Abstract
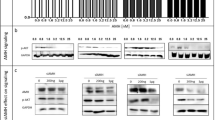
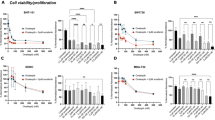
The enzyme iodothyronine deiodinase type 3 (DIO3) contributes to cancer proliferation by inactivating the tumor-suppressive actions of thyroid hormone (T3). We recently established DIO3 involvement in the progression of high-grade serous ovarian cancer (HGSOC). Here we provide a link between high DIO3 expression and lower survival in patients, similar to common disease markers such as Ki67, PAX8, CA-125, and CCNE1. These observations suggest that DIO3 is a logical target for inhibition. Using a DIO3 mimic, we developed original DIO3 inhibitors that contain a core of dibromomaleic anhydride (DBRMD) as scaffold. Two compounds, PBENZ-DBRMD and ITYR-DBRMD, demonstrated attenuated cell counts, induction in apoptosis, and a reduction in cell proliferation in DIO3-positive HGSOC cells (ES-2 and KURAMOCHI), but not in DIO3-negative normal ovary cells (CHOK1) and ES-2 depleted for DIO3 or its substrate, T3. Potent tumor inhibition with a high safety profile was further established in HGSOC xenograft model, with no effect in DIO3-depleted tumors. The antitumor effects are mediated by downregulation in an array of pro-cancerous proteins, the majority of which known to be repressed by T3. To conclude, using small molecules that specifically target the DIO3 enzyme we present a new treatment paradigm for ovarian cancer and potentially other DIO3-dependent malignancies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
15 March 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41388-023-02629-2
References
Luongo C, Dentice M, Salvatore D. Deiodinases and their intricate role in thyroid hormone homeostasis. Nat Rev Endocrinol. 2019;15:479–88.
Goemann IM, Marczyk VR, Romitti M, Wajner SM, Maia AL. Current concepts and challenges to unravel the role of iodothyronine deiodinases in human neoplasias. Endocr Relat Cancer. 2018;25:R625–R645.
Goemann IM, Romitti M, Meyer ELS, Wajner SM, Maia AL. Role of thyroid hormones in the neoplastic process: an overview. Endocr Relat Cancer. 2017;24:R367–R385.
Catalano V, Dentice M, Ambrosio R, Luongo C, Carollo R, Benfante A, et al. Activated thyroid hormone promotes differentiation and chemotherapeutic sensitization of colorectal cancer stem cells by regulating Wnt and BMP4 signaling. Cancer Res. 2016;76:1237–44.
Dentice M, Luongo C, Ambrosio R, Sibilio A, Casillo A, Iaccarino A, et al. beta-Catenin regulates deiodinase levels and thyroid hormone signaling in colon cancer cells. Gastroenterology. 2012;143:1037–47.
Dentice M, Luongo C, Huang S, Ambrosio R, Elefante A, Mirebeau-Prunier D, et al. Sonic hedgehog-induced type 3 deiodinase blocks thyroid hormone action enhancing proliferation of normal and malignant keratinocytes. Proc Natl Acad Sci USA. 2007;104:14466–71.
Di Girolamo D, Ambrosio R, De Stefano MA, Mancino G, Porcelli T, Luongo C, et al. Reciprocal interplay between thyroid hormone and microRNA-21 regulates hedgehog pathway–driven skin tumorigenesis. J Clin Investig. 2016;126:2308–20.
Romitti M, Wajner SM, Ceolin L, Ferreira CV, Ribeiro R, Rohenkohl HC, et al. MAPK and SHH pathways modulate type 3 deiodinase expression in papillary thyroid carcinoma. Endocr Relat Cancer. 2016;23:135–46.
Nappi A, De Stefano MA, Dentice M, Salvatore D. Deiodinases and cancer. Endocrinology. 2021;162:bqab016.
Moskovich D, Alfandari A, Finkelshtein Y, Weisz A, Katzav A, Kidron D, et al. DIO3, the thyroid hormone inactivating enzyme, promotes tumorigenesis and metabolic reprogramming in high grade serous ovarian cancer. Cancer Lett. 2021;501:224–33.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7–33.
Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393:1240–53.
Renko K, Schäche S, Hoefig CS, Welsink T, Schwiebert C, Braun D, et al. An improved nonradioactive screening method identifies genistein and xanthohumol as potent inhibitors of iodothyronine deiodinases. Thyroid. 2015;25:962–8.
Steegborn C, Schweizer U. Structure and mechanism of iodothyronine deiodinases–what we know, what we don’t know, and what would be nice to know. Exp Clin Endocrinol Diabetes. 2020;128:375–8.
Manna D, Mugesh G. A chemical model for the inner‐ring deiodination of thyroxine by iodothyronine deiodinase. Angew Chem Int Ed. 2010;49:9246–9.
Manna D, Mugesh G. Deiodination of thyroid hormones by iodothyronine deiodinase mimics: does an increase in the reactivity alter the regioselectivity? J Am Chem Soc. 2011;133:9980–3.
Manna D, Mugesh G. Regioselective deiodination of thyroxine by iodothyronine deiodinase mimics: an unusual mechanistic pathway involving cooperative chalcogen and halogen bonding. J Am Chem Soc. 2012;134:4269–79.
Mondal S, Mugesh G. Biomimetic deiodination of thyroid hormones and iodothyronamines–a structure–activity relationship study. Org Biomolecular Chem. 2016;14:9490–9500.
Raja K, Mugesh G. Remarkable effect of chalcogen substitution on an enzyme mimetic for deiodination of thyroid hormones. Angew Chem Int Ed. 2015;54:7674–8.
Győrffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208.
Schweizer U, Schlicker C, Braun D, Köhrle J, Steegborn C. Crystal structure of mammalian selenocysteine-dependent iodothyronine deiodinase suggests a peroxiredoxin-like catalytic mechanism. Proc Natl Acad Sci USA. 2014;111:10526–31.
Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126.
Yaginuma Y, Westphal H. Abnormal structure and expression of the p53 gene in human ovarian carcinoma cell lines. Cancer Res. 1992;52:4196–9.
Bargonetti J, Prives C. Gain-of-function mutant p53: history and speculation. J Mol Cell Biol. 2019;11:605–9.
Cole AJ, Dwight T, Gill AJ, Dickson K-A, Zhu Y, Clarkson A, et al. Assessing mutant p53 in primary high-grade serous ovarian cancer using immunohistochemistry and massively parallel sequencing. Sci Rep. 2016;6:1–12.
Simpkins F, Jang K, Yoon H, Hew KE, Kim M, Azzam DJ, et al. Dual Src and MEK inhibition decreases ovarian cancer growth and targets tumor initiating stem-like cells. Clin Cancer Res. 2018;24:4874–86.
Nguyen VHL, Hough R, Bernaudo S, Peng C. Wnt/β-catenin signalling in ovarian cancer: Insights into its hyperactivation and function in tumorigenesis. J Ovarian Res. 2019;12:1–17.
Ghannam-Shahbari D, Jacob E, Kakun RR, Wasserman T, Korsensky L, Sternfeld O, et al. PAX8 activates a p53-p21-dependent pro-proliferative effect in high grade serous ovarian carcinoma. Oncogene. 2018;37:2213–24.
Dentice M, Antonini D, Salvatore D. Type 3 deiodinase and solid tumors: an intriguing pair. Expert Opin Ther Targets. 2013;17:1369–79.
Hernandes MZ, Cavalcanti SMT, Moreira DRM, de Azevedo Junior WF, Leite ACL. Halogen atoms in the modern medicinal chemistry: hints for the drug design. Curr Drug Targets. 2010;11:303–14.
Wilcken R, Zimmermann MO, Lange A, Joerger AC, Boeckler FM. Principles and applications of halogen bonding in medicinal chemistry and chemical biology. J Med Chem. 2013;56:1363–88.
Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E–mutated colorectal cancer. N. Engl J Med. 2019;381:1632–43.
Dhillon S. Lonafarnib: first approval. Drugs. 2021;81:283–9.
Smith BD, Kaufman MD, Lu W-P, Gupta A, Leary CB, Wise SC, et al. Ripretinib (DCC-2618) is a switch control kinase inhibitor of a broad spectrum of oncogenic and drug-resistant KIT and PDGFRA variants. Cancer Cell. 2019;35:738–51.e739.
Andrea VR. The role of halogen bonding in inhibitor recognition and binding by protein kinases. Curr Top Med Chem. 2007;7:1336–48.
Wasik R, Łebska M, Felczak K, Poznański J, Shugar D. Relative role of halogen bonds and hydrophobic interactions in inhibition of human protein kinase CK2α by tetrabromobenzotriazole and some C (5)-substituted analogues. J Phys Chem B. 2010;114:10601–11.
Contreras-Jurado C, Alonso-Merino E, Saiz-Ladera C, Valiño AJ, Regadera J, Alemany S, et al. The thyroid hormone receptors inhibit hepatic interleukin-6 signaling during endotoxemia. Sci Rep. 2016;6:1–12.
Suarez J, Scott BT, Suarez-Ramirez JA, Chavira CV, Dillmann WH. Thyroid hormone inhibits ERK phosphorylation in pressure overload-induced hypertrophied mouse hearts through a receptor-mediated mechanism. Am J Physiol Cell Physiol. 2010;299:C1524–C1529.
Flores-Morales A, Gullberg H, Fernandez L, Ståhlberg N, Lee NH, Vennström B, et al. Patterns of liver gene expression governed by TRβ. Mol Endocrinol. 2002;16:1257–68.
García-Silva S, Aranda A. The thyroid hormone receptor is a suppressor of ras-mediated transcription, proliferation, and transformation. Mol Cell Biol. 2004;24:7514–23.
Punchihewa C, Inoue A, Hishiki A, Fujikawa Y, Connelly M, Evison B, et al. Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication. J Biol Chem. 2012;287:14289–300.
Perez-Juste G, Aranda A. The cyclin-dependent kinase inhibitor p27Kip1 is involved in thyroid hormone-mediated neuronal differentiation. J Biol Chem. 1999;274:5026–31.
Pérez-Juste G, Garcı́a-Silva S, Aranda A. An element in the region responsible for premature termination of transcription mediates repression of c-myc gene expression by thyroid hormone in neuroblastoma cells. J Biol Chem. 2000;275:1307–14.
Gagne R, Green JR, Dong H, Wade MG, Yauk CL. Identification of thyroid hormone receptor binding sites in developing mouse cerebellum. BMC Genomics. 2013;14:1–14.
Nappi A, Murolo M, Sagliocchi S, Miro C, Cicatiello AG, Di Cicco E, et al. Selective inhibition of genomic and non-genomic effects of thyroid hormone regulates muscle cell differentiation and metabolic behavior. Int J Mol Sci. 2021;22:7175.
Porlan E, Vidaurre OG, Rodríguez-Peña A. Thyroid hormone receptor-β (TRβ1) impairs cell proliferation by the transcriptional inhibition of cyclins D1, E and A2. Oncogene. 2008;27:2795–800.
Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31:139–70.
Shinderman-Maman E, Cohen K, Weingarten C, Nabriski D, Twito O, Baraf L, et al. The thyroid hormone-alphavbeta3 integrin axis in ovarian cancer: regulation of gene transcription and MAPK-dependent proliferation. Oncogene. 2016;35:1977–87.
Cohen K, Ellis M, Khoury S, Davis PJ, Hercbergs A, Ashur-Fabian O. Thyroid hormone is a MAPK-dependent growth factor for human myeloma cells acting via alphavbeta3 integrin. Mol Cancer Res. 2011;9:1385–94.
Acknowledgements
The work of DM was done in partial fulfillment of the requirements for a PhD degree from the Sackler Faculty of Medicine, Tel Aviv University, Israel. OA-F and BL received support from the Israel Innovation Authority, Nofar Program for Applied Research in Academia, Ministry of Economics (Project 59435).
Author information
Authors and Affiliations
Contributions
DM preformed, analyzed, and interpreted the experimental data. AA, AR, and YF performed some of the methods. AW, AK, and DK assisted in the IHC assays. GM, TL, and BL designed and developed the DIO3 inhibitors. SM and HU performed enzymatic inhibition assays. OA-F designed, analyzed, and interpreted the experimental data. DM, GM, BL, ME, and OA-F wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
BL, GM, TL, and OA-F hold a patent for DIO3 inhibitors or are involved in patent commercialization. The other authors declare no potential conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Moskovich, D., Finkelshtein, Y., Alfandari, A. et al. Targeting the DIO3 enzyme using first-in-class inhibitors effectively suppresses tumor growth: a new paradigm in ovarian cancer treatment. Oncogene 40, 6248–6257 (2021). https://doi.org/10.1038/s41388-021-02020-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02020-z
This article is cited by
-
Expression of decitabine-targeted oncogenes in meningiomas in vivo
Neurosurgical Review (2022)