Abstract
Uncovering the mechanisms that underpin how tumor cells adapt to microenvironmental stress is essential to better understand cancer progression. The HACE1 (HECT domain and ankyrin repeat-containing E3 ubiquitin-protein ligase) gene is a tumor suppressor that inhibits the growth, invasive capacity, and metastasis of cancer cells. However, the direct regulatory pathways whereby HACE1 confers this tumor-suppressive effect remain to be fully elucidated. In this report, we establish a link between HACE1 and the major stress factor, hypoxia-inducible factor 1 alpha (HIF1α). We find that HACE1 blocks the accumulation of HIF1α during cellular hypoxia through decreased protein stability. This property is dependent on HACE1 E3 ligase activity and loss of Ras-related C3 botulinum toxin substrate 1 (RAC1), an established target of HACE1 mediated ubiquitinylation and degradation. In vivo, genetic deletion of Rac1 reversed the increased HIF1α expression observed in Hace1–/– mice in murine KRasG12D-driven lung tumors. An inverse relationship was observed between HACE1 and HIF1α levels in tumors compared to patient-matched normal kidney tissues, highlighting the potential pathophysiological significance of our findings. Together, our data uncover a previously unrecognized function for the HACE1 tumor suppressor in blocking HIF1α accumulation under hypoxia in a RAC1-dependent manner.
Similar content being viewed by others
Introduction
The E3 ubiquitin-protein ligase HACE1 gene was first identified as a tumor suppressor gene in sporadic Wilms’ tumor (WT), based on its location at a WT translocation breakpoint and loss of expression compared to the normal kidney [1, 2]. Full-length HACE1 protein consists of six N-terminal ankyrin repeats responsible for protein–protein interactions and a C-terminal HECT domain responsible for its E3 ligase activity [1]. In addition to WT, HACE1 inactivation has been reported in multiple other tumor types, including non-Hodgkin’s lymphoma, as well as lung, ovarian, pancreatic, and prostate carcinomas [1, 3,4,5,6,7,8,9,10,11,12,13,14]. Consistent with this, genetic inactivation of HACE1 in mice leads to the development of multiple late-onset tumors, including sarcomas, breast, lung, and other carcinomas, as well as lymphomas [3]. Recently, HACE1 was described as a potential tumor suppressor gene in osteosarcoma [13], where it was found to inhibit growth, as well as invasive and metastatic capacity in vivo [13]. However, the mechanism whereby HACE1 elicits these anti-tumorigenic effects remain incompletely characterized.
The most well-characterized E3 ligase target of HACE1 is RAC1 [10, 15,16,17,18]. We previously reported that HACE1 targets RAC1 for degradation when the latter is localized to the membrane nicotinamide adenine dinucleotide phosphate (NADPH) oxidase holoenzyme, inhibiting the generation of de novo reactive oxygen species (ROS) by RAC1-dependent NADPH oxidases [15]. RAC1 was originally identified as an oncogene promoting cancer cell survival and metastasis and essential for transformation by mutant KRAS [19]. Small GTPases (20–25 kDa) including the RAC1 cycle between their inactive inert GDP-bound and active GTP-bound states [20, 21]. Upon activation, interaction with effector proteins leads to stimulation of downstream signaling pathways that can impact cell migration and invasion [22,23,24].
Hypoxia-inducible factor-1alpha (HIF1α) is a transcription factor that is a key regulator of metastasis [25, 26]. HIF1α is induced following the onset of hypoxic stress, where it transcriptionally activates a wide array of genes that are important for the re-programming of multiple pathways that impact cell survival under hypoxia, such as angiogenesis [27,28,29]. Enhanced HIF1α levels have been reported in both low-grade and advanced tumors, suggesting a crucial role for HIF1α in oncogenesis [30,31,32,33,34]. Previous reports have shown that RAC1 is activated by stress stimuli including hypoxia [35, 36]. In addition, HIF1α induction under hypoxia requires activation of RAC1 [36,37,38,39]. Despite these established relationships between HACE1 and RAC1, and RAC1 and HIF1α, potential links between HACE1 and HIF1α have not been investigated.
In this study, we explore whether HACE1 influences HIF1α levels. We establish that HACE1 is induced at the mRNA and protein level under hypoxia. HACE1 reduces HIF1α accumulation in an E3 ligase dependent manner by reducing active RAC1 levels. In line with its putative tumor-suppressive role, the absence of HACE1 correlated with enhanced HIF1α protein levels in WT tissues compared to the patient-matched normal kidneys. Together, these data provide additional mechanistic insights into the tumor suppressor activity of HACE1, namely through the regulation of HIF1α.
Results
HACE1 expression is induced under diverse forms of cell stress
It has previously been suggested that HACE1’s tumor suppressor activity may be linked to cellular stress responses [3]. To further investigate this possibility, we examined HACE1 expression in HEK293 cells subjected to prototypical stress forms, namely short-term nutrient deprivation, hypoxia, endoplasmic reticulum (ER) stress, and γ-irradiation, as described in “Experimental procedures”. HACE1 mRNA expression was increased by each of these stress forms, as shown in Fig. 1A. Since we previously studied HACE1 functions under oxidative stress [15], we explored its role during hypoxia for the present study. Under hypoxia, HACE1 mRNA expression increased significantly and in a temporal manner (Fig. 1B). This was recapitulated at the protein level, whereby HACE1 increased in a time-dependent manner in hypoxia-exposed HEK293 and MCF7 breast cancer cells (Fig. 1C, D). We, therefore, further probed the consequences of HACE1 induction in response to hypoxia. We performed Affymetrix gene expression profiling to compare vector alone control cells (MSCV) and HEK293 cells overexpressing HACE1 under normoxic versus hypoxic conditions (Fig. 1E, F, see Fig. S1 for relative HACE1 expression). In control cells exposed to normoxia and hypoxia, clear patterns of differential gene expression were observed (n = 327 genes with fold change > 2 and FDR < 0.05). This response was largely muted in HACE1 overexpressing cells, where only a subset of 26 of the above 327 genes met the same differential expression criteria (Fig. 1E and Table S1). Overlap of these differential expression sets revealed that only 14 genes (3 upregulated; 11 downregulated) were shared, suggesting that HACE1 overexpression mitigates the cellular response to hypoxia. This effect was clearly visible when comparing gene expression profiles under hypoxia vs. normoxia, where differentially expressed clusters in control cells under stress were reduced in cells overexpressing HACE1 (Fig. 1F). Together, these data indicate that HACE1 is induced in response to diverse cellular stress and mitigates the transcriptional response under hypoxic conditions.
A Relative expression levels of HACE1 mRNA under control and the indicated cellular stress conditions, as determined by qPCR. Data shown represent mean ± SD, n = 2 (*P < 0.05). B HEK293 cells were exposed to 1% O2 for the indicated times, followed by qPCR to determine HACE1 mRNA levels. Data shown represent mean ± SD, n = 3 (*P < 0.05; **P < 0.01). C Endogenous levels of HACE1 protein were examined in HEK293 cells cultured under 1% O2 for the indicated times by Western blotting. Actin was used as a loading control. D Protein levels of HACE1 increase under hypoxia. Endogenous levels of HACE1 protein were examined in MCF7 cells cultured under 1% O2 for the indicated times and lysates analyzed by Western blotting. Actin was used as a loading control. E UpSet plot shows a number of differentially expressed genes in HEK293 cells expressing HA-HACE1 or vector alone cells cultured under hypoxia versus normoxia. Differentially regulated genes were defined as having a fold difference of two or greater and an FDR < 0.05. Bars show the number of genes upregulated (red) and downregulated (black) during hypoxia, for HACE1 and vector alone cells. F The heatmap depicts the Z-score expression of 339 genes that show differential expression (fold difference of two or greater and an FDR < 0.05) in HA-HACE1 or vector alone cells during hypoxia.
HACE1 reduces HIF1α accumulation under hypoxia
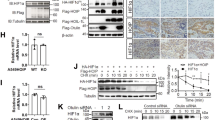
Since HIF1α is a key transcription factor induced under hypoxia, transcriptionally activating a wide array of genes that are important for the cytoprotective re-programming of tumor cells under hypoxia [30, 40], including many in Table S1, we decided to examine potential relationships between HIF1α and HACE1. We first ectopically expressed HACE1 in HEK293 and MCF7 cells and examined temporal HIF1α protein levels under hypoxia (1% O2). Overexpression of HACE1 reduced HIF1α accumulation under hypoxia (Fig. 2A, B). Knockdown of HACE1 using siRNAs reversed this trend, whereby HIF1α levels increased under hypoxia (Fig. 2C). Next, wild type (wt) and Hace1–/– mouse embryonic fibroblasts (MEFs) were exposed to hypoxia and cellular HIF1α levels were determined by Western blotting. Hace1 genetic inactivation led to a dramatic accumulation of HIF1α over 24 h compared to controls (Fig. 2D). Reintroduction of hemagglutinin (HA)-tagged HACE1 into Hace1–/– MEFs repressed HIF1α accumulation (Fig. 2E). In agreement with these observations, siRNAs targeting HACE1 in HEK293 cells increased HIF1α accumulation compared to scrambled control siRNAs (Fig. 2F). In HEK293 cells with stable shRNA HACE1 knockdown, HIF1α accumulation was also increased under hypoxia compared to controls (Fig. 2G). Together, these findings indicate that HACE1 regulates the level of the critical stress response and pro-oncogenic proteins, HIF1α under hypoxia. To determine if HACE1 requires its E3 ligase activity to reduce HIF1α levels, we ectopically expressed the previously described ligase dead HACE1-C876S mutant (Fig. S2) [1, 3] in HEK293 cells and Hace1–/– MEFs. However, HACE1-C876S was unable to reduce HIF1α levels in HEK293 cells and Hace1–/– MEFs (Fig. 2H, I). This indicates that the observed effects are dependent on the E3 ligase activity of HACE1.
A HA-tagged HACE1 was overexpressed in HEK293 cells, and cells were exposed to 1% O2 for the indicated times. HIF1α and HA-HACE1 protein levels were examined by Western blotting. B HA-tagged HACE1 was overexpressed in breast cancer MCF7 cells, and cells were exposed to 1% O2 for the indicated times. HIF1α and HA-HACE1 protein levels were examined by Western blotting. C Transient knockdown of HACE1 using two individual siRNAs in MCF7 cells. Cells were exposed to 1% O2 for 4-h. HIF1α and HACE1 protein levels were examined by Western blotting. D Hace1–/– and wild-type MEFs were exposed to 1% O2 for various times as indicated. Whole protein lysates were collected for immunoblotting with HIF1α and HACE1 antibodies. Actin was used as a loading control. E HA-tagged HACE1 was expressed in Hace1–/– MEFs using a retrovirus system. Cells were exposed to 1% O2 for 2- or 24-h. HIF1α protein levels were examined by Western blotting. F Transient knockdown of HACE1 using two individual siRNAs in HEK293 cells. Cells were exposed to 1% O2 for the times as indicated. HIF1α and HACE1 protein levels were examined by Western blotting. G Stable knockdown of HACE1 using shRNAs in HEK293 cells. Cells were exposed to 1% O2 for the times indicated. HIF1α and HACE1 protein levels were examined by Western blotting. H HEK293 overexpressing HA-tagged wild-type HACE1 or ligase dead HACE1-C876S were exposed to 1% O2 for 4-h and analyzed by Western blotting for HIF1α and HA-HACE1. I MEFs overexpressing HA-tagged WT HACE1 or ligase dead HACE1-C876S were exposed to 1% O2 for 4-h and analyzed by Western blotting for HIF1α and HA-HACE1.
HACE1 reduces the half-life of HIF1α under hypoxia
HIF1α is an unstable protein that is strictly regulated by oxygen levels [41, 42]. To examine whether HACE1 affects the half-life of HIF1α, Hace1–/– MEFs were exposed to hypoxia to allow HIF1α accumulation and subsequently treated with cycloheximide (CHX) to block new protein synthesis. The observed half-life of HIF1α in wt MEFs was short, with virtually all protein lost after just 20 min following CHX treatment (Fig. 3A). However, in Hace1–/– MEFs, HIF1α protein signals were retained at both the 20- and 40-min time points, demonstrating an extended half-life (Fig. 3A). Re-expression of HACE1 in knockout MEFs reversed this observation, restoring the HIF1α half-life to that found in wt cells (Fig. 3B). In agreement with these results, stable shRNA knockdown of HACE1 in HEK293 cells also increased the stability of HIF1α compared to scrambled control cells (Fig. 3C).
A Hace1–/– and wild-type MEFs were cultured under 1% O2 for 4-h to accumulate HIF1α protein (0 min). Cells were then treated with cycloheximide (CHX) for the indicated time points to block protein synthesis, and lysates were analyzed for HIF1α levels by Western blotting. The graph shows quantified HIF1α protein amounts normalized to actin levels. B HACE1 was expressed in Hace1–/– MEFs by retroviral transduction. Cells were then treated under hypoxia with CHX as in Fig. 2A and similarly analyzed for HIF1α protein levels. C HEK293 cells stably transduced with HACE1 shRNAs (KD) or the scrambled control (Scr) were exposed to 1% O2 for 4-h and were treated with CHX for the indicated times. D HEK293 cells stably overexpressing HA-tagged HACE1 or vector alone were grown under 21% (normoxia) or 1% O2 for 2-h followed by treatment with the proteasome inhibitor MG132 for a further 2-h. Total, as well as the hydroxylated HIF1α levels, were examined by Western blotting using anti-HIF1α or anti-hydroxylated HIF1α (HIF1α-OH) antibodies, respectively. E Wild-type V5-tagged HIF1α, as well as mutant V5-HIF1α (P402A/P564A) that cannot be hydroxylated, was transiently transfected into HEK293 cells. Cells were culture under 1% O2 for 4-h to allow HIF1α protein to accumulate (0 min), followed by CHX treatment for the indicated times. Levels of exogenous HIF1α protein were analyzed by Western blotting using anti-V5 antibodies. F GFP-ubiquitin was transfected into HEK293 cells overexpressing HA-HACE1 or vector alone. Cells were cultured under 1% O2 for 3-h in the presence of MG132, followed by immunoprecipitation (IP) from whole-cell lysates using anti-GFP antibodies. Anti-HIF1α immunoblotting was performed to detect ubiquitin associated HIF1α proteins. IgG was used as an IP control antibody. G HA-HACE1 or vector alone constructs were stably overexpressed in RCC4 cells lacking VHL or RCC4 cells re-expressing wild-type VHL (RCC4 + VHL). Cells were exposed to 1% O2 for the indicated times and whole protein lysates were immunoblotted with anti-HIF1α or anti-HA antibodies for HIF1α and HACE1 levels.
To assess whether proteasome activity is required for HACE1-mediated reduction of HIF1α, Hace1–/– MEFs stably expressing HA-Hace1 or vector alone (MSCV) were treated with the proteasome inhibitor, MG132, following pre-incubation in normoxic versus hypoxic conditions for 2 h. MG132 completely blocked the ability of HACE1 to reduce HIF1α levels under hypoxia (Fig. 3D). Proteasomal degradation requires hydroxylation on proline residues 402 and 564 of HIF1α by prolyl-hydroxylases (PHDs) [43]. We therefore examined the hydroxylation status of HIF1α and found that MG132 treatment led to the clear accumulation of the hydroxylated form of this protein under hypoxia in both MEFs and HEK293 cells (Fig. 3D). HACE1 over-expression resulted in enhanced levels of hydroxylated HIF1α in both cell types (Fig. 3D). Antibody specificity for hydroxylated HIF1α was confirmed using a PHD inhibitor, CoCl2 (Fig. S3), and these observations were further validated with a V5-tagged mutant HIF1α (P402A/P564A) that cannot be hydroxylated by PHDs nor degraded via the Elongin BC/Cul2/von Hippel Lindau factor (VHL) complex (ECV), an E3 ubiquitin ligase complex which targets the alpha subunit of HIF1α for ubiquitin-mediated degradation. [44,45,46]. V5-tagged wt or mutant HIF1α plasmids were transiently transfected into HEK293 cells stably expressing HA-HACE1 and exposed to hypoxia (1% O2 for 4h) followed by a time-course of CHX treatment. HACE1 overexpression reduced V5-HIF1α levels in wt cells but did not affect the mutant form (Fig. 3E), suggesting that hydroxylation is required for HACE1- mediated degradation.
To further investigate the link between HIF1α degradation and HACE1, we expressed GFP-tagged ubiquitin in HEK293 cells and examined the ubiquitylation status of HIF1α. Affinity purification using anti-GFP antibodies pulled down increased amounts of poly-ubiquitylated HIF1α in HACE1 over-expressing cells compared to controls under 1% O2 (Fig. 3F). These findings indicate that HACE1 facilitates hydroxylation of HIF1α under hypoxia and triggers its ubiquitylation and degradation through the proteasome. Similar to HACE1-associated degradation of cyclin D1 [3], we found no evidence of direct ubiquitylation of HIF1α by Hace1 by in vitro assays, indicating that this process is likely indirect. Loss of function mutations of VHL protein, a key ECV component in renal clear cell carcinoma (RCC) results in elevated HIF1α levels [47, 48]. HA-HACE1 expression in RCC4 cells that express mutant VHL failed to reduce HIF1α levels when exposed to hypoxia (Fig. 3G). However, the effects of HACE1 on HIF1α were rescued in RCC4 cells re-expressing wt VHL (RCC4 + VHL), suggesting that HIF1α protein levels are regulated by HACE1 in a proteasome- and ECV-dependent manner. Together, these findings indicate that the impact of HACE1 on protein stability under hypoxia is due to increased protein degradation mediated by the hydroxylation and ubiquitylation status of HIF1α.
HACE1 regulates HIF1α activity under hypoxia
HIF1α binds to hypoxia response element (HRE) sequences in promoters of its target genes and regulates their expression [40, 49, 50]. To examine whether HACE1 reduces HIF1α functional activity under hypoxia, we transfected HEK293 control cells and cells stably expressing HA-HACE1 with an additional construct encoding GFP driven by an HRE-containing promoter (HRE-GFP) and cultured the resulting populations under hypoxia for 16-h. As a transfection control, a mCherry protein-encoding plasmid was co-transfected. Overexpression of HACE1 significantly reduced HRE-GFP signals under hypoxia (Fig. 4A, B). Similarly, HACE1 knockdown enhanced HRE-GFP expression compared to scrambled controls (Fig. 4C, D). The results of the HACE1 overexpression and knockdown were confirmed by Western blotting (Fig. 4E, F), indicating that HACE1-mediated reduction of HIF1α reduces its ability to regulate transcription of its target genes. Using cDNA hybridization, we probed the effects of HACE1-mediated control on the expression of known HIF1α target genes in RCC4 + VHL cells (i.e., HACE1 overexpression vs. vector alone), which were exposed to hypoxia for 3 h. HACE1 inhibited expression of many HIF1α target genes in the array; specifically, expression of genes involved in glucose metabolism (PDK1, Glut-1, and Glut-3) metastasis (MMP1, MMX1, and PAI-1), or signal transduction (PDGF) were all reduced by HACE1 (Fig. 4G). Notably, VHL mRNA expression was also reduced under hypoxia in HACE1 overexpressing cells, even though no changes in VHL protein levels were observed in our previous experiments (see Fig. 3G), potentially reflecting negative feedback regulation at the mRNA level. Together, these results demonstrate that HACE1-mediated regulation of HIF1α levels results in dynamic transcriptional changes in HIF1α targets regulated in response to hypoxia.
A HRE-GFP plasmids were transfected into HEK293 cells overexpressing HA-HACE1 or vector alone, along with a mCherry-encoding plasmid as a transfection control. Cells were then cultured under 1% O2 for 16-h. GFP and mCherry signals were examined by fluorescence microscopy. B Quantification of the relative levels of GFP positive cells in comparison to vector alone cells. (*P < 0.05; **P < 0.01). C An identical experiment as in (A) was performed using HRE-GFP expressing HEK293 after transfection with HACE1-targeting (KD) or the scrambled control shRNAs (Scr). D Quantification of the relative levels of GFP positive cells in comparison to vector alone cells (*P < 0.05; **P < 0.01). E HRE-GFP plasmids were transfected into HEK293 cells overexpressing HA-HACE1 or vector alone, along with a mCherry-encoding plasmid as a transfection control. Cells were cultured under 1% O2 for 16-h. HIF1α, HRE-GFP, and HA-HACE1 protein levels were examined by Western blotting. F HRE-GFP plasmids were transfected into HEK293 cells after transfection with HACE1-targeting (KD) or the scrambled shRNAs (Scr), along with a mCherry-encoding plasmid as a transfection control. Cells were then cultured under 1% O2 for 16-h. HIF1α, HRE-GFP, and HA-HACE1 protein levels were examined by Western blotting. G RCC4 + VHL cells expressing HA-HACE1 or vector alone were exposed to 1% O2 for 2-h. Total RNA was isolated, converted to cDNA in the presence of biotin-dUTP, and hybridized to gene-specific oligonucleotides in a 96-well plate. The captured cDNA was then detected with streptavidin-HRP on a microplate luminometer. Fold induction under hypoxia (compared to normoxia) is shown on the y-axis.
Loss of HACE1 expression correlates with increased HIF1α expression in WT and sarcoma tissues
To investigate whether HACE1 decreases HIF1α levels in vivo, we analyzed formalin-fixed paraffin-embedded (FFPE) tissues obtained from nude mice bearing xenografted HEK293 cells with or without stable HACE1 knockdown, by immunohistochemistry (IHC) for HIF1α expression, as described previously [3]. In agreement with our in vitro observations, HACE1 knockdown led to increased levels of HIF1α levels in tumor tissues (Fig. 5A, B). To probe the pathophysiological significance, we investigated the relative levels of HACE1 and HIF1α protein in WT cases compared with patient-matched normal kidney adjacent to tumor tissues. Consistent with our previous findings [3], HACE1 protein levels were reduced in all tumor cases tested (n = 3) compared to the patient-matched normal kidney, while HIF1α levels were correspondingly increased (Fig. 5C). To confirm that HACE1 tumor suppressor activity is linked to its ability to reduce HIF1α levels, we performed soft agar colony assays using the SKNEP1 human Ewing sarcoma cell line that lacks endogenous expression of HACE1 [1]. Ectopic expression of HACE1 in these cells was previously shown to significantly reduce in vitro soft agar colony formation [3]. As shown in Fig. 5D, HACE1 also significantly reduced colony formation in wt HIF1α expressing cells compared to parental SKNEP1 cells, while HACE1-C876S failed to do so. In cells expressing V5-tagged mutant HIF1α (P402A/P564A) in which HIF1α cannot be hydroxylated nor degraded by ECV, no changes were observed in colony formation in the presence of HACE1. To further assess the link between HACE1 and HIF1α in human tumors, tissue microarrays (TMAs) consisting of WT (9 cases) and different childhood sarcomas (18 cases, including 5 Ewing sarcoma cases, 3 alveolar rhabdomyosarcoma cases, 4 embryonal rhabdomyosarcoma cases, and 6 synovial sarcoma cases, which were analyzed collectively due to the limited case numbers) were subjected to IHC for HACE1 and HIF1α expression. A significant correlation was observed between high HIF1α and low HACE1 expression in both tumor cohorts (Fig. 5E, F), highlighting the inverse relationship between HACE1 and HIF1α levels in vivo. Together, these findings validate the role of HACE1 as a negative regulator of HIF1α activity in vivo.
A Immunohistochemistry for HIF1α conducted on the paraffin blocks from nude mice injected with HEK293 cells with stable HACE1 knockdown or non-targeting shRNA used in this study, generated as described previously [3]. The scale bar is 100 μm. B Quantification of A was conducted respectively using ImageJ software and data represented as average value ± SEM for n = 15 HPFs in 3 tumors/group (*P < 0.05; **P < 0.01). C Whole-cell lysates from three cases of Wilms’ tumor (WT) with paired normal kidney (N) were subjected to Western blotting with antibodies endogenous HACE1 and HIF1α as indicated. D Constructs encoding wild type (wt) or HIF1α-P402A/P564A (non-OH-HIF1α) were expressed in SKNEP1 Ewing tumor cells stably expressing wt HACE1 or ligase dead HACE1-C876S as indicated. Cells were seeded in soft agar and grown for 14 days. Colonies were then counted as a percentage of seeded cells, using an inverted microscope. Data shown represent means ± SD, n = 3. (*P < 0.05; **P < 0.01). E Immunohistochemistry (IHC) of HIF1α and HACE1 were conducted on serial sections of TMAs consistent with Wilms’ tumors (9 cases) and sarcomas (18 cases) provided by Children Hospital of Philadelphia. Representative IHC images from matched patient samples on serial histological sections were shown. Scale bar = 100 μm. F Box plot showing H‐scores (staining intensity × percentage) for HIF1α and HACE1, along with relative sample sizes for each group based on the IHC analysis in (E) was shown. The cohort is grouped according to the tumor type. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
HACE1 blocks HIF1α accumulation under hypoxia through degradation of RAC1
Since RAC1 is required for activation of HIF1α [36,37,38], and RAC1 is the best-characterized ubiquitylation target of the HACE1 E3 ligase [10, 15,16,17,18], we wondered whether HACE1’s effects on HIF1α are mediated through RAC1. We found that RAC1 activation was higher under hypoxia than ambient conditions in HEK293 cells (Fig. 6A). While HACE1 overexpression reduced RAC1 activation under both ambient and hypoxic conditions, this reduction was proportionally greater under hypoxia, indicating that HACE1 can also inhibit RAC1 activation under hypoxia (Fig. 6A). To further test this connection, we incubated HEK293 cells ± ectopic HACE1 overexpression with EHT1864, a selective inhibitor of RAC1 activation that traps RAC1 in an inactive state [51]. EHT1864 has been shown to inhibit RAC1 downstream signaling and cellular transformation by guanine nucleotide displacement, impairing RAC1 mediated functions in vivo [52]. EHT1864 treatment effectively prevented RAC1 activation in HEK293 cells, regardless of HACE1 overexpression, as expected (Fig. 6A). Moreover, EHT1864 blocked HIF1α accumulation under hypoxia in both control and HACE1 overexpressing cells (Fig. 6B), indicating that RAC1 activity is required for HIF1α induction under hypoxia in these cells.
A GTP bound RAC1 levels are shown using RAC1 G-LISA Activation Assay Kit in the cells stably expressing HACE1 or the empty vector controls under normoxia or hypoxia in the presence or absence of EHT 1864. Total RAC1 levels are shown by Western blotting. (*P < 0.05; **P < 0.01). B HEK293 cells stably expressing HACE1 or the empty vector alone were treated (or not treated) with 20 μM EHT 1864 for 1-h, then subjected to hypoxia for 3-h. Protein levels of HIF1α were analyzed by Western blotting. C HEK293 cells with stable HACE1 KD were transfected with control shRNA (Scr) or one of two independent shRNAs targeting RAC1 (shRAC1). Rescue was generated by re-introducing pHAGE-RAC1 plasmid to the cells using a lentiviral transduction system. Cells were exposed to 1% O2 for 4-h and subjected to Western blotting. D HEK293 cells stably expressing HACE1 or the empty vector alone were transfected with control siRNA (Scr) or a siRNA targeting RAC1 (siRAC1), then subjected to hypoxia for 4-h before analysis of HIF1α expression by Western blotting. E MCF7 cells were transfected with control siRNA (Scr) or two individual siRNAs targeting RAC1 (siRAC1), then subjected to hypoxia for 4-h before being analyzed for HIF1α expression by Western blotting. F HEK293 cells stably expressing HACE1 or the empty vector alone were transfected with combinations of wild-type RAC1, RAC1-K147R, and vector alone, then subjected to hypoxia for 4-h before analysis of HIF1α and RAC1 expression by Western blotting. G Quantification of (F) was conducted respectively using ImageJ software and data represented as average value ± SEM.
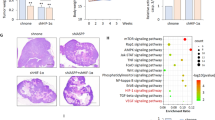
Stable RAC1 knockdown using two independent shRNAs markedly reduced HIF1α levels under hypoxia, which could be rescued by ectopic RAC1 re-expression (Fig. 6C). Moreover, in HEK293 cells stably expressing ectopic HACE1, RAC1 knockdown further attenuated HIF1α accumulation (Fig. 6D). HIF1α accumulation was also reduced in MCF7 breast cancer cells under hypoxia when RAC1 was knocked down (Fig. 6E). Finally, to verify a link to HACE1 in these observations, we used a RAC1-K147R mutant that is resistant to HACE1 degradation, due to substitution of the RAC1 lysine residue targeted for ubiquitylation by HACE1 [10, 15]. In hypoxic HEK293 cells stably co-expressing ectopic HACE1 along with vector alone, RAC1-K147R, or wt RAC1, HACE1 was able to block HIF1α accumulation in control and wt RAC1 expressing cells (Fig. 6F, G; lanes 2 and 4). In contrast, cells expressing HACE1-resistant RAC1-K147R retained HIF1α accumulation under hypoxia even in the presence of HACE1 overexpression (Fig. 6F, G; lanes 5 and 6). To investigate this process in vivo, we used a described previously mouse lung cancer model of Hace1 inactivation in which deletion of Rac1 and expression of oncogenic KRasG12D can be simultaneously induced by Adeno-Cre administration [3, 53]. IHC was performed to assess HIF1α expression in FFPE lung tumor tissues obtained from 8 and 16-week-old KRasG12DHace1+/+Rac1+/+, KRasG12DHace1–/–Rac1+/+, KRasG12DHace1+/+Rac1fl/fl, and KRasG12DHace1–/–Rac1fl/fl mice. In agreement with our in vitro observations, deletion of Hace1 alone (i.e. KRasG12DHace1–/–Rac1+/+) resulted in elevated HIF1α levels compared to KRasG12DHace1+/+Rac1+/+ controls, while genetic inactivation of Rac1 alone (i.e. KRasG12DHace1+/+Rac1fl/fl) decreased HIF1α levels compared to controls (Fig. 7A–C). KRasG12DHace1–/–Rac1fl/fl mice showed a significant reduction in HIF1α levels compared to KRasG12DHace1–/–Rac1+/+ mice at both weeks 8 and 16 post-induction, and comparable HIF1α levels to tumors of KRasG12DHace1+/+Rac1fl/fl mice at both time points (Fig. 7A–C). Together, these data support a model in which HACE1 blocks HIF1α accumulation in a RAC1-dependent manner.
A Immunohistochemistry for HIF1α conducted on the paraffin blocks from the KRasG12D-driven lung adenocarcinoma mouse model at weeks 8 and 16 after lung cancer induction for KRasG12DHace1+/+Rac1+/+, KRasG12DHace1–/–Rac1+/+, KRasG12DHace1+/+Rac1fl/fl, and KRasG12DHace1–/–Rac1fl/fl mice, generated as described previously [53]. The scale bar is 100 μm. B Quantification of (A) (left panel) for 8 weeks after post-induction was conducted respectively using ImageJ software and data represented as average value ± SEM for n = 15 HPFs in three tumors/group. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). C Quantification of (A) (right panel) for 16 weeks after post-induction was conducted respectively using ImageJ software and data represented as average value ± SEM for n = 15 HPFs in three tumors/group. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Discussion
To better understand the regulatory mechanisms that underpin stress adaptation in cancer cells, we examined how the previously identified tumor suppressor HACE1 might function in such adaptive processes, given its previous links to cell stress [1, 3]. We observed consistent increases in HACE1 mRNA expression under diverse stress conditions, highlighting HACE1 as a potentially broad stress response factor, in keeping with in vivo consequences of its genetic inactivation in mice, in which Hace1 loss leads to the development of multiple late-onset tumors [3]. Here we show that hypoxia leads to marked increases in HACE1 at both the mRNA and protein levels, and surprisingly, that this is associated with loss of HIF1α protein. HIF1α induction in response to hypoxia is a well-established hallmark of cancer cell responses to stress, driving the transcription of factors essential for cell survival and ultimately dissemination under low oxygen conditions [27, 54, 55]. The ability to reduce HIF1α levels provides a plausible mechanism for the anti-tumorigenic activity of HACE1, including potential roles in blocking metastasis, as it was recently shown that HACE1 overexpression completely eliminated detectable lung metastases of osteosarcoma cells in mice [13].
We found that HACE1 did not appreciably alter HIF1A mRNA expression, but instead reduced HIF1α protein stability. Ligase dead HACE1-C876S was unable to reduce HIF1α levels in the models tested, indicating that this activity is dependent on HACE1 E3 ligase activity. However, we were unable to show that HACE1 directly targets HIF1α for ubiquitylation and degradation. Notably, HACE1 was unable to block the accumulation of a HIF1α mutant with substitution of proline residues 402 and 564, sites of HIF1α hydroxylation by PHDs [43], and critical for HIF1α degradation by the canonical ECV E3 ligase complex that targets HIF1α [44,45,46]. We found that HIF1α degradation still requires the ECV complex, as overexpression of HACE1 in RCC4 cells with mutant VHL was unable to block HIF1α accumulation unless the cells were engineered to re-express wt VHL. This suggests indirect targeting of HIF1α by HACE1. Interestingly, the kinetics of HIF1α accumulation following ectopic expression of HACE1 varied among the different cell lines tested. This might reflect differences in levels or isoforms of PHDs that hydroxylate HIF1α, or members of the ECV E3 ligase complex, that are expressed in each cell line. Moreover, levels of Fe2+, 2-oxyglutarate, ascorbate, and other metabolites can also influence HIF1α degradation, potentially also perturbing the kinetics of HIF1α degradation [56, 57].
A number of studies report that the upregulation of HIF1α in response to cellular stress is dependent on activation of the small Rho GTPase, RAC1 [36,37,38]. RAC1 has previously been identified as a proto-oncogene, where its activation leads to increased cell migration and invasion [22, 23]. In addition, RAC1 can be activated by stress stimuli such as hypoxia [35, 36]. HACE1 controls cell migration in different cell types through degradation of active RAC1 at NADPH oxidase complexes [15], including in breast cancer cells, where HACE1 inhibits cell migration and invasion by targeting active RAC1 [58]. Therefore, we searched for links between HACE1, RAC1, and HIF1α, and found that HACE1 blocks HIF1α accumulation under hypoxia in a RAC1 dependent manner. RAC1 is a primary target of the E3 ligase functionality of HACE1, suggesting a potential regulation point for mediation of HIF1α regulation by HACE1. This observation is consistent with previous findings indicating that HACE1 binds to RAC1 with higher affinity under hypoxia than normoxia, leading to its proteasomal dependent degradation, counteracting RAC1-mediated cell migration and invasion in breast cancer [58]. Moreover, we found that a RAC1-K147R mutant that is resistant to HACE1-mediated degradation rescued the accumulation of HIF1α in response to hypoxia in HACE1 overexpressing cells.
We also tested EHT1864, a relatively selective RAC1 activation inhibitor, and found that it abolished HIF1α accumulation under hypoxia. RAC1 and Cdc42 GTPases are key signaling intermediates with important roles in cancer initiation and progression, and inhibitors of these proteins, including EHT1864, have shown promising preclinical efficacy [59]. While none of the inhibitors targeting these GTPases have been approved for cancer therapy to date, RAC1/Cdc42 inhibition as a strategy to overcome therapy resistance has been validated previously [59,60,61,62]. Consistently, stable co-knockdown of RAC1 in HACE1 deficient cells inhibits HIF1α accumulation in these cells, suggesting targeting of RAC1 is a potential mechanistic intervention point for targeted therapy in diverse cancer types. In vivo, we found that loss of Hace1 markedly enhances HIF1α expression in KRasG12D-driven lung tumors, while in contrast, Rac1 loss alone decreases HIF1α expression in this model. Simultaneous deletion of Hace1 and Rac1 was similar to the loss of Rac1 alone and showed significantly reduced HIF1α levels compared to the loss of Hace1 alone, further suggesting that the ability of HACE1 to reduce HIF1α accumulation is dependent on RAC1.
HACE1 was originally identified as a tumor suppressor based on loss of expression in WT. In the present study, we found that HACE1 expression was also lost in WT specimens but retained in matched normal kidney tissues derived from WT nephrectomy specimens. Moreover, we detected HIF1α protein expression only in tumor tissues from these paired specimens, verifying the clinical relevance of our findings and consistent with previous observations [3]. These findings were corroborated by IHC staining of TMAs containing additional WTs as well as childhood sarcomas, also demonstrating a strong inverse relationship between HACE1 HIF1α protein levels. Further mechanistic details of how the HACE1-RAC1 axis regulates HIF1α stability, and whether other components are involved in this regulation, remains to be elucidated and warrants additional investigation. For example, previous studies of HIF1α suggest a crucial role for ROS generation might be involved in the stabilization of HIF1α under hypoxia [29, 36, 63]. Since RAC1 increases de novo ROS generation, and is targeted by HACE1 at NADPH oxidases [15], regulation of HIF1α by RAC1 might involve deregulated ROS generation. Our work illuminates a previously unrecognized link between HACE1, its primary target RAC1, and the oncogenic driver HIF1α. As HIF1α is well-established to facilitate oncogenesis in a variety of cancers, the HACE1-RAC1 axis provides an attractive potential target for therapeutic intervention.
Experimental procedures
Reagents
Cycloheximide, MG132, and cobalt chloride (CoCl2) were obtained from Calbiochem (San Diego, CA). Antibodies and plasmids used in this study are described in Supplemental Experimental Procedures.
Cell culture
RCC4 and RCC4-VHL cells were generous gifts from Dr. Michael Ohh (University of Toronto). Culture conditions for each cell line are described in Supplemental Experimental Procedures. The cell lines stably expressing MSCV, HA-HACE1, or the mutant HA-HACE1-C876S were generated as described [1]. Stable knockdown HACE1 cell lines were generated using the Block-iT Lentiviral RNAi kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols as described [3]. Nutrient deprivation was performed using HBSS/HEPES solution for the indicated time points. Hypoxia treatment consisted of culturing in a hypoxia chamber (COY Laboratory Products, INC. Grass Lake, Michigan) at 1% O2, 5% CO2, and balanced with N2 at 37 °C. Cells were seeded in equal numbers and incubated under the same conditions, then randomized and categorized into control and experimental groups in all experiments conducted unless stated otherwise.
Transfections
Transient plasmid transfections were performed using FuGene six reagent (Roche, Penzberg, DE) in a six-well plate, according to the user manual. Transfections of siRNAs were performed with 25 nM siRNA using RNAiMax (Invitrogen, Waltham, MA, USA). The following siRNAs were used in the study: control siRNA (C): (5′–3′) AUAUCGGCUAGGUCUAACA; Hace1-1 (H1): Hs_Hace1_1 (FlexiTube, Qiagen, Hilden, DE); Hace1–2 (H2): Hs_Hace1_4 (FlexiTube, Qiagen, Hilden, DE); Human Rac1 (R): Hs_Rac1_6 (FlexiTube, Qiagen, Hilden, DE); and FlexiTube GeneSolution GS5879 (FlexiTube, Qiagen, Hilden, DE).
Immunoprecipitation and immunoblotting
Immunoprecipitation and immunoblotting were conducted using standard methods. Detailed procedures are described in the Supplemental Experimental Procedures.
Immunohistochemistry
IHC on tissue sections obtained from the paraffin blocks generated previously by Kogler et al. [53] and Zhang et al. [3] was conducted on a Ventana Discovery XT system. All IHC analysis was conducted blindly by two independent pathologists. Detailed procedures are described in the Supplemental Experimental Procedures.
RAC1 G-LISA activation assay
The G-LISA RAC1 activation kit (Cat# BK126 Cytoskeleton, Denver, CO, USA) was performed according to the user manual to determine active (GTP bound) RAC1 levels. Detailed procedures are described in the Supplemental Experimental Procedures.
Clinical patients
The three cases of WT with matching normal kidney were collected from the Children’s Oncology Group (COG) WT tissue bank, and generously supplied by Dr. Paul Grundy, Cross Cancer Center, Edmonton, Alberta.
Statistical analysis
All statistical analysis was conducted using a Student’s two-tailed T-test, unless otherwise indicated, with p-values < 0.05 being considered as being statistically significant.
References
Anglesio MS, Evdokimova V, Melnyk N, Zhang L, Fernandez CV, Grundy PE, et al. Differential expression of a novel ankyrin containing E3 ubiquitin-protein ligase, Hace1, in sporadic Wilms’ tumor versus normal kidney. Hum Mol Genet. 2004;13:2061–74.
Fernandez CV, Lestou VS, Wildish J, Lee CL, Sorensen PH. Detection of a novel t(6;15)(q21;q21) in a pediatric wilms tumor. Cancer Genet Cytogenet. 2001;129:165–7.
Zhang L, Anglesio MS, O’Sullivan M, Zhang F, Yang G, Sarao R, et al. The E3 ligase HACE1 is a critical chromosome 6q21 tumor suppressor involved in multiple cancers. Nat Med. 2007;13:1060–9.
Hibi K, Sakata M, Sakuraba K, Shirahata A, Goto T, Mizukami H, et al. Aberrant methylation of the HACE1 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2008;28(3A):1581–4.
Stewenius Y, Jin Y, Ora I, Panagopoulos I, Moller E, Mertens F, et al. High-resolution molecular cytogenetic analysis of Wilms tumors highlights diagnostic difficulties among small round cell kidney tumors. Genes Chromosom Cancer. 2008;47:845–52.
Thelander EF, Ichimura K, Corcoran M, Barbany G, Nordgren A, Heyman M, et al. Characterization of 6q deletions in mature B cell lymphomas and childhood acute lymphoblastic leukemia. Leuk Lymphoma. 2008;49:477–87.
Sakata M, Kitamura YH, Sakuraba K, Goto T, Mizukami H, Saito M, et al. Methylation of HACE1 in gastric carcinoma. Anticancer Res. 2009;29:2231–3.
Huang Y, de Reynies A, de Leval L, Ghazi B, Martin-Garcia N, Travert M. et al. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood. 2010;115:1226–37.
Slade I, Stephens P, Douglas J, Barker K, Stebbings L, Abbaszadeh F, et al. Constitutional translocation breakpoint mapping by genome-wide paired-end sequencing identifies HACE1 as a putative Wilms tumour susceptibility gene. J Med Genet. 2010;47:342–7.
Goka ET, Lippman ME. Loss of the E3 ubiquitin ligase HACE1 results in enhanced Rac1 signaling contributing to breast cancer progression. Oncogene 2015;34:5395–405.
Castillo-Lluva S, Tan CT, Daugaard M, Sorensen PHB, Malliri A. The tumour suppressor HACE1 controls cell migration by regulating Rac1 degradation. Oncogene. 2013;32:1735–42.
Bouzelfen A, Kora H, Alcantara M, Bertrand P, Latouche J-B, Jardin F. Heterogeneous epigenetic regulation of HACE1 in Burkitt–Lymphoma-derived cells. Leuk Res. 2017;60:53–7.
El-Naggar AM, Clarkson PW, Negri GL, Turgu B, Zhang F, Anglesio MS, et al. HACE1 is a potential tumor suppressor in osteosarcoma. Cell Death Dis. 2019;10:21.
Yu Z, Li Y, Han T, Liu Z. Demethylation of the HACE1 gene promoter inhibits the proliferation of human liver cancer cells. Oncol Lett. 2019;17:4361–8.
Daugaard M, Nitsch R, Razaghi B, McDonald L, Jarrar A, Torrino S, et al. Hace1 controls ROS generation of vertebrate Rac1-dependent NADPH oxidase complexes. Nat Commun. 2013;4:2180.
Mettouchi A, Lemichez E. Ubiquitylation of active Rac1 by the E3 ubiquitin-ligase HACE1. Small GTPases. 2012;3:102–6.
Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, et al. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev Cell. 2011;21:959–65.
Acosta MI, Urbach S, Doye A, Ng Y-W, Boudeau J, Mettouchi A, et al. Group-I PAKs-mediated phosphorylation of HACE1 at serine 385 regulates its oligomerization state and Rac1 ubiquitination. Sci Rep. 2018;8:1410.
Qiu R-G, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature. 1995;374:457–9.
Cardama GA, Gonzalez N, Maggio J, Menna PL, Gomez DE. Rho GTPases as therapeutic targets in cancer (review). Int J Oncol. 2017;51:1025–34.
Day GJ, Mosteller RD, Broek D. Distinct subclasses of small GTPases interact with guanine nucleotide exchange factors in a similar manner. Mol Cell Biol. 1998;18:7444–54.
Katoh H, Hiramoto K, Negishi M. Activation of Rac1 by RhoG regulates cell migration. J Cell Sci. 2006;119:56
Lawson CD, Ridley AJ. Rho GTPase signaling complexes in cell migration and invasion. J Cell Biol. 2018;217:447–57.
Duran RV, Hall MN. Regulation of TOR by small GTPases. EMBO Rep. 2012;13:121–8.
Rohwer N, Lobitz S, Daskalow K, Jöns T, Vieth M, Schlag PM, et al. HIF-1α determines the metastatic potential of gastric cancer cells. Br J Cancer. 2009;100:772–81.
Liao D, Corle C, Seagroves TN, Johnson RS. Hypoxia-inducible factor-1α is a key regulator of metastasis in a transgenic model of cancer initiation and progression. Cancer Res. 2007;67:563–72.
Van Welden S, Selfridge AC, Hindryckx P. Intestinal hypoxia and hypoxia-induced signalling as therapeutic targets for IBD. Nat Rev Gastroenterol Hepatol. 2017;14:596–611.
Karakashev SV, Reginato MJ. Progress toward overcoming hypoxia-induced resistance to solid tumor therapy. Cancer Manag Res. 2015;7:253–64.
Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309.
Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32.
Zhong H, Semenza GL, Simons JW, De Marzo AM. Up-regulation of hypoxia-inducible factor 1alpha is an early event in prostate carcinogenesis. Cancer Detect Prev. 2004;28:88–93.
Kimbro KS, Simons JW. Hypoxia-inducible factor-1 in human breast and prostate cancer. Endocr Relat Cancer. 2006;13:739–49.
Rundqvist H, Johnson RS. Hypoxia and metastasis in breast cancer. Curr Top Microbiol Immunol. 2010;345:121–39.
El-Naggar AM, Veinotte CJ, Cheng H, Grunewald TG, Negri GL, Somasekharan SP, et al. Translational activation of HIF1alpha by YB-1 promotes sarcoma metastasis. Cancer Cell. 2015;27:682–97.
Du J, Xu R, Hu Z, Tian Y, Zhu Y, Gu L, et al. PI3K and ERK-induced Rac1 activation mediates hypoxia-induced HIF-1α expression in MCF-7 breast cancer cells. PLoS ONE. 2011;6:e25213–e.
Turcotte S, Desrosiers RR, Béliveau R. HIF-1α mRNA and protein upregulation involves Rho GTPase expression during hypoxia in renal cell carcinoma. J Cell Sci. 2003;116:2247–60.
Hirota K, Semenza GL. Rac1 activity is required for the activation of hypoxia-inducible factor 1. J Biol Chem. 2001;276:21166–72.
Wang Y, Bibi M, Min P, Deng W, Zhang Y, Du J. SOX2 promotes hypoxia-induced breast cancer cell migration by inducing NEDD9 expression and subsequent activation of Rac1/HIF-1α signaling. Cell Mol Biol Lett. 2019;24:55.
Diebold I, Petry A, Djordjevic T, Belaiba RS, Fineman J, Black S. et al. Reciprocal regulation of Rac1 and PAK-1 by HIF-1alpha: a positive-feedback loop promoting pulmonary vascular remodeling. Antioxid Redox Signal. 2010;13:399–412.
Kenneth NS, Rocha S. Regulation of gene expression by hypoxia. Biochem J. 2008;414:19–29.
Berra E, Roux D, Richard DE, Pouyssegur J. Hypoxia-inducible factor-1 alpha (HIF-1 alpha) escapes O(2)-driven proteasomal degradation irrespective of its subcellular localization: nucleus or cytoplasm. EMBO Rep. 2001;2:615–20.
Moroz E, Carlin S, Dyomina K, Burke S, Thaler HT, Blasberg R, et al. Real-time imaging of HIF-1alpha stabilization and degradation. PLoS ONE. 2009;4:e5077.
Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW. et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279:38458–65.
D’Angelo G, Duplan E, Boyer N, Vigne P, Frelin C. Hypoxia up-regulates prolyl hydroxylase activity: a feedback mechanism that limits HIF-1 responses during reoxygenation. J Biol Chem. 2003;278:38183–7.
Masson N, Ratcliffe PJ. HIF prolyl and asparaginyl hydroxylases in the biological response to intracellular O(2) levels. J Cell Sci. 2003;116(Pt 15):3041–9.
Sufan RI, Ohh M. Role of the NEDD8 modification of Cul2 in the sequential activation of ECV complex. Neoplasia. 2006;8:956–63.
Ohh M. Ubiquitin pathway in VHL cancer syndrome. Neoplasia. 2006;8:623–9.
Shuin T, Yamasaki I, Tamura K, Okuda H, Furihata M, Ashida S. Von Hippel-Lindau disease: molecular pathological basis, clinical criteria, genetic testing, clinical features of tumors and treatment. Jpn J Clin Oncol. 2006;36:337–43.
Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci Singling. 2005;2005:re12
Kaluz S, Kaluzova M, Stanbridge EJ. Does inhibition of degradation of hypoxia-inducible factor (HIF) alpha always lead to activation of HIF? Lessons learnt from the effect of proteasomal inhibition on HIF activity. J Cell Biochem. 2008;104:536–44.
Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J Biol Chem. 2007;282:35666–78.
Onesto C, Shutes A, Picard V, Schweighoffer F, Der CJ. Characterization of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. Methods Enzymol. 2008;439:111–29.
Kogler M, Tortola L, Negri GL, Leopoldi A, El-Naggar AM, Mereiter S, et al. HACE1 prevents lung carcinogenesis via inhibition of RAC-family GTPases. Cancer Res. 2020;80:3009.
Wigerup C, Pahlman S, Bexell D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol Ther. 2016;164:152–69.
Dengler VL, Galbraith M, Espinosa JM. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. 2014;49:1–15.
Kohn KW, Riss J, Aprelikova O, Weinstein JN, Pommier Y, Barrett JC. Properties of switch-like bioregulatory networks studied by simulation of the hypoxia response control system. Mol Biol Cell. 2004;15:3042–52.
Qutub AA, Popel AS. A computational model of intracellular oxygen sensing by hypoxia-inducible factor HIF1 alpha. J Cell Sci. 2006;119(Pt 16):3467–80.
Kim I, Shin S-H, Lee JE, Park J-W. Oxygen sensor FIH inhibits HACE1-dependent ubiquitination of Rac1 to enhance metastatic potential in breast cancer cells. Oncogene. 2019;38:3651–66.
Maldonado MDM, Dharmawardhane S. Targeting Rac and Cdc42 GTPases in cancer. Cancer Res. 2018;78:3101–11.
Chen HY, Yang YM, Stevens BM, Noble M. Inhibition of redox/Fyn/c-Cbl pathway function by Cdc42 controls tumour initiation capacity and tamoxifen sensitivity in basal-like breast cancer cells. EMBO Mol Med. 2013;5:723–36.
Rosenblatt AE, Garcia MI, Lyons L, Xie Y, Maiorino C, Desire L. et al. Inhibition of the Rho GTPase, Rac1, decreases estrogen receptor levels and is a novel therapeutic strategy in breast cancer. Endocr Relat Cancer. 2011;18:207–19.
Gonzalez N, Cardama GA, Comin MJ, Segatori VI, Pifano M, Alonso DF. et al. Pharmacological inhibition of Rac1-PAK1 axis restores tamoxifen sensitivity in human resistant breast cancer cells. Cell Signal. 2017;30:154–61.
Jung SN, Yang WK, Kim J, Kim HS, Kim EJ, Yun H. et al. Reactive oxygen species stabilize hypoxia-inducible factor-1 alpha protein and stimulate transcriptional activity via AMP-activated protein kinase in DU145 human prostate cancer cells. Carcinogenesis. 2008;29:713–21.
Acknowledgements
The authors thank Joan Mathers, Michelle Pollard and Dr Karl Mechtler for helpful discussions; Dr Michael Ohh (University of Toronto) for providing the VHL-null RCC4 cell line; Dr. Thilo Hagen (National University of Singapore) for mutant HIF1α plasmids; and Dr. Peggy Olive (British Columbia Cancer Research Center) for HRE-GFP plasmids. FZ was supported by fellowships from the Canada Institutes of Health Research (CIHR) and the Michael Smith Foundation for Health Research. This work is supported in part by a grant to PHS from CIHR Foundation [Grant#: FDN-143280], and by the British Columbia Cancer Foundation through generous donations from Team Finn and other riders in the Ride to Conquer Cancer. JMP is supported by grants from IMBA, the Austrian Ministry of Sciences, the Austrian Academy of Sciences, a European Research Council (ERC) Advanced Grant, an Era of Hope Innovator award, and a Canada 150 Grant. LT is supported by the Swiss National Science Foundation [Grant#: PBEZP3_145993]. MD is supported by the Canadian Institutes of Health Research (CIHR) [Grant#: 377771].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Turgu, B., Zhang, F., El-Naggar, A. et al. HACE1 blocks HIF1α accumulation under hypoxia in a RAC1 dependent manner. Oncogene 40, 1988–2001 (2021). https://doi.org/10.1038/s41388-021-01680-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01680-1
This article is cited by
-
Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1
Nature Structural & Molecular Biology (2024)