 2011;12:489.
Liu C, Lou W, Yang JC, Liu L, Armstrong CM, Lombard AP, et al. Proteostasis by STUB1/HSP70 complex controls sensitivity to androgen receptor targeted therapy in advanced prostate cancer. Nat Commun. 2018;9:4700.
Kwegyir-Afful AK, Ramalingam S, Purushottamachar P, Ramamurthy VP, Njar VC. Galeterone and VNPT55 induce proteasomal degradation of AR/AR-V7, induce significant apoptosis via cytochrome c release and suppress growth of castration resistant prostate cancer xenografts in vivo. Oncotarget. 2015;6:27440–60.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14.
Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–81.
Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66.
Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8.
Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19.
Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939.
Zhang H, Diab A, Fan H, Mani SK, Hullinger R, Merle P, et al. PLK1 and HOTAIR accelerate proteasomal degradation of SUZ12 and ZNF198 during hepatitis B virus-induced liver carcinogenesis. Cancer Res. 2015;75:2363–74.
Yang R, Liu N, Chen L, Jiang Y, Shi Y, Mao C, et al. GIAT4RA functions as a tumor suppressor in non-small cell lung cancer by counteracting Uchl3-mediated deubiquitination of LSH. Oncogene. 2019;38:7133–45.
Davey RA, Grossmann M. Androgen receptor structure, function and biology: from bench to bedside. Clin Biochem Rev. 2016;37:3–15.
Taplin ME, Balk SP. Androgen receptor: a key molecule in the progression of prostate cancer to hormone independence. J Cell Biochem. 2004;91:483–90.
Seruga B, Ocana A, Tannock IF. Drug resistance in metastatic castration-resistant prostate cancer. Nat Rev Clin Oncol. 2011;8:12–23.
Yoshida T, Kinoshita H, Segawa T, Nakamura E, Inoue T, Shimizu Y, et al. Antiandrogen bicalutamide promotes tumor growth in a novel androgen-dependent prostate cancer xenograft model derived from a bicalutamide-treated patient. Cancer Res. 2005;65:9611–6.
Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharm. 2008;8:440–8.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22.
Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–13.
Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–30.
Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, et al. Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin Cancer Res. 2014;20:3198–210.
Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–9.
Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–62.
Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125:531–7.
Qi J, Tripathi M, Mishra R, Sahgal N, Fazli L, Ettinger S, et al. The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell. 2013;23:332–46.
Geng C, Rajapakshe K, Shah SS, Shou J, Eedunuri VK, Foley C, et al. Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 2014;74:5631–43.
Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21:4037–48.
Sarkar S, Brautigan DL, Parsons SJ, Larner JM. Androgen receptor degradation by the E3 ligase CHIP modulates mitotic arrest in prostate cancer cells. Oncogene. 2014;33:26–33.
Li B, Lu W, Yang Q, Yu X, Matusik RJ, Chen Z. Skp2 regulates androgen receptor through ubiquitin-mediated degradation independent of Akt/mTOR pathways in prostate cancer. Prostate. 2014;74:421–32.
Shipley WU, Seiferheld W, Lukka HR, Major PP, Heney NM, Grignon DJ, et al. Radiation with or without antiandrogen therapy in recurrent prostate cancer. N Engl J Med. 2017;376:417–28.
Lu X, Horner JW, Paul E, Shang X, Troncoso P, Deng P, et al. Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature. 2017;543:728–32.
Acknowledgements
This work was supported by National Natural Science Foundation of China (grants 81872100 and 81772756), Natural Science Foundation of Tianjin (18PTLCSY00030 and 19JCYBJC24900), and Tianjin Research Innovation Project for Postgraduate Students.
Author information
Authors and Affiliations
Contributions
BZ: project design, data collection, high-throughput data analysis, in vitro experiments, in vivo experiments, article writing; MZ: project design, partial data collection, in vitro experiments, in vivo experiments, article writing; QL: patient tissue collection, cell culture, virus transfection, animal feeding; YY: case collection, cell culture, animal breeding, nude mice euthanized; JY: provide antibodies, part of the project design; SZ: provided clinical information; YN: project design, experimental guidance; ZS: project design, experimental guidance, and reagents provided.
Corresponding author
Ethics declarations
Ethics approval
All related experiments were approved by the Ethics Committee of the Second Hospital of Tianjin Medical University.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1038/s41388-023-02790-8
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, B., Zhang, M., Yang, Y. et al. RETRACTED ARTICLE: Targeting KDM4A-AS1 represses AR/AR-Vs deubiquitination and enhances enzalutamide response in CRPC. Oncogene 41, 387–399 (2022). https://doi.org/10.1038/s41388-021-02103-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02103-x
This article is cited by
-
Targeting epigenetic regulators to overcome drug resistance in cancers
Signal Transduction and Targeted Therapy (2023)
-
Post-transcriptional modification of m6A methylase METTL3 regulates ERK-induced androgen-deprived treatment resistance prostate cancer
Cell Death & Disease (2023)
-
E2F1-mediated KDM4A-AS1 up-regulation promotes EMT of hepatocellular carcinoma cells by recruiting ILF3 to stabilize AURKA mRNA
Cancer Gene Therapy (2023)
-
The crosstalk between ubiquitination and endocrine therapy
Journal of Molecular Medicine (2023)